Abstract
Histone mRNAs are naturally intronless and accumulate efficiently in the cytoplasm. To learn whether there are cis-acting sequences within histone genes that allow efficient cytoplasmic accumulation of RNAs, we made recombinant constructs in which sequences from the mouse H2a gene were cloned into a human β-globin cDNA. By using transient transfection and RNase protection analysis, we demonstrate here that a 100-bp sequence within the H2a coding region permits efficient cytoplasmic accumulation of the globin cDNA transcripts. We also show that this sequence appears to suppress splicing and can functionally replace Rev and the Rev-responsive element in the cytoplasmic accumulation of unspliced HIV-1-related mRNAs. Like the Rev-responsive element, this sequence acts in an orientation-dependent manner. We thus propose that the sequence identified here may be a member of the cis-acting elements that facilitate the cytoplasmic accumulation of naturally intronless gene transcripts.
In eukaryotic cells, most transcripts synthesized by RNA polymerase II contain introns that are removed by splicing before transport to the cytoplasm. Unspliced mRNAs are usually excluded from the cytoplasm. Although the molecular basis for the nuclear retention of unspliced transcripts is generally unclear, it seems likely that either the process of splicing somehow positively affects RNA transport or the presence of spliceosomes on transcripts negatively affects RNA export from the nucleus. Based on experiments in a yeast system, it has been proposed that intron-containing mRNAs are retained in the nucleus due to association with spliceosomes (1). In mammalian systems, similar observations have been obtained. Chang and Sharp (2) reported enhanced cytoplasmic accumulation of unspliced mRNAs when splice sites present in nascent transcripts were incapable of efficient spliceosome formation. In a recent study with the mouse polyoma virus, it was found that the strength of the virus late 5′-splice site inversely correlates with cytoplasmic levels of unspliced viral messages (3)
Splicing is not always a prerequisite for efficient cytoplasmic accumulation of mRNAs. In retroviruses, for example, alternative splicing of a single viral pre-mRNA generates multiple RNA products, a significant fraction of which are partially spliced or fully unspliced. These intron-containing mRNAs encode viral structural proteins and their cytoplasmic accumulation is essential for the viral life cycle. In the case of HIV-1, efficient cytoplasmic accumulation of singly spliced and unspliced viral mRNAs requires a viral regulatory protein, called Rev, which interacts with the Rev-responsive element (RRE) present in the target transcripts (reviewed in ref. 4). For simple retroviruses, such as the Mason–Pfizer monkey virus, cytoplasmic localization of unspliced viral mRNAs involves the interaction of a cis-acting RNA element with an as-yet-unidentified cellular factor(s) (5). Thus, specific interactions between positive cis-acting RNA elements and appropriate viral or cellular factors appear to facilitate the cytoplasmic accumulation of intron-containing retroviral mRNAs.
On the other hand, we have reported that cytoplasmic accumulation of intron-containing polyoma virus late messages does not seem to require positive cis-acting elements (3). Rather, it is likely that inefficiently utilized splicing signals within viral transcripts allow the cytoplasmic accumulation of unspliced viral mRNAs. Influenza virus expresses a message whose unspliced form encodes for a viral regulatory protein NS1 during infection (6). The NS1 protein has been reported to block the nucleocytoplasmic transport of all poly(A)-containing mRNAs by binding to the poly(A) sequence of these mRNAs (7). However, it is not yet clear how the unspliced message for the NS1 protein accumulates in the cytoplasm.
Some viral genes naturally lack introns. Examples include hepatitis B virus (reviewed in ref. 8) and herpes simplex virus thymidine kinase (9) genes. Unlike the intronless variants of the highly intron-dependent gene transcripts that usually fail to accumulate in the cytoplasm (10–20), these intronless viral transcripts can efficiently accumulate in the cytoplasm without undergoing the process of splicing (8, 9, 21). Recent studies have indicated that the cytoplasmic accumulation of unspliced hepatitis B virus transcripts is facilitated by a specific cis-acting RNA element that interacts with cellular factors (22–24).
In contrast to viral intronless gene expression, much less is known about the expression of cellular intronless genes, which include the genes coding for histone proteins (25), β-adrenergic receptor (26), α-interferon (27), and c-jun (28). The histone proteins are among the most abundant cellular proteins and are essential for cell viability (25, 29). Most histone mRNAs are not polyadenylated; instead, they end in highly conserved stem–loop structures (30). However, there are histone genes that produce polyadenylated histone mRNAs (31), and some that usually end in the histone stem–loop can, under some situations, also be processed by polyadenylation (32–34). Because histone mRNAs are able to efficiently accumulate in the cytoplasm without being spliced (29), we wanted to know whether there are cis-acting sequences within histone genes that allow them to do so. In this study, we used the mouse histone H2a gene to address this question. We have found that a 100-bp sequence within the H2a coding region can activate the cytoplasmic accumulation of a human β-globin cDNA transcript. We have also demonstrated that this sequence can functionally mimic Rev and RRE for the cytoplasmic accumulation of unspliced HIV-1-related mRNA. We thus postulate that this sequence may belong to a class of cis-acting elements that enable efficient cytoplasmic accumulation of naturally intronless gene transcripts.
MATERIALS AND METHODS
Constructs.
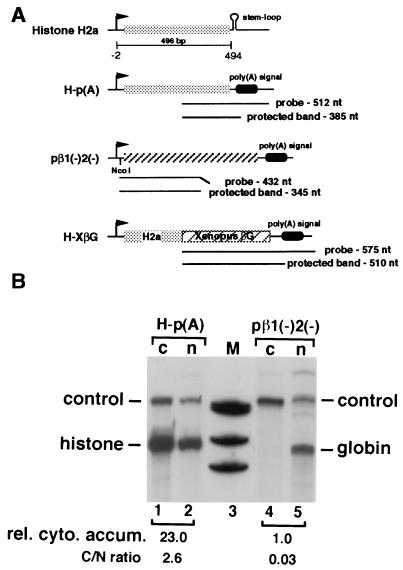
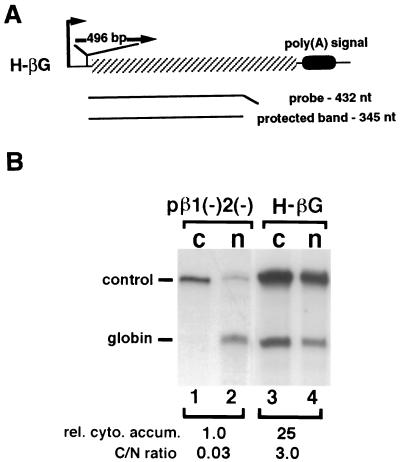
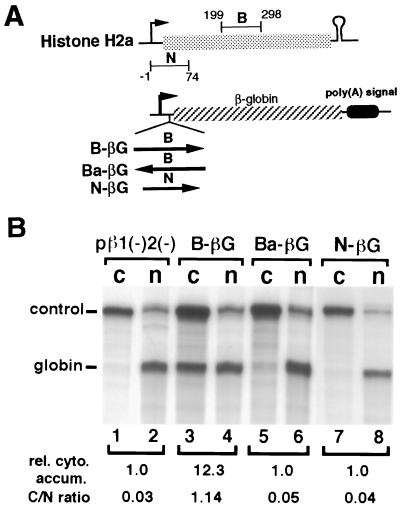
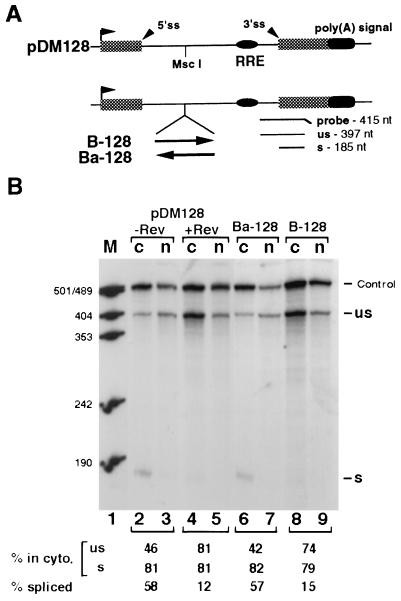
The mouse histone H2a gene-containing plasmid, MM614 (35), and plasmids pβ1(−)2(−) (8, 20), pDM128, and pRSV-Rev (36, 37) have been described. H-p(A) was created by inserting the 496-bp H2a PCR fragment (nt −2 to 494 relative to the H2a transcription start site) into a pBluescript-based expression vector made in our laboratory (see Fig. 1A). Transcription is driven by the cytomegalovirus immediate early promoter and the 3′ end of RNA is formed by the rabbit β-globin poly(A) signal. H-βG was constructed by cloning the 496-bp fragment into a unique NcoI site at the globin 5′-untranslated region in pβ1(−)2(−) (see Fig. 2A). B-βG and Ba-βG were built by inserting the 100-bp H2a PCR fragment (fragment B, nt +199 to +298, relative to the transcription start site) into pβ1(−)2(−) at the NcoI site in the sense and antisense orientations, respectively (see Fig. 3A). N-βG was created by cloning the 75-bp PCR fragment (fragment N, nt −1 to +74 relative to the transcription start site) into pβ1(−)2(−) at the NcoI site in the sense orientation. B-128 was made by cloning fragment B into pDM128 at the unique MscI site. Ba-128 is identical to B-128 except that fragment B was inserted in the antisense orientation (see Fig. 4A). H-XβG was created by placing a Xenopus laevis β-globin cDNA immediately 3′ of the histone sequence in H-p(A) (see Fig. 1A). This chimeric gene expresses messages that partition between the nucleus and the cytoplasm in a reproducible manner. This construct was used in all experiments as an internal control both for transfection efficiency and for subcellular fractionation.
Figure 1.
Polyadenylated H2a mRNA accumulates efficiently in the cytoplasm. (A) Schematic diagram of the plasmids. Shaded box represents the coding region of the H2a gene. Hatched boxes indicate either human or Xenopus β-globin genes. Arrows indicate transcriptional start sites. The thin line below the histone H2a construct marks the region used to build H-p(A). The numbers beneath the line represent nucleotides relative to the transcription start site. The probes used in RNase protection assays and the protected bands are indicated. The tilted portions of the probes depict nonhomologous sequences derived from plasmid vectors. In pβ1(−)2(−) the unique NcoI site is marked. The control construct H-XβG is described in Materials and Methods. Sizes are not to scale. (B) Autoradiograms of RNase protection assays of the globin and histone RNAs expressed in cells transfected with the indicated plasmids. M, molecular size markers (501, 404, 353, and 242 bp, respectively, from the top); c, cytoplasmic RNA; n, nuclear RNA; histone, mRNA from H-p(A); globin, RNA from pβ1(−)2(−); control, RNA from the cotransfected control plasmid pH-XβG. rel. cyto. accum., relative cytoplasmic mRNA accumulation obtained by setting the amount of cytoplasmic level of the globin RNA (lane 4) to 1, with normalization to the internal control RNA. C/N ratio, cytoplasmic and nuclear RNA distribution ratio after being normalized using the internal control RNA.
Figure 2.
Sequences contained within the H2a gene facilitate the cytoplasmic accumulation of cDNA transcripts. (A) Structure of construct H-βG. The 496-bp histone fragment inserted at the unique NcoI site is shown, with its orientation indicated. Other symbols are the same as those described for Fig. 1A. (B) Results from an RNase protection assay of the globin and the histone-globin chimeric RNAs accumulated in the cells transfected with the indicated constructs. control, same as for Fig. 1B; globin, RNAs from pβ1(−)2(−) (lanes 1 and 2) and H-βG (lanes 3 and 4), respectively. rel. cyto. accum., relative cytoplasmic globin-related mRNA accumulation obtained as described in the legend to Fig. 1 after setting the amount of cytoplasmic level of the globin RNA (lane 1) to 1, with normalization to the internal control RNA. C/N ratio, cytoplasmic and nuclear RNA distribution ratio after being normalized using the internal control RNA.
Figure 3.
A small element contained within the H2a coding region is sufficient to promote the cytoplasmic accumulation of the globin cDNA transcripts. (A) Structures of the plasmid constructs used. Shown at the top are the relative positions of the 100-bp (B) and 75-bp (N) fragments within the H2a gene. The numbers above or below indicate nucleotides relative to the transcription initiation site. Shown at the bottom are the three histone-globin chimeric gene constructs. The inserted fragments are indicated by thick lines, with arrows depicting their orientations. Other symbols are the same as for Fig. 1A. (B) Results from RNase protection assays of RNAs prepared from the cells transfected with the indicated plasmids. control, same as in Fig. 1B; globin, mRNAs from the indicated constructs. rel. cyto. accum., relative cytoplasmic globin mRNA accumulation obtained as described in the legend to Fig. 1 after arbitrarily setting the amount of cytoplasmic level of the globin RNA (lane 1) to 1, with normalization to the internal control RNA. C/N ratio, cytoplasmic and nuclear RNA distribution ratio after being normalized using the internal control RNA.
Figure 4.
The histone element functionally mimics the Rev/RRE system. (A) Structures of the plasmid constructs. Shaded boxes and thin lines denote the exon and the intron sequences from the HIV-1 gene, respectively. The RRE sequence and the unique MscI site are also marked. In B-128 and Ba-128 the inserted histone fragments are indicated, with the arrows depicting the orientations. 5′ss, 5′-splice site; 3′ss, 3′-splice site; us, unspliced RNA; s, spliced RNA. (B) RNase protection assays of RNAs prepared from cells transfected with the indicated plasmids. +Rev, with cotransfection of the Rev-expressing vector; −Rev, without cotransfection of the Rev-expression vector. Other labels are identical to those for Fig. 1B. At the bottom is the quantitation of the fraction of either unspliced or spliced RNA that was detected in the cytoplasm of transfected cells, as well as the percentage of total RNA that was spliced.
Cells and Transfections.
COS7 cells were maintained and propagated as described (38). Transfections were carried out with a modified CaPO4 DNA coprecipitation method (39). Approximately 6 hr prior to transfection, cells were diluted 2-fold and replated in 150-mm dishes. Five micrograms of reporter DNA [pβ1(−)2(−), H-p(A), H-βG, B-βG, Ba-βG, N-βG, pDM128, B-128, or Ba-128], 5 μg of the control DNA H-XβG, and 25 μg of pBluescript DNA or of Rev-expressing plasmid pRSV-Rev were used per plate for transfection.
RNA Preparation.
Nuclear and cytoplasmic RNAs were isolated 48 hr after transfection. For the preparation of cytoplasmic RNA, cells were rinsed free of media with ice-cold PBS and were then disrupted with an Nonidet P-40 (NP-40) lysis buffer (10 mM Hepes, pH 7.6/10 mM NaCl/3 mM MgCl2/0.5% NP-40) on ice for 30 sec. Cytoplasmic lysates were collected in new tubes and appropriate amounts of guanidinium thiocyanate crystals were added to give a final concentration of 4 M. Cytoplasmic RNA was then purified through 5 M CsCl step gradients. For nuclear RNA, the above intact cell nuclei, which were still attached to the plates, were rinsed with ice-cold NP-40 buffer followed by lysis in 4 M guanidinium isothiocyanate, 20 mM sodium acetate (pH 5.2), 0.1 mM DTT, and 0.5% N-lauryl sarcosine. The RNA was then pelleted through cesium chloride as described above.
RNase Protection Assays.
Internally labeled RNA probes were made by in vitro transcription by T3 or T7 RNA polymerase in the presence of [α-32P]UTP. DNA templates were removed by RQ1 DNase digestion followed by phenol/chloroform extraction. Internally labeled riboprobes were hybridized to target RNAs at 60°C overnight as described previously (40). The hybridization products were digested with a T1/T2 mixture (41) at 37°C for 1.5 hr and the resulting samples were resolved on 6% denaturing polyacrylamide gels. Routinely, 50% volume of total nuclear and cytoplasmic RNA samples were used for each RNase protection assay.
Quantitation of RNase Protection Data.
Protected bands were quantitated using a Packard Instant Imager. Background was subtracted using regions of identical size located immediately below each of the experimental bands. When bands within the same lane were compared, values were adjusted for length and uridine content, because the radioactive probes used were internally labeled with [32P]UTP.
To accurately determine the subcellular distribution of various RNAs, the cytoplasmic and nuclear RNA distribution ratio of RNA expressed from the internal control plasmid H-XβG was used to normalize all experimental results. In different experiments, this ratio varied slightly. We arbitrarily set 0.50 as a standard ratio for normalization purposes, and values for other RNAs were adjusted by the same amount. For example, the experimental radioactive counts of the control RNA in Fig. 4B (lane 3) were reduced by 12% to achieve a ratio of 0.50. This same 12% reduction was then applied to the values for the corresponding reporter RNA (us and s) bands in the same lane. Similar corrections were performed for other RNase protection data.
RESULTS
Polyadenylated H2a mRNA Accumulates Efficiently in the Cytoplasm, but Human β-Globin cDNA Transcripts Do Not.
To determine whether there are sequences within the histone H2a gene that function to help in cytoplasmic RNA accumulation, we first made a plasmid construct H-p(A) that expresses polyadenylated histone mRNAs (Fig. 1A). H-p(A) contains the mouse histone H2a transcribed region (nt −2 to +494 relative the histone transcription start site) but lacks the 3′-end stem–loop and other histone 3′-end processing sequences. Transcription is driven by the cytomegalovirus promoter and mRNA 3′ ends are generated by polyadenylation. This clone allowed us to uncouple expression of the H2a gene from cell cycle regulation, since transcription and histone 3′-end processing (not polyadenylation) have been shown to be the key targets for the cell cycle regulation (42, 43). To analyze the intracellular distribution of polyadenylated histone mRNAs, H-p(A) was transfected into COS7 cells. Nuclear and cytoplasmic RNAs were extracted 48 hr after transfection and examined by quantitative RNase protection assays. As a control, the level of mRNA produced from a human β-globin cDNA expression vector was examined in a parallel experiment.
As shown in Fig. 1B, the cytoplasmic accumulation of the globin cDNA transcripts is extremely low (lane 4), suggesting that nuclear export of these RNAs is poor. In contrast, a large fraction of the mRNAs transcribed from the H-p(A) construct accumulate in the cytoplasm (lane 1), indicating that export of these transcripts is much more efficient compared with that of the globin cDNA transcripts. The size of the protected band indicates that the histone mRNA is polyadenylated at the predicted site (lanes 1 and 2). RNase protection assays using probes complementary to other regions of the histone gene revealed that no cryptic splicing had occurred (data not shown). The above results therefore raised the possibility that the H2a transcribed region may contain cis-acting sequences that allow efficient cytoplasmic accumulation of intronless or unspliced mRNAs.
Sequences Within the H2a Gene Permit Efficient Cytoplasmic Accumulation of Human β-Globin cDNA Transcripts.
To identify putative cis-acting sequences responsible for intronless mRNA transport, we created a chimeric gene H-βG, in which the 496-bp H2a transcribed region was inserted into the 5′-untranslated region of a human β-globin cDNA clone (Fig. 2A). We then asked whether this sequence could rescue the defects in cytoplasmic accumulation of the globin cDNA transcripts. The H-βG plasmid was introduced into COS7 cells by transfection and the subcellular distribution of RNAs was measured by RNase protection assays. Results shown in Fig. 2B reveal that inclusion of the histone sequences elevated the cytoplasmic accumulation of the globin RNA by 25-fold (compare lane 3 with lane 1). Insertion of the same histone sequences upstream of the β-globin promoter region did not have such an effect (data not shown). These results strongly suggested the presence of cis-acting sequences within the H2a transcribed region that enable efficient cytoplasmic accumulation of unspliced mRNAs.
Identification of a Functional Element That Facilitates Cytoplasmic Accumulation of Intronless Transcripts.
Based on the findings described above, we next searched for a smaller cis-acting sequence capable of enhancing the cytoplasmic accumulation of unspliced mRNAs. With PCR and cloning techniques we were able to define a 100-bp sequence (fragment B) within the H2a coding region that has such a function, whereas a fragment from another region does not. As shown in Fig. 3A, three constructs were made by inserting PCR fragments into the 5′-untranslated region of the globin cDNA gene. In B-βG and Ba-βG, fragment B was inserted in the sense and antisense orientations, respectively. Fragment B does not contain any AUG codon that might confuse results by inducing nonsense-mediated mRNA decay within the nucleus. In N-βG, the 75-bp histone sequence (fragment N) was inserted in the sense orientation. Results from an RNase protection assay with RNAs prepared from the transfected cells are shown in Fig. 3B. Fragment B in the sense orientation increased the cytoplasmic accumulation of the globin RNA by approximately 12-fold (compare lanes 3 and 1). This effect is orientation-dependent, since the same sequence inserted in the opposite direction had no effect on the intracellular distribution of the globin RNA (compare lanes 5 and 1). This effect is also specific, since insertion of the sequence from another region of the H2a gene did not change the cytoplasmic accumulation of the globin RNA (compare lanes 7 and 1).
The Histone Sequence Can Replace Rev and RRE for the Cytoplasmic Accumulation of Unspliced HIV-1-Related mRNA.
To further understand how this sequence might work, fragment B was inserted into the intron region of an HIV-1-based reporter construct pDM128 (Fig. 4A). In B-128 and Ba-128, the fragment was inserted in the sense and antisense orientations, respectively. It has been shown that efficient cytoplasmic accumulation of unspliced mRNAs transcribed from subgenomic HIV-1 constructs is generally dependent on the presence of a functional Rev protein (37, 42–44). Consistent with this, the cytoplasmic accumulation of the unspliced species transcribed from the reporter construct is significantly augmented when Rev is expressed (Fig. 4B, compare lanes 4 and 2). In addition, the increased level of unspliced species is accompanied by a decreased level of spliced species (compare lanes 4 and 5 with lanes 2 and 3). These results are in agreement with observations from other groups (5, 37, 45). Intriguingly, in the absence of Rev, the presence of the 100-bp sequence in the reporter construct leads to increased cytoplasmic accumulation of the unspliced species and a concomitant decrease in the level of the spliced species (compare lanes 8 and 9 with lanes 2 and 3). These suggest that the histone sequence may act to enhance the nuclear export of the unspliced RNAs and to suppress splicing as well. Like RRE, this sequence is functional only when placed in the sense orientation (compare lanes 8 and 9 with lanes 6 and 7). Because the presence of this sequence in the globin cDNA or in the HIV-1-related gene construct did not significantly alter mRNA half-lives in either the nucleus or the cytoplasm (data not shown), it is thus unlikely that it acts at the RNA stability level. That the histone sequence can functionally mimic RRE and acts in an orientation-dependent manner is consistent with the view that it might function at the posttranscriptional level.
DISCUSSION
We have described here an 100-bp cis-acting sequence contained within the mouse histone H2a gene, which allows efficient cytoplasmic accumulation of intronless gene transcripts. We have (i) located this novel cellular sequence to nt 199–298 within the H2a coding region, (ii) demonstrated that this sequence facilitates cytoplasmic accumulation of human β-globin cDNA transcripts, (iii) provided evidence that this sequence appears to be functionally equivalent to the RRE in promoting cytoplasmic accumulation of unspliced HIV-1-related mRNA, and (iv) shown that this sequence functions in an orientation-dependent manner.
Increased cytoplasmic accumulation of mRNAs often results from enhanced nucleocytoplasmic transport. Our finding that the 100-bp histone sequence permits efficient cytoplasmic accumulation of unspliced mRNAs suggests that this sequence might function as a cis-acting RNA transport element. Our finding also supports the view that naturally intronless transcripts may contain positive cis-acting RNA transport elements. Importantly, the sequence identified here is the first well-characterized example of a cellular sequence that has such a function. We do note that the existence of such an element in the c-jun message has been suggested (21). Sequences with a similar function have also been described in two viral systems (21, 23, 24). It has been recently reported that an RNA sequence (called the posttranscriptional regulatory element or PRE) located at the 3′ region of intronless hepatitis B virus transcripts promotes efficient cytoplasmic accumulation of the viral mRNAs (23, 24). Similarly, a cis-acting sequence (called the posttranscriptional processing element or PPE) present in the coding region of the herpes simplex virus thymidine kinase transcript has been shown to have the same effects (21). Like the histone sequence, both viral elements can functionally replace introns for the expression of intron-dependent genes and appear to rely on cellular factors for their function. It is thus extremely likely that it may be a general situation for other intronless genes to contain positive cis-acting transport elements that facilitate their nuclear export.
The precise mechanism by which the histone sequence facilitates cytoplasmic accumulation of intronless gene transcripts is not yet known. One hypothesis is that this sequence might function by directly promoting histone mRNA nuclear export. As shown in this study, the histone sequence acts constitutively to transport unspliced HIV-1-related mRNA to the cytoplasm in the absence of Rev (Fig. 4B, compare lanes 8 and 9 with lanes 2 and 3). Additionally, it appears to have a negative effect on splicing (Fig. 4B, compare lanes 8 and 9 with lanes 2 and 3). Furthermore, these functions are orientation-dependent (Fig. 4B, compare lanes 8 and 9 with lanes 6 and 7). Because Rev/RRE also has been reported to inhibit splicing as well as promote RNA transport, the histone sequence functionally mimics Rev/RRE. Accumulated data have suggested that Rev may facilitate the nuclear export of unspliced HIV-1 mRNA by directly interacting with the cellular export machinery (45–53). It is thus plausible that the histone sequence might function by a similar mechanism, i.e., by interacting with a cellular factor(s) whose function is analogous to that of Rev.
3′ end processing has been shown to play an essential role in mRNA nuclear export. Studies have indicated that polyadenylation promotes mRNA transport from the nucleus to the cytoplasm (38, 54). Likewise, histone 3′ end processing has been implicated in the nuclear export of histone mRNAs (54, 55). Thus, the formation of a mature 3′ end, either by polyadenylation or by the histone processing machinery, might direct the messages along a pathway that leads to the nuclear pore. Evidence exists that the presence of introns can activate polyadenylation (18, 56–61). Recent results from our laboratory (unpublished work) also suggest that the histone sequence facilitates polyadenylation. Based on these data, we therefore hypothesize that part of the function of the histone sequence might mimic introns that act to enhance polyadenylation, which in turn might lead to efficient RNA nuclear export.
Inhibition of splicing by the histone sequence (Fig. 4B, lanes 8 and 9) suggests that it might be a natural function of this sequence to block potential spliceosome formation on histone mRNAs. This might be a general aspect of transport elements for intronless gene transcripts, since these transcripts may contain cryptic or nonconsensus splice sites.
The identification of the histone sequence suggests that messages can have more than one positive element that signal nuclear export. In the case of the H2a mRNA, these positive elements include the sequence identified in this work, sequences involved in 3′ end processing, and, perhaps, the 5′ cap structure as well (62–64). These elements may act in concert to allow for efficient nucleocytoplasmic transport of H2a mRNA. Although the histone sequence maintains the ability to activate the cytoplasmic accumulation of intronless gene transcripts, it does not appear to be fully functional compared with the 496-bp fragment (compare Fig. 2B, lane 3 with Fig. 3B, lane 3). Therefore, we speculate that the H2a coding region might contain more than one such sequence.
An interesting aspect of this study is that the histone sequence we define here overlaps with the histone promoter enhancer region identified (65–68). However, the sequence we used does not display significant transcription enhancement activity, perhaps due to the different promoters we used and the different experimental context. Additionally, when the entire H2a transcribed region was inserted upstream of the promoter region in some of our constructs, it did not have such a function (data not shown). Nevertheless, it remains possible that this histone sequence can function both at the DNA (for transcription enhancement) and RNA (for mRNA export) levels.
It has been well established that the histone 3′ end processing (not polyadenylation) and the histone promoters are the targets for the cell cycle regulation (42, 43). Replacement of the histone 3′ processing sequences with polyadenylation signals led to cell cycle-independent histone expression (31, 33, 69). Based on these data, we postulate that the function of the histone sequence in mRNA transport is probably not cell cycle regulated.
In summary, we have identified a novel cellular sequence in the mouse histone H2a gene that appears to function constitutively to facilitate the cytoplasmic accumulation of intronless gene transcripts. This sequence can also functionally mimic the Rev/RRE system in an HIV-1-based construct; it can both replace Rev and RRE for the efficient cytoplasmic accumulation of unspliced HIV-1-related mRNA and appears to suppress splicing. Moreover, it is found in the rather abundant and ubiquitous messages for histone H2a and is thus not likely to be cell-type or species-dependent in its action. Therefore, we speculate that all histone messages have a similar element and suggest that the natural abundance of these messages should make it possible to identify and isolate the factor(s) involved. As such, it might be of interest not only in furthering our understanding of mRNA transport, but also might prove of value in applications such as the expression of cDNAs using recombinant vectors or as an aid in gene therapy.
Acknowledgments
We thank M. Kumar, X. Li, M. Szlachetka, K. Wimler, and S. Young for helpful comments on the manuscript. We gratefully acknowledge W. Marzluff for the histone H2a-containing plasmid, T. Hope for plasmid pDM128, and J. Mertz for plasmid pβ1(−)2(−), as well as for helpful comments on the manuscript. We especially thank K. Wimler for excellent technical assistance. This work is supported by Grant CA45382 from the National Cancer Institute.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: RRE, Rev-responsive element.
References
- 1.Legrain P, Rosbash M. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 2.Chang D D, Sharp P A. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 3.Huang Y, Carmichael G G. Mol Cell Biol. 1996;16:6046–6054. doi: 10.1128/mcb.16.11.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen B R. Microbiol Rev. 1992;56:375–394. doi: 10.1128/mr.56.3.375-394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M L. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alonso-Caplen F V, Nemeroff M E, Qiu Y, Krug R M. Genes Dev. 1992;6:255–267. doi: 10.1101/gad.6.2.255. [DOI] [PubMed] [Google Scholar]
- 7.Qiu Y, Krug R M. J Virol. 1994;68:2425–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yen T S B. Semin Virol. 1993;4:33–42. [Google Scholar]
- 9.McKnight S L. Nucleic Acids Res. 1980;8:5949–5964. doi: 10.1093/nar/8.24.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buchman A R, Berg P. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Callis J, Fromm M, Walbot V. Genes Dev. 1987;1:1183–1200. doi: 10.1101/gad.1.10.1183. [DOI] [PubMed] [Google Scholar]
- 12.Chung S, Perry R P. Mol Cell Biol. 1989;9:2075–2082. doi: 10.1128/mcb.9.5.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng T, Li Y, Johnson L F. Nucleic Acids Res. 1989;17:645–658. doi: 10.1093/nar/17.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasser C S, Simonsen C C, Schilling J W, Schimke R T. Proc Natl Acad Sci USA. 1982;79:6522–6526. doi: 10.1073/pnas.79.21.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruss P, Lai C-J, Dhar R, Khoury G. Proc Natl Acad Sci USA. 1979;76:4317–4321. doi: 10.1073/pnas.76.9.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamer D, Leder P. Cell. 1979;17:737–747. doi: 10.1016/0092-8674(79)90280-0. [DOI] [PubMed] [Google Scholar]
- 17.Jonsson J J, Foresman M D, Wilson N, McIvor R S. Nucleic Acids Res. 1992;20:3191–3198. doi: 10.1093/nar/20.12.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nesic D, Cheng J, Maquat L E. Mol Cell Biol. 1993;13:3359–3369. doi: 10.1128/mcb.13.6.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neuberger M S, Williams G T. Nucleic Acids Res. 1988;16:6713–6724. doi: 10.1093/nar/16.14.6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryu W S, Mertz J E. J Virol. 1989;63:4386–4394. doi: 10.1128/jvi.63.10.4386-4394.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Mertz J. Genes Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- 22.Donello J E, Beeche A A, Smith G J, III, Lucero G R, Hope T J. J Virol. 1996;70:4345–4351. doi: 10.1128/jvi.70.7.4345-4351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Liang T J. Mol Cell Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Z-M, Yen T S B. Mol Cell Biol. 1995;15:3864–3869. doi: 10.1128/mcb.15.7.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kedes L H. Annu Rev Biochem. 1979;48:837–870. doi: 10.1146/annurev.bi.48.070179.004201. [DOI] [PubMed] [Google Scholar]
- 26.Koilka B K, Frielle T, Collins S, Yang-Feng T, Kobilka T S, Francke U, Lefkowitz R J, Caron M G. Nature (London) 1987;329:75–79. doi: 10.1038/329075a0. [DOI] [PubMed] [Google Scholar]
- 27.Nagata S, Mantei N, Weissmann C. Nature (London) 1980;287:401–408. doi: 10.1038/287401a0. [DOI] [PubMed] [Google Scholar]
- 28.Hattori K, Angel P, Beau M M L, Karin M. Proc Natl Acad Sci USA. 1988;85:9148–9152. doi: 10.1073/pnas.85.23.9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hentschel C C, Birnstiel M X. Cell. 1981;25:301–313. doi: 10.1016/0092-8674(81)90048-9. [DOI] [PubMed] [Google Scholar]
- 30.Marzluff W F. Gene Expression. 1992;2:93–97. [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng G, Nandi A, Clerk S, Skoultchi A I. Proc Natl Acad Sci USA. 1989;86:7002–7006. doi: 10.1073/pnas.86.18.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Challoner P B, Moss S B, Groudine M. Mol Cell Biol. 1989;9:902–913. doi: 10.1128/mcb.9.3.902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirsch A L, Groudine M, Challoner P B. Genes Dev. 1989;3:2172–2179. doi: 10.1101/gad.3.12b.2172. [DOI] [PubMed] [Google Scholar]
- 34.Mannironi C, Bonner W M, Hatch C L. Nucleic Acids Res. 1989;17:9113–9126. doi: 10.1093/nar/17.22.9113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Graves R A, Wellman E, Chiu I M, Marzluff W F. J Mol Biol. 1985;183:179–194. doi: 10.1016/0022-2836(85)90211-6. [DOI] [PubMed] [Google Scholar]
- 36.Hope T J, Huang X, McDonald D, Parslow T G. Proc Natl Acad Sci USA. 1990;87:7787–7791. doi: 10.1073/pnas.87.19.7787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterlin B M, Luciq P A, Barr P J, Walker M D. Proc Natl Acad Sci USA. 1986;83:9734–9738. doi: 10.1073/pnas.83.24.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y, Carmichael G G. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cahill K B, Carmichael G G. J Virol. 1989;63:3634–3642. doi: 10.1128/jvi.63.9.3634-3642.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adami G R, Marlor C W, Barrett N L, Carmichael G G. J Virol. 1989;63:85–93. doi: 10.1128/jvi.63.1.85-93.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lichtler A, Barrett N L, Carmichael G G. BioTechniques. 1992;12:231–232. [PubMed] [Google Scholar]
- 42.Harris M E, Bohni R, Schneiderman M H, Ramamurthy L, Schumperli D, Marzluff W F. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sittman D B, Graves R A, Marzluff W F. Proc Natl Acad Sci USA. 1983;80:1849–1853. doi: 10.1073/pnas.80.7.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malim M H, Hauber J, Le S-Y, Maizel J V, Cullen B R. Nature (London) 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 45.Malim M H, Cullen B R. Mol Cell Biol. 1993;13:6180–6189. doi: 10.1128/mcb.13.10.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bogerd H P, Fridell R A, Madore S, Cullen B R. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 47.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 48.Fritz C C, Zapp M L, Green M R. Nature (London) 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 49.Kalland K H, Szilvay A M, Brokstad K A, Saetrevik W, Haukenes G. Mol Cell Biol. 1994;14:7436–7444. doi: 10.1128/mcb.14.11.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malim M H, Bohnlein S, Hauber J, Cullen B R. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 51.Meyer B E, Malim M H. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 52.Pfeifer K, Weiler B E, Ugarkovic D, Bachmann M, Schroder H C, Muller W E. Eur J Biochem. 1991;199:53–64. doi: 10.1111/j.1432-1033.1991.tb16091.x. [DOI] [PubMed] [Google Scholar]
- 53.Stutz F, Neville M, Rosbash M. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- 54.Eckner R, Ellmeier W, Birnstiel M L. EMBO J. 1991;10:3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams A S, Ingledue T R, Kay B K, Marzluff W F. Nucleic Acids Res. 1994;22:4660–4666. doi: 10.1093/nar/22.22.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Collis P, Antoniou M, Grosveld F. EMBO J. 1990;9:233–240. doi: 10.1002/j.1460-2075.1990.tb08100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang M T F, Gorman C M. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X, Mertz J E. Nucleic Acids Res. 1993;21:5256–5263. doi: 10.1093/nar/21.22.5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X, Mertz J E. Nucleic Acids Res. 1996;24:1765–1774. doi: 10.1093/nar/24.9.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Niwa M, Rose S D, Berget S M. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 61.Pandey N B, Chodchoy N, Liu T J, Marzluff W F. Nucleic Acids Res. 1990;18:3161–3170. doi: 10.1093/nar/18.11.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dargemont C, Kühn L C. J Cell Biol. 1992;118:1–9. doi: 10.1083/jcb.118.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamm J, Mattaj I W. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 64.Izaurralde E, Stepinski J, Darzynkiewicz E, Mattaj I W. J Cell Biol. 1992;118:1287–1295. doi: 10.1083/jcb.118.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bowman T L, Hurt M M. Nucleic Acids Res. 1995;23:3083–3092. doi: 10.1093/nar/23.16.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hurt M M, Pandey N B, Marzluff W F. Proc Natl Acad Sci USA. 1989;86:4450–4454. doi: 10.1073/pnas.86.12.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hurt M M, Bowman T L, Marzluff W F. Mol Cell Biol. 1991;11:2929–2936. doi: 10.1128/mcb.11.6.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalukov N K, Pena-Pabon L, Hurt M M. Nucleic Acids Res. 1996;24:523–531. doi: 10.1093/nar/24.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levine B J, Chodchoy N, Marzluff W F, Skoultchi A I. Proc Natl Acad Sci USA. 1987;84:6189–6193. doi: 10.1073/pnas.84.17.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]