Abstract
Albicidin phytotoxins are pathogenicity factors in a devastating disease of sugarcane known as leaf scald, caused by Xanthomonas albilineans. A gene (albD) from Pantoea dispersa has been cloned and sequenced and been shown to code for a peptide of 235 amino acids that detoxifies albicidin. The gene shows no significant homology at the DNA or protein level to any known sequence, but the gene product contains a GxSxG motif that is conserved in serine hydrolases. The AlbD protein, purified to homogeneity by means of a glutathione S-transferase gene fusion system, showed strong esterase activity on p-nitrophenyl butyrate and released hydrophilic products during detoxification of albicidins. AlbD hydrolysis of p-nitrophenyl butyrate and detoxification of albicidins required no complex cofactors. Both processes were strongly inhibited by phenylmethylsulfonyl fluoride, a serine enzyme inhibitor. These data strongly suggest that AlbD is an albicidin hydrolase. The enzyme detoxifies albicidins efficiently over a pH range from 5.8 to 8.0, with a broad temperature optimum from 15 to 35°C. Expression of albD in transformed X. albilineans strains abolished the capacity to release albicidin toxins and to incite disease symptoms in sugarcane. The gene is a promising candidate for transfer into sugarcane to confer a form of disease resistance.
Keywords: phytotoxin resistance, leaf scald disease, serine hydrolase, pathogenicity factor
Many phytopathogenic bacteria and fungi produce phytotoxins that interfere with various functions of plant cells, resulting in symptoms including chlorosis, wilting, necrosis, watersoaking, and growth abnormalities (1, 2). The toxins may weaken or suppress constitutive and inducible plant defense mechanisms (3, 4). Toxin production is commonly associated with greater pathogen multiplication and disease severity, and in some cases the toxins are essential for initial colonization or systemic invasion by the pathogen (4, 5). In many cases, a close relationship exists between toxin tolerance and resistance of plants to disease (6). The first cloned plant disease resistance gene was the maize HM1 gene encoding a reductase that inactivates HC toxin produced by the pathogenic fungus Cochliobolus carbonum race 1 (7). Resistance based on insensitivity to toxins is expected to be stable in cases that can only be overcome by a gain of function in the pathogen. Phytotoxin resistance provides a paradigm indicating the need for greater emphasis on understanding of compatibility factors as a basis for engineering stable resistance to plant diseases (8).
Leaf scald is one of the most devastating diseases of sugarcane in many countries, and it continues to spread into new areas (9–11). The causal agent, Xanthomonas albilineans, is a systemic, xylem-invading bacterium that may be present at low population sizes in symptom-less plants (12). The pathogen produces a family of antibiotics and phytotoxins that block DNA replication in bacteria and sugarcane proplastids (13, 14). The major toxin, named albicidin, has been partially characterized as a low Mr compound with several aromatic rings. Because albicidin is rapidly bactericidal to a range of Gram-positive and Gram-negative bacteria at concentrations as low as 1 ng ml−1, it is also of interest as a potential clinical antibiotic (15).
Symptoms of leaf scald disease include the emergence of chlorotic leaves, wilting, necrosis, and sometimes rapid death of plants, often after a prolonged latent period. An early diagnostic symptom is the presence of white pencil lines on emerging leaves after inoculation of the pathogen onto sugarcane plants decapitated above the apical meristem (9). Chlorosis results from blocked chloroplast development after inhibition of plastid DNA replication by albicidins produced by the pathogen in invaded xylem (14, 16). Albicidin phytotoxins appear to be important in systemic disease development, because tox− mutants of X. albilineans fail to induce any disease symptoms in inoculated sugarcane (17). However, rapid attenuation of the pathogen in culture has prevented critical experiments comparing the capacity of tox− mutants and tox+ revertants to induce systemic disease in the field. Albicidin resistance genes are therefore of great interest as a critical tool to assess the role of albicidin phytotoxins in leaf scald disease development and potentially to confer leaf scald disease resistance in transgenic sugarcane.
Several mechanisms of albicidin resistance recently have been identified, including elimination of active antibiotic uptake in Escherichia coli (18); production of a protein that reversibly binds the antibiotic in Klebsiella oxytoca (19) and in Alcaligenes denitrificans (20); and irreversible detoxification of albicidin in Pantoea dispersa (21, 22). Here we show that a single gene encoding an esterase from P. dispersa is responsible for albicidin detoxification. Expression of the gene in transformed X. albilineans abolished the capacity to release albicidin and to incite symptoms in inoculated sugarcane. The enzyme is active under conditions that indicate potential to destroy albicidin toxins in transgenic sugarcane plants.
MATERIALS AND METHODS
Bacteria and Cultivation.
E. coli strain DH5α was used to bioassay albicidin and also as the host strain for DNA cloning and subcloning. X. albilineans strain XA3 and P. dispersa strain SB1403 were isolated from diseased sugarcane from Queensland, Australia (21). E. coli was grown at 37°C in Luria–Bertani medium (23), and the other bacteria were grown at 28°C in sucrose and peptone medium (13). Broth cultures were aerated by shaking at 180 rpm on an orbital shaker.
Preparation of Albicidin and Cell-Free Extracts.
Albicidins produced in culture by X. albilineans XA3 were prepared and quantified as described (13, 21). Unless stated otherwise, the mixture of albicidins obtained after HW-40(s) chromatography was used in experiments reported here. Cell-free extracts were prepared from E. coli and X. albilineans strains as described for P. dispersa strains (21), except the X. albilineans strains were grown for 3 days before harvesting.
Cell-free extracts from sugarcane leaves were prepared by grinding 1 g of young leaves in liquid nitrogen and continued grinding to a fine homogenate after addition of 4 ml of protein extraction buffer (100 mM potassium phosphate/10 mM DTT/1 mM EDTA/0.1% Triton X-100, pH 7.0). Cell debris was removed by centrifugation at 11,000 × g for 20 min at 4°C. Protein concentrations in the cell-free extracts were measured by the Bradford dye-binding method (24), using BSA for calibration.
Cloning and Sequencing of the albD Gene.
A genomic library was constructed by partial digestion of DNA from P. dispersa SB1403 with BamHI restriction endonuclease, ligation into the BamHI site of the cosmid cloning vector pLAFR3, and transfection into E. coli DH5α (25, 26). Albicidin-resistant (Albr) recombinant clones were selected by patching transformants onto Luria–Bertani agar medium containing 10 μg/ml tetracycline and 200 ng/ml albicidin. Subcloning into pBluescript II SK(+) was carried out by routine techniques (27).
Albr clone pQZE533 and four ExoIII deletion albicidin-sensitive (Albs) clones (pQZE456, pQZE457, pQZE540, and pQZE560) were used to obtain the complete DNA sequence from both strands of the cloned albD gene, based on the dideoxynucleotide chain termination method (28) using the PRISM Cycle Sequencing Kit and 373A DNA Sequencer (Applied Biosystems).
Purification of AlbD Enzyme.
The albD coding region was amplified by PCR with forward primer 5′-TTAAG CGGGA TCCGT TTTGA TGGAC-3′ and reverse primer 5′-GATTG AATCG TATCA GCTGG AAGAG-3′, then digested by BamHI and PvuII, and ligated to the BamHI- and SmaI-digested glutathione S-transferase (GST) gene fusion vector pGEX-2T (Pharmacia). The resultant construct (pGSTAld) contained the chimeric albD gene fused in frame to the GST gene, which is under the control of isopropyl-β-d-thiogalactoside (IPTG)-inducible tac promoter. The IPTG-induced E. coli DH5α (pGSTAld) culture was pelleted by centrifugation, and cell-free extracts were prepared by ultrasonification and applied to a glutathione Sepharose 4B affinity column (Pharmacia) to bind the GST–AlbD fusion protein. AlbD protein was released by digestion with the protease thrombin for 16 h at room temperature and then analyzed by SDS/PAGE to confirm purity. The purified enzyme was stored at −20°C in PBS buffer (140 mM NaCl/2.7 mM KCl/10 mM Na2HPO4/1.8 mM KH2PO4, pH 7.3) plus 20% glycerol.
AlbD Enzymatic Assays.
Albicidin detoxification assay mixtures included AlbD enzyme (2 ng/μl) and albicidin (15 ng/μl) in 0.2 M phosphate buffer, pH 7.0. The reaction mixtures were incubated in water baths at selected temperatures for 30 min, then stopped by adding 10% SDS to a final concentration of 1% and bioassayed for remaining albicidin. AlbD activity is presented as the percentage of albicidin detoxified by the enzyme. The effect of pH on AlbD activity was determined by the same procedure except that the reactions were conducted at 28°C in 0.2 M phosphate buffers of different pH values.
For HPLC analysis of detoxification products, albicidin purified by reverse phase HPLC on preparative reverse phase 1 and octadecyldimethysilane preparative columns (13), and the β peak (15) was stored at −20°C. This albicidin was dissolved in 0.2 M phosphate buffer (pH 7.0) containing 5% methanol and was incubated with purified AlbD (5 ng/μl) for 1 h at 28°C. AlbD enzyme then was precipitated by increasing the methanol concentration to 80%. The supernatant after centrifugation was freeze-dried and dissolved in the desired mobile phase for HPLC through a C-18 analytical column and spectral diode array detector (Perkin–Elmer LC 250/235).
Sensitivity to phenylmethylsulfonyl fluoride (PMSF) was tested by incubating AlbD enzyme (250 ng/μl) with PMSF (2 mM) at 25°C for 30 min before enzyme activity assays. For esterase activity assay, 5 μl of PMSF-treated or untreated AlbD solution was added to 200 μl of esterase assay mixture (10 mM potassium phosphate buffer/0.2% Triton X-100/3 mM p-nitrophenyl butyrate, pH 7.0), incubated at 25°C for 15 min, and then stopped by SDS. The reaction mixture was made up to 1 ml by adding water, and the concentration of p-nitrophenol released was determined by measuring OD410 nm.
The tributyrin diffusion agar method was used for the detection of lipase activity (29), using a clone containing the apl-1 lipase gene from Aeromonas hydrophila as a positive control (30).
Genetic Transformation of X. albilineans with AlbD.
A 906-bp SphI–PvuII fragment (including 156 bp of upstream promoter region, the coding region, and 50 bp downstream of the stop codon of the albD gene) was blunt-ended by treatment with DNA polymerase Klenow fragment in the presence of a dNTP mixture, and ligated into the HincII site of the suicide vector pJP5603 (31). The ligation products were used to transform E. coli JM109 (λ-pir). A recombinant clone, pJPAldSP, was identified by restriction enzyme digestion and agarose gel electrophoresis, then transferred via the mobilizing strain S17–1 (λ-pir) into X. albilineans strain XA3, which is resistant to ampicillin. Transconjugant colonies were selected on sucrose and peptone agar medium containing 50 μg ml−1 kanamycin and 100 μg ml−1 ampicillin, then tested for albicidin production. Those colonies that failed to produce albicidin were further tested for AlbD production and for ability to utilize glucose, sucrose, fructose, and 20 amino acids as a sole carbon or nitrogen source (13).
Plant Material and Bacterial Inoculation.
Single-node cuttings from healthy sugarcane plants (variety Q44) were potted for ≈2 months before inoculation as described (21). The actively growing bacteria were centrifuged for 1 min at 11,000 × g, resuspended with sterilized water to OD600 = 1.68 (4 × 109 cfu/ml), and kept on ice until inoculation. A 200-μl volume of bacterial inoculum was added to the freshly cut surface of sugarcane plants decapitated above the apical meristem. Disease symptoms were recorded 2 weeks after inoculation.
RESULTS
Cloning and Characterization of the Albicidin Detoxification Region.
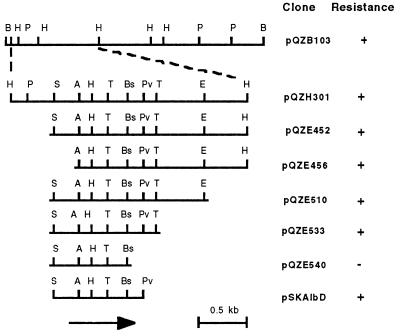
Five Albr clones were identified from 1600 tetracycline-resistant cosmid clones from a genomic DNA library of Albr P. dispersa SB1403. Dose response analysis showed that these Albr clones conferred more than 200-fold increased albicidin resistance in E. coli strain DH5α. Restriction analysis of four clones revealed a common band of 8 kb released by digestion with BamHI. Subclone pQZB103 containing this 8-kb fragment in vector pBluescript conferred albicidin detoxification activity 5-fold that of the Albr cosmid clones, probably because of the higher copy number of the subcloned plasmid.
Clone pQZH301 with an insert size of 2.5 kb was derived by partial HincII digestion of pQZB103. A series of exonuclease III directional deletions narrowed the region that confers Albr to the 1.1 kb of DNA insert in clone pQZE533 (Fig. 1). Cell-free extracts of all of the Albr clones showed strong albicidin detoxification activity.
Figure 1.
Restriction map of the P. dispersa albicidin resistance region. Clone pQZB103 contains the 8-kb BamHI fragment identified by restriction enzyme analysis of overlapping cosmid clones. Subclones were produced by restriction enzyme digestion and exonuclease III directional deletion, except pG5ZAlbD, which was obtained by ligating the SphI and PvuII fragment from pQZE533 into SphI- and EcoRV-digested cloning vector pGEM-5Zf (+). The location and direction of transcription of the albD ORF is shown by an arrow. Albicidin resistance of the clones is shown on the right: +, resistant to albicidin >1000 ng/ml; −, sensitive to albicidin at 100 ng/ml. Restriction enzymes: A, AatII; B, BamHI; Bs, BspHI; H, HincII; P, PstI; Pv, PvuII; S, SphI; and T, TaqI.
Sequence of the Albicidin Detoxification Gene.
The DNA sequence and the inferred amino acid sequence of the albD gene in pQZE533 is shown in Fig. 2. A sequence (TAAGCGGG) with six of eight bases complementary to the 3′ end of the E. coli 16S rRNA is centered 14 nt upstream from the ATG initiation codon leading the only ORF and may serve as a ribosome binding site (32). The best sequence match (TAATAT) to the E. coli consensus-10 promoter region (TATAAT) occurs 33 bp upstream of the initiation codon but no significant homology to the consensus −35 region was found at the expected location. Two TCTT boxes and a TGTG box that closely resemble the TCTG consensus sequence characteristic of factor-independent termination sites (33) occur downstream of the termination codon of albD.
Figure 2.
Nucleotide sequence and predicted translation of the albD gene cloned from P. dispersa. The putative ribosome binding (SD) region and the −10 region are underlined, and the putative transcription termination codon is indicated by an asterisk. The GXSXG region conserved in serine hydrolases is shaded. The putative factor-independent termination sites are double underlined.
The product of albD is predicted to be a 235-amino acid protein with a calculated Mr of 25,411 and an isoelectric point at 6.23 due to 32 acidic and 27 basic residues. It shows no significant similarity to other proteins in the major databases (GenBank, European Molecular Biology Laboratory, Protein Information Resource, and Swiss-Prot) by fasta and blast analysis or to consensus profiles of more than 600 different groups of proteins and enzymes tested using the Genetics Computer Group (Madison, WI) profilescan and motif programs. However, as described in the next section, the purified AlbD enzyme showed apparent hydrolase activity against albicidin. By comparison with various hydrolases, a GxSxG motif, which is a common feature of serine hydrolases, was found near the middle of the AlbD peptide (Figs. 2 and 3).
Figure 3.
The GXSXG regions of various hydrolytic enzymes. Rml, Rhizomucor miehei lipase; hpl, human pancreatic lipase; Pcl, Penicillium camembertii lipase; Pfl, Pseudomonas fragi lipase; Fsc, Fusarium solani cutinase; Vmt, Vibrio harveryi myristoyl transferase (38). Bse, Bacillus stearothermophilus esterase (40); Pfe, Pseudomonas fluorescens esterase A (41); mce, murine carboxylesterase (42); hce, human pancreatic cholesterol esterase (43); hpf, human protease factor XII (44); and hpt, human prothrombin (44).
Purification and Properties of the AlbD Enzyme.
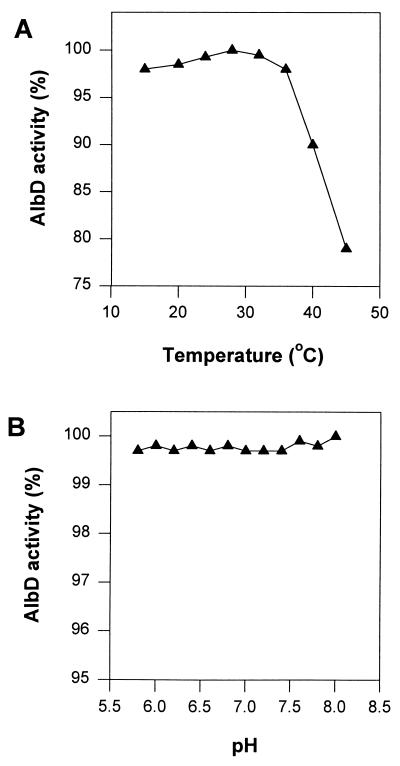
Recombinant AlbD protein purified using the GST gene fusion system (34) has four extra amino acids (glycine, serine, valine, and leucine) at the N terminus of the predicted AlbD peptide. The SDS/PAGE analysis showed the purified AlbD enzyme as a band at ≈26 kDa, which is consistent with the predicted Mr of 25,411 from DNA sequence analysis. Purified AlbD detoxified albicidin in a phosphate buffer and was stable for several months in 20% glycerol buffer at −20°C. Detoxification was progressive and irreversible, as demonstrated for crude cell extracts from P. dispersa strain SB1403 (21) in contrast to nonenzymatic albicidin binding proteins (19, 20). Albicidin detoxification activity of purified AlbD was insensitive to pH in the range from pH 5.8 to 8.0 and showed a broad temperature optimum between 15 and 35°C and >75% activity at 45°C (Fig. 4).
Figure 4.
Effect of temperature (A) and pH (B) on AlbD enzyme activity.
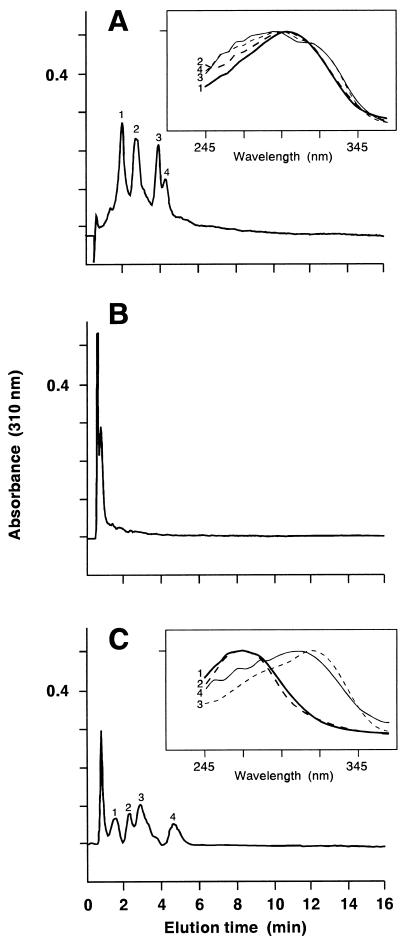
The stored albicidin β peak from preparative HPLC (15) was resolved into four peaks (with retention times ranging from 1.87 to 4.01 min) on an analytical reverse phase HPLC column (Fig. 5A). After AlbD treatment, antibiotic activity was abolished, and the reaction products were eluted as a single peak (with a retention time of 0.49 min) under the same HPLC conditions (Fig. 5B). The reaction products were resolved into four major peaks when a more polar eluent was used for HPLC (Fig. 5C), indicating that detoxification of albicidins by AlbD yields more hydrophilic products. The unreacted albicidins show similar absorption spectra, with a peak at 290–300 nm (Fig. 5A) whereas the reaction products fall into two classes with absorption maxima at 270 nm or 310–320 nm (Fig. 5C).
Figure 5.
HPLC profiles of albicidins before and after the AlbD enzymatic reaction, analyzed by isocratic elution with (A and B) 60% methanol in water containing 1% (vol/vol) acetic acid or (C) 50% methanol in water containing 1% acetic acid at a flow rate of 1 ml/min through a C18 analytical HPLC column [40 × 4.6 mm (Beckman)]. (A) Untreated albicidins; (B, C) products from treatment of albicidins with AlbD enzyme. The first peak in the HPLC profiles is the injected solvent front. Insets in A and C are the overlaid absorption spectra of the separated peaks.
AlbD showed strong esterase activity when p-nitrophenyl butyrate was used as substrate. Hydrolysis of p-nitrophenyl butyrate and detoxification of albicidin were inhibited by 97–98% when AlbD was pretreated with PMSF, a serine enzyme inhibitor. Cell extracts from X. albilineans strains and sugarcane, including leaf scald-susceptible and -resistant varieties, showed strong esterase activity on p-nitrophenyl butyrate but did not detoxify albicidin. E. coli clones DH5α (pQZE301) and DH5α (pQZE533) did not produce clear zones around colonies on tributyrin agar plates, indicating lack of lipase activity by AlbD.
Genetic Transformation of X. albilineans with albD and Pathogenesis of Recombinant X. albilineans Strains.
Mobilization of the R6K-based suicide vector pJP5603 (31) or the albD derivative pJPAldSP into X. albilineans resulted in kanamycin-resistant, ampicillin-resistant transconjugant colonies at a frequency of 2 × 10−8. All 18 tested transformants with the control vector pJP5603 produced the same level of albicidin as their parental strain XA3. Of 30 tested transconjugants from 6 separate matings with the albD derivative pJPAldSP, 16 failed to release detectable toxin. Albicidin detoxification activity was detected from all 16 tox− colonies but not from the 14 transconjugants that produced albicidin. Strains Xald5, Xald21, and Xald24, which express albD, were selected for pathogenicity testing. These strains are not different from the parental strain XA3 in morphology or in ability to metabolize carbohydrates and amino acids.
These AlbD+ X. albilineans strains failed to induce any white pencil line symptoms on leaf scald-susceptible sugarcane variety Q44. The parent strain XA3 and control strains Xkn1 and Xkn2, which were transformed with suicide vector pJP5603, produced the same level of albicidin toxin in culture and induced severe pencil line symptoms in sugarcane (Table 1). Two weeks after inoculation, AlbD+ strains and controls reached similar population sizes in inoculated leaves, but the AlbD+ strains either failed to invade or reached lower population sizes in young stem tissues of the inoculated plants (Table 1).
Table 1.
Effect of albD expression in X. albilineans on albicidin release and pathogenicity
| Strain | AlbD activity | Release of albicidin,* ng/ml | White pencil lines† | Bacterial population, cfu/g dry weight‡
|
|
|---|---|---|---|---|---|
| Inoculated leaves | Young stems | ||||
| XA3 | − | 200 | 108 | 7 × 106 | 6 × 105 |
| Xkn1 | − | 190 | 111 | 7 × 106 | 1 × 105 |
| Xkn2 | − | 200 | 115 | 2 × 106 | 4 × 105 |
| Xald5 | + | 0 | 0 | 1 × 106 | <2 × 102 |
| Xald21 | + | 0 | 0 | 1 × 106 | 4 × 104 |
| Xald24 | + | 0 | 0 | 3 × 106 | <2 × 102 |
Maximum albicidin concentration in the culture supernatant within 5 days; means of two replicates.
Totals from three inoculated leaves on six sugarcane (Q44) plants per treatment.
Two weeks after inoculation of sugarcane (Q44) plants; means of two replicates.
DISCUSSION
Our results show that the capacity of P. dispersa SB1403 to detoxify albicidin is determined by a gene designated albD. The albD coding region, 5′ promoter, and 3′ terminator regions were defined in a DNA fragment of 1023 bp by deletion analysis and were fully sequenced. Expression of the cloned gene in transformed E. coli conferred Albr and the capacity to detoxify albicidin. We have shown elsewhere that site-directed mutagenesis of albD in P. dispersa eliminates its ability to detoxify albicidin (22). Thus, expression of albD is necessary and sufficient for albicidin detoxification.
The AlbD protein is very likely an enzyme in the serine hydrolase superfamily (35), with a central GXSXG motif (surrounding serine 105) and aspartic acid (D 169) and histidine (H 125) residues appropriately spaced to form the catalytic triad of the enzyme. As in the case of several lipases (35), these three amino acids are located at loop regions of the enzyme. The finding that the serine enzyme inhibitor PMSF strongly inhibits hydrolysis of p-nitrophenyl butyrate and detoxification of albicidin provides strong evidence that the GXSXG motif in AlbD is of catalytic importance.
The serine hydrolase superfamily includes serine proteases, lipases, and esterases, and some other hydrolytic enzymes such as cutinase. AlbD is likely to be a member of the lipase/esterase family. Secondary structure analysis using PHD, a profile-based neural network system (36, 37), indicates α/β protein structural features consisting of six α-helix and six β strands. The region containing the GXSXG motif consists of a five-residue β strand, a six-residue turn with a serine in the middle, and a buried α-helix of eight residues that is very similar to the β-ɛSer-α structural motif in lipases and esterases (35, 38). Hydrolysis of the ester bond of p-nitrophenyl butyrate, but not tributyrin, indicates that the AlbD is not a true lipase (triacylglyceride ester hydrolase, EC 3.1.1.3) but is likely to be an arylesterase (EC 3.1.1.2). This is consistent with the evidence that detoxification by AlbD of albicidins, which are aromatic compounds (13, 15), requires no cofactors and generates two classes of more hydrophilic products. However, more work needs to be done on albicidin structure, AlbD substrate specificity, and enzyme kinetics to fully characterize AlbD as an albicidin hydrolase.
AlbD enzyme detoxified all of the albicidin derivatives produced in culture by X. albilineans, indicating a probable common ester linkage that is crucial for biological activity. Esterases in cell extracts from sugarcane and X. albilineans did not detoxify albicidin. The albD gene was cloned from a P. dispersa strain isolated from X. albilineans-infected sugarcane on a site used continuously for leaf scald disease resistance trials. Comparison of the albicidin detoxification and esterase activities of AlbD with that of its homologues from P. dispersa strains unlikely to have been exposed to albicidin may be revealing with respect to the structural determinants and evolution of this specificity.
Purified AlbD effectively detoxified all albicidins, across a broad range of temperatures (16–45°C) and pH (5.8 to 8.0), without any complex cofactors for enzymatic activity. These properties make albD an attractive candidate for transfer into sugarcane (39) to confer albicidin resistance and to determine the role of albicidin phytotoxins in systemic disease development.
As an interim test, we introduced albD into the genome of X. albilineans. No albicidin toxins were detected from the culture supernatants of the genetically modified X. albilineans strains that produce AlbD enzyme. These strains failed to induce any white pencil line symptom when inoculated on leaf scald-susceptible sugarcane variety Q44 whereas albicidin-producing strains, transformed with a control vector lacking albD, induced severe pencil line symptoms diagnostic of leaf scald disease on sugarcane. Furthermore, the strains producing AlbD enzyme showed diminished capability for invasion of young stems, a major determinant in the development of economically damaging disease. The data strongly support the involvement of albicidins in leaf scald disease and the potential for the albD gene to confer leaf scald disease resistance when appropriately expressed in sugarcane. We are proceeding to test these effects in transgenic sugarcane expressing albD by inoculation with wild-type strains of the pathogen under field conditions.
Acknowledgments
Portions of this work were supported by grants from the Australian Research Council and the Sugar Research and Development Corporation and by fellowships from the University of Queensland and the Australian Research Council to L.Z.
ABBREVIATIONS
- Albr
albicidin-resistant
- Albs
albicidin-sensitive
- PMSF
phenylmethylsulfonyl fluoride
- GST
glutathione S-transferase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database [1023-bp insert in pQZE533 (accession no. U96453)].
References
- 1.Durbin R D, editor. Toxins in Plant Disease. New York: Academic; 1981. [Google Scholar]
- 2.Graniti A. Experientia. 1991;47:751–754. [Google Scholar]
- 3.Atkinson M M. Adv Plant Pathol. 1993;10:35–64. [Google Scholar]
- 4.Rudolph K W E. In: Pathogenesis and Host Specificity in Plant Diseases: I. Prokaryotes. Singh U S, Singh R P, Kohmoto K, editors. Oxford: Pergamon/Elsevier; 1995. pp. 47–138. [Google Scholar]
- 5.Scheffer R P. Experientia. 1991;47:804–811. [Google Scholar]
- 6.Buiatti M, Ingram D S. Experientia. 1991;47:811–819. [Google Scholar]
- 7.Johal G S, Briggs S P. Science. 1992;258:985–987. doi: 10.1126/science.1359642. [DOI] [PubMed] [Google Scholar]
- 8.Briggs S P, Johal G S. Trends Genet. 1994;10:12–16. doi: 10.1016/0168-9525(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 9.Ricaud C, Ryan C C. In: Diseases of Sugarcane: Major Diseases. Ricaud C, Egan B T, Gillaspie A G Jr, Hughes C G, editors. Amsterdam: Elsevier; 1989. pp. 39–53. [Google Scholar]
- 10.Grisham M P, Legendre B L, Comstock J C. Plant Dis. 1993;77:537. [Google Scholar]
- 11.Autrey L J C. In: Proceedings of the International Society of Sugarcane Technologists XXII Congress, Cartegena, September, 1995. Cock J H, Brekelbaum T, editors. Vol. 2. Cali, Colombia: Tecnicana; 1996. pp. 469–470. [Google Scholar]
- 12.Rott P, Ael M, Soupa D, Feldmann P, Letourmy P. Plant Dis. 1994;78:241–247. [Google Scholar]
- 13.Birch R G, Patil S S. J Gen Microbiol. 1985;131:1069–1075. doi: 10.1099/00221287-131-5-1069. [DOI] [PubMed] [Google Scholar]
- 14.Birch R G, Patil S S. Physiol Mol Plant Pathol. 1987;30:207–214. [Google Scholar]
- 15.Birch, R. G. & Patil, S. S. (1985) U.S. Patent 4,525,354.
- 16.Birch R G, Patil S S. Phytopathology. 1983;73:1368–1374. [Google Scholar]
- 17.Birch R G, Patil S S. Physiol Mol Plant Pathol. 1987;30:199–206. [Google Scholar]
- 18.Birch R G, Pemberton J M, Basnayake W V S. J Gen Microbiol. 1990;136:51–58. doi: 10.1099/00221287-136-1-51. [DOI] [PubMed] [Google Scholar]
- 19.Walker M J, Birch R G, Pemberton J M. Microbiology. 1988;2:443–454. doi: 10.1111/j.1365-2958.1988.tb00050.x. [DOI] [PubMed] [Google Scholar]
- 20.Basnayake W V S, Birch R G. Microbiology. 1995;141:551–560. doi: 10.1099/13500872-141-3-551. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Birch R G. Lett Appl Microbiol. 1996;22:132–136. [Google Scholar]
- 22.Zhang L, Birch R G. J Appl Microbiol. 1997;82:389–398. doi: 10.1046/j.1365-2672.1997.00135.x. [DOI] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 24.Bradford M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Priefer U, Simon R, Pühler A. In: Advanced Molecular Genetics. Pühler A, Timmis K N, editors. Berlin: Springer; 1984. pp. 190–201. [Google Scholar]
- 26.Staskawicz B J, Dahlbeck D, Keen N, Napoli C. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lawbence R C, Frayer T F, Reiter B. Nature (London) 1967;25:1264–1265. [Google Scholar]
- 30.Ingham A B, Pemberton J M. Curr Microbiol. 1995;31:28–33. doi: 10.1007/BF00294630. [DOI] [PubMed] [Google Scholar]
- 31.Penfold R J, Pemberton J M. Gene. 1992;118:145–146. doi: 10.1016/0378-1119(92)90263-o. [DOI] [PubMed] [Google Scholar]
- 32.Shine J, Dalgarno L. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Platt T. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 34.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 35.Derewenda Z S, Derewenda U. Biochem Cell Biol. 1991;69:842–851. doi: 10.1139/o91-125. [DOI] [PubMed] [Google Scholar]
- 36.Rost B, Sander C, Schneider R. Comput Appl Biol Sci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 37.Rost B. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 38.Derewenda Z S, Sharp A M. Trends Biochem Sci. 1993;18:20–25. doi: 10.1016/0968-0004(93)90082-x. [DOI] [PubMed] [Google Scholar]
- 39.Bower R, Elliott A R, Potier B A M, Birch R G. Mol Breed. 1996;2:239–249. [Google Scholar]
- 40.Kugimiya W, Otani Y, Kohno M, Hashimoto Y. Biosci Biotechnol Biochem. 1992;56:716–719. doi: 10.1271/bbb.56.716. [DOI] [PubMed] [Google Scholar]
- 41.Shaw J F, Chang R C, Chuang K H, Yen Y T, Wang Y J, Wang F G. Biochem J. 1994;298:675–680. doi: 10.1042/bj2980675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ovnic M, Tepperman K, Medda S, Elliott R W, Stephen D A, Grant S G, Ganschow R E. Genomics. 1991;9:344–354. doi: 10.1016/0888-7543(91)90263-e. [DOI] [PubMed] [Google Scholar]
- 43.Baba T, Downs D, Jackson K W, Tang J, Wang C S. Biochemistry. 1991;30:500–510. doi: 10.1021/bi00216a028. [DOI] [PubMed] [Google Scholar]
- 44.Sasaki M, Ohkubo I. In: Biological Functions of Proteases and Inhibitors. Katanuma N, Suzuki K, Travis J, Fritz H, editors. Tokyo: Japan Scientific Societies; 1994. pp. 61–71. [Google Scholar]