Abstract
The gene for the maturation protein of the single-stranded RNA coliphage MS2 is preceded by an untranslated leader of 130 nt, which folds into a cloverleaf, i.e., three stem–loop structures enclosed by a long distance interaction (LDI). This LDI prevents translation because its 3′ moiety contains the Shine–Dalgarno sequence of the maturation gene. Previously, several observations suggested that folding of the cloverleaf is kinetically delayed, providing a time window for ribosomes to access the RNA. Here we present direct evidence for this model. In vitro experiments show that ribosome binding to the maturation gene is faster than refolding of the denatured cloverleaf. This folding delay appears related to special properties of the leader sequence. We have replaced the three stem–loop structures by a single five nt loop. This change does not affect the equilibrium structure of the LDI. Nevertheless, in this construct, the folding delay has virtually disappeared, suggesting that now the RNA folds faster than ribosomes can bind. Perturbation of the cloverleaf by an insertion makes the maturation start permanently accessible. A pseudorevertant that evolved from an infectious clone carrying the insertion had overcome this defect. It showed a wild-type folding delay before closing down the maturation gene. This experiment reveals the biological significance of retarded cloverleaf formation.
Keywords: RNA phage, RNA structure, ribosome binding/RNA evolution/gene expression
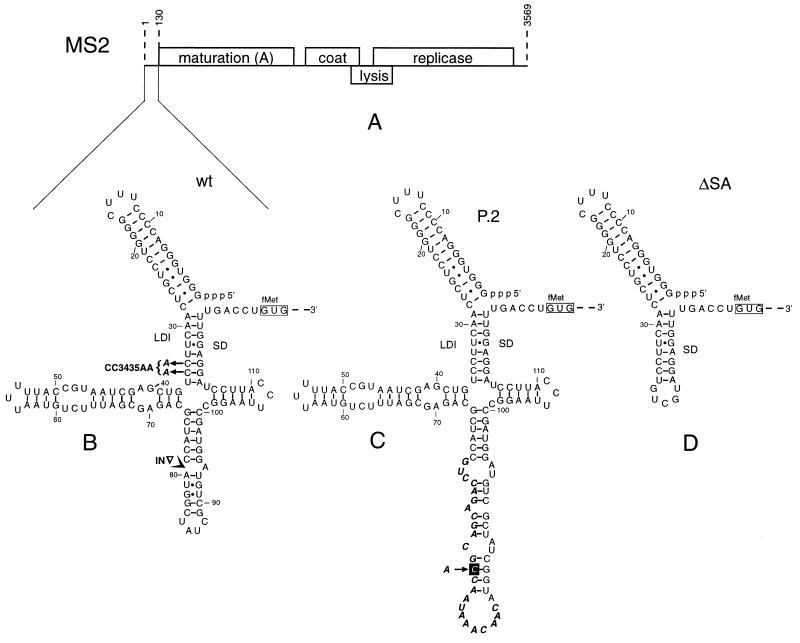
The single-stranded RNA genome of bacteriophage MS2 is 3,569 nt long and contains 4 genes (Fig. 1A). Their products are necessary for phage maturation, encapsidation, lysis of the host, and phage RNA replication, respectively. In RNA bacteriophages, gene expression is controlled at the level of translation. The timing and quantity of expression of coat, lysis, and replicase have been extensively studied (reviewed in ref. 1). Restricted access of ribosomes to the translation–initiation region of the genes by RNA secondary structure turns out to be the major control design.
Figure 1.
(A) Genetic map of phage MS2. (B) Secondary structure model for the 5′ leader as deduced from phylogenetic comparison, computer prediction, and structure probing (see ref. 2). Mutants CC3435AA and IN are indicated in boldface. (C) Proposed secondary structure for pseudorevertant P.2. Nucleotides remaining from the original insert are shown in boldface italics. The boxed A → C substitution found in the pseudorevertant is supportive for the proposed folding, which is also predicted by MFOLD. (D) Probable structure of the mutant ΔSA in which the three arms of the cloverleaf have been replaced by five nt.
The maturation gene is preceded by a 130 nt leader that was shown to fold into a 5′ terminal hairpin followed by three stem–loops enclosed by a long distance interaction (LDI), thereby forming a cloverleaf (Fig. 1B). The downstream part of the LDI is the Shine–Dalgarno sequence of the maturation gene. Previously, we have presented a model in which translation of the maturation gene is facilitated by the delay in the folding of this cloverleaf structure (2). During plus-strand synthesis, the maturation start will only be effective as long as the Shine–Dalgarno sequence has not found its upstream complement. We have ascribed this delay to trapping of the chain in metastable alternative structures. The model was based on several observations. First, the stimulation of translation by mismatches introduced in the LDI was much less than expected, were translation controlled by the equilibrium structure (3). Second, stabilizing the LDI did not have the predicted inhibitory effect. Third, replacing the three arms of the cloverleaf by a five nt loop (Fig. 1D) led to a drastic reduction of translation, presumably due to the fact that the structure search process is shortened by the reduced number of alternatives.
In this paper, we ask whether the folding time of the cloverleaf is indeed slow enough to allow the binding of ribosomes. In addition, we analyze whether the short version (Fig. 1D) takes a shorter time to fold than wild type. Finally, we perform an evolutionary experiment. An insertion mutant lacking retarded folding is passaged to examine whether it adopts a structure in which the maturation start is again temporarily accessible.
MATERIALS AND METHODS
MS2 RNA Isolation.
MS2 phages were grown on Escherichia coli strain GM-1 (4) and purified as described (5). RNA was isolated from phage particles by extraction with phenol in the presence of 0.1% SDS, and was further purified by extraction with chloroform, precipitation with ethanol, followed by Sephadex G25 (medium) chromatography.
Synthesis of MS2 cDNA.
MS2 cDNA, used as a template for T7 transcription, was amplified from plasmid pPlaMS22 or its mutant derivatives, SA, CC3435AA, and IN2 (2) using Taq polymerase (Promega) and the primer 5′-TGTAATACGACTCACTATAGGGTGGGACCCCTTTC, containing the MS2 RNA 5′-end sequence fused to the promoter sequence for the T7 RNA polymerase. The other primer, 5′-GAATGAGTTATCTTCAGTCTCACC, was complementary to nucleotides 199–222 in MS2 RNA. cDNA fragments were purified for T7 transcription by extraction with phenol and chloroform, precipitation with ethanol, and passage through Sephadex G25 (medium). MS2 cDNA of revertant P.2 was prepared after reverse transcription of the relevant part of P.2 phage RNA.
T7 Transcription.
T7 transcripts encoding nucleotides 1–222 of the wild-type or the mutant MS2 sequences P.2, CC3435AA, SA, or IN2 (2), in this paper referred to as ΔSA and IN, respectively, were obtained from the appropriate MS2 cDNA fragments using T7 RNA polymerase from Pharmacia. For use in toeprint experiments, T7 transcripts were synthesized and purified as recommended by the manufacturers.
Toeprint Analysis.
Primer extension inhibition (or toeprinting) on partial or full-length MS2 RNA was performed essentially as described (6). A primer complementary to residues 199–222 was used for extension. (This primer did not work very well on intact MS2 RNA. Therefore, an oligonucleotide complementary to nucleotides 250–277 was used for full-length phage RNA.) Three microliters of mixture A, containing 270 nM T7 transcript or 100 nM full-length MS2 RNA and 300 nM 32P-labeled primer in 10 mM Tris acetate (pH 7.5), 60 mM NH4Cl, 0.1 mM EDTA was incubated for 5 min at 70°C, frozen for 2 min in liquid nitrogen, and thawed on ice for 15 min. To this mixture 2.25 μl of mixture B (0°C) was added containing 1.5 mM dNTPs and 1.7 units/μl RNase inhibitor (RNAguard, Pharmacia) in 5 mM Tris acetate (pH 7.5), 20 mM magnesium acetate, 30 mM NH4Cl. The resulting sample was incubated in a bath at 37°C for the times indicated, frozen for 2 min in liquid nitrogen, and thawed on ice for 15 min; thereafter, 2 μl of 2 μM 30S subunits, isolated as described (7), and 2 μl 20 μM tRNAfMet (Boehringer Mannheim), both in standard buffer (10 mM Tris acetate, pH 7.5/10 mM magnesium acetate/60 mM NH4Cl/0.1 mM EDTA) at 0°C, were added, followed by incubation for 10 min on ice and 7 min at 37°C. (The incubation times on ice have been introduced for practical purposes.) Then, 1 μl of 0.5 unit/μl avian myeloblastosis virus reverse transcriptase (Promega) in standard buffer was added and reverse transcription was carried out for 15 min at 37°C. Reaction products were separated on an 8% polyacrylamide/8 M urea gel. The relative toeprinting efficiency was determined by measuring the radioactivity of inhibited and uninhibited extension products in the gel, using a Betascope 603 Blot Analyzer (Betagen, Waltham, MA) and defined as the percentage of total extension that was inhibited by 30S initiation complexes (8). The relative toeprinting efficiency is therefore the fraction of the RNA population that contains a 30S initiation complex on the start of the maturation gene.
RESULTS
The model to be supported proposes that the 5′-untranslated leader of bacteriophage MS2 folds so slowly that, after its synthesis, there is a short time window during which ribosomes can engage the RNA before the structure becomes permanently closed. This delay is presumably caused by the presence of metastable intermediates. Accordingly, mutations in the cloverleaf are expected to change the folding time and to result in a different expression.
To measure the capacity of ribosomes to interfere with RNA folding we designed the following experiment. MS2 RNA was heat denatured and allowed to renature for various times at 37°C. Then, 30S ribosomal subunits and tRNAfMet were added and binding to the start of the maturation gene was measured by toeprinting. A scheme of the experiment is shown in Fig. 2. An assumption, which turned out to be correct, is that intramolecular base pairing in the MS2 RNA cannot displace an initiating ribosome.
Figure 2.
Scheme showing the experimental design for measuring the capacity of ribosomes to bind the denatured maturation start after varying renaturation times. Solid boxes signify the Shine–Dalgarno sequence and the start codon. The two hairpins in the third stage of the scheme symbolize putative metastable intermediary structures.
The analysis was carried out with a T7 transcript containing the MS2 RNA sequence from position 1 to 222. As a reference, we used the ΔSA construct also extending from position 1 to 222, but here the three arms of the cloverleaf are replaced by a 5 nt loop (Fig. 1D). Previously, we measured that in vivo the ΔSA construct has a 10 times lowered translation efficiency, consistent with a predicted quicker folding (2).
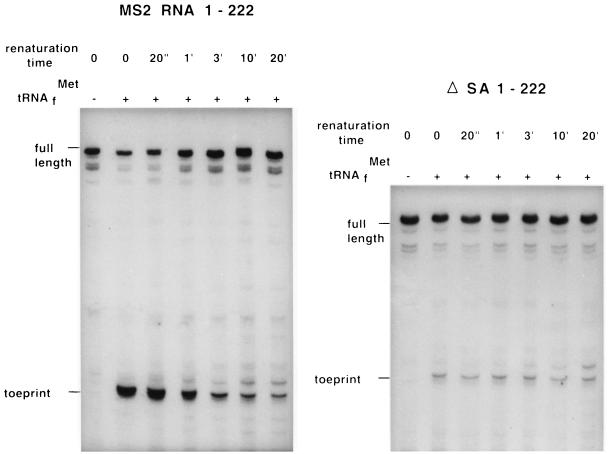
In Fig. 3 (Left) we show the autoradiograph of a primer extension inhibition (toeprinting) experiment using the first 222 nt of MS2 RNA. The toeprint reflects the presence of an initiating ribosome, while the full-length band derives from MS2 RNA not complexed with 30S subunits and bound initiator tRNA. As expected, no toeprint is found in the absence of initiator tRNA (first lane). As can be seen, the fraction of MS2 RNA bound to ribosomes decreases with increasing renaturation time, illustrating that in time the ribosome binding site (RBS) becomes inaccessible due to formation of secondary structure. If we do the same experiment with RNA from the ΔSA construct the picture is quite different (Fig. 3 Right). Here, even at zero renaturation time, there is hardly any binding of ribosomes showing that the single hairpin (Fig. 1D) folds more quickly than ribosomes can bind. In Fig. 4 (Left), the results are presented in a graphical way. It is noteworthy that the extent of ribosome binding to MS2 and ΔSA is the same when the structures are at equilibrium, i.e., after 20 min renaturation time. Therefore, the 10-fold difference in translation that we measured in vivo cannot be ascribed to differences in the thermodynamic stability of the LDI but is the likely result of retarded folding.
Figure 3.
Progression in RNA folding as revealed by the decreasing accessibility of the maturation start to ribosomes. Full-length transcripts result from primer extension on 30S-free RNA. Toeprint refers to primer extension stopped by the presence of a 30S initiation complex on the maturation start. Parallel sequence lanes, not present in this audioradiogram, show the reverse transcription to stop at position +16 relative to the first G of the start codon (6). (Left) MS2 RNA covering nucleotides 1–222. (Right) The same MS2 RNA, but the arms of the cloverleaf are replaced by a 5 nt loop (as shown in Fig. 1D). In the absence of tRNAfMet, the 30S subunit is displaced by reverse transcriptase. Full length in ΔSA 1–222 is 72 nt shorter than in MS2 RNA 1–222. When MS2 RNA 1–222 was renatured by slowly cooling from 70°C to 37°C, the toeprint intensity was the same as measured here after 20 min of renaturation.
Figure 4.
Accessibility of the maturation start to 30S subunits in several RNAs as a function of the renaturation time. Curves represent the average of two toeprint experiments. IN contains a 99 nt insertion derived from M13 in the NcoI site at position 79. In previous work, the insertion construct was denoted as IN2 (2) and P.0 (9), respectively. Measurements were performed on T7 transcripts covering the phage sequence from nucleotide 1 up to map position 222. Full-length MS2 contains the complete genomic RNA.
It may also be interesting to look at full-length MS2 RNA. Such an experiment can provide some insight into the functional folding of a large molecule. As shown in Fig. 4 (Left), the results are qualitatively the same as for the short chain (1–222), but quantitatively there is a distinct reproducible difference; at equilibrium, full-length MS2 RNA is even more inaccessible to ribosomes than the 222 nt long fragment. A possible reason will be given in the Discussion.
To further support the statement that the toeprint curves as shown in Fig. 4 indeed reflect the folding of the cloverleaf we analyzed mutant CC3435AA. Here, the C residues at positions 34 and 35 have been changed to A residues, replacing two C⋅G pairs by A⋅G pairs and leading to a weakening of the LDI (Fig. 1B). In vivo, these two changes cause about a 5-fold increase in maturation protein synthesis (2). This we have ascribed to the fact that the destabilized LDI causes translation to be leaky even after cloverleaf folding. Accordingly, in our toeprint analysis, one would expect this mutant to display about the same kinetics of folding, but to stay at a higher level of ribosome binding after reaching the equilibrium structure. Fig. 4 (Left) shows that this is precisely what we find.
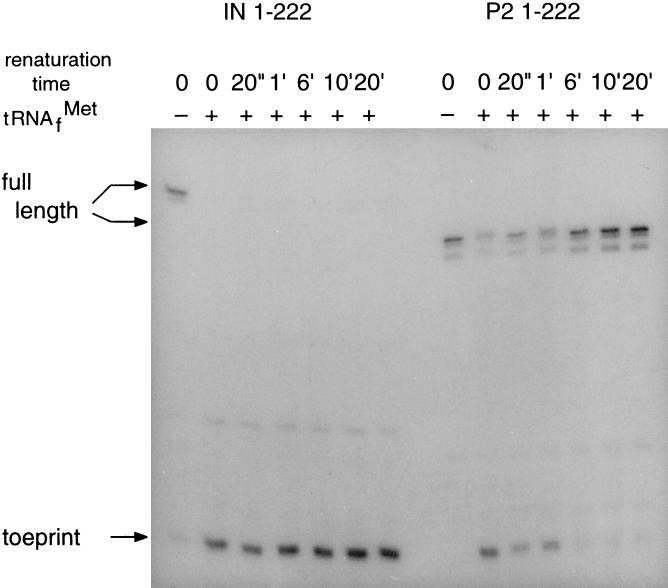
A further analysis was devoted to mutant IN containing a 99 nt insertion, derived from phage M13, in the NcoI site (nt 79) (see Fig. 1B). The toeprint analysis presented in Fig. 5 (Left) and Fig. 4 (Right) shows that this RNA fails to fold into a ribosome-inaccessible structure. In accordance, in vivo expression studies showed mutant IN to produce 10 times as much maturation protein as wild type (2).
Figure 5.
Progression in RNA folding as revealed by the decreasing accessibility of the maturation start to ribosomes. (Left) MS2 RNA with a 99 nt insertion at position 79 (IN). (Right) Pseudorevertant P.2 that evolved from mutant IN. Measurements were performed on T7 transcripts from position 1 to 222 of the wild-type MS2 genome.
From this and previous work (2), the strong suggestion has always been that delayed folding is a biological necessity, i.e., the maturation gene must only transiently be accessible to the translational machinery. We have now performed an evolutionary experiment to lend further credence to this idea.
Mutation IN, causing uncontrolled maturation synthesis, was incorporated into an infectious MS2 cDNA clone where it caused a 106-fold decrease in infectivity (10). Pseudorevertants with an infectivity equal to wild type could be recovered. Of these pseudorevertants, five were sequenced and found to have removed between 74 and 96 nt of the original insert (10). Pseudorevertant P.2 was selected for further analysis. Its sequence and probable structure, as presented in Fig. 1C, shows that most of the original insert has been deleted, whereas the remaining 25 nt have incorporated an additional suppressor mutation (solid box). Surprisingly, pseudorevertant P.2 shows again a transiently accessible maturation protein start (Figs. 4 and 5 Right). Clearly, there is strong selection pressure on this type of translational control.
DISCUSSION
It has been known for a long time that the maturation protein gene of single-stranded RNA phages like R17 and MS2 is only accessible to ribosomes after formaldehyde treatment or after degradation (11, 12). Now that we have much more advanced techniques it has become possible to understand the molecular basis and the kinetic component of this feature.
In a previous paper (2) we suggested a model in which translation of the maturation gene of RNA phage MS2 could only start on nonequilibrated RNA transcripts. We proposed that by the time the RBS is synthesized, pairing with its upstream complementary sequence is delayed leaving a short time window for ribosome attachment. In this paper we have analyzed the folding time of the 5′-untranslated region of wild-type and four mutant sequences. To examine folding, one needs a tool to arrest intermediate stages. Some authors have successfully used chemical modification (13) or hybridization with deoxyoligonucleotides (14) combined with cutting by RNase H (15). We have employed the binding of 30S ribosomes to stop the reaction from going to completion. In our case this method is quite appropriate, since the biological aim of the delay is precisely to permit ribosome binding.
The results obtained support our model. A delay of several minutes can be measured before the RNA becomes inaccessible. This is actually an unexpected result. The folding of tRNA, which is about the size of our cloverleaf, is in the millisecond range (16). To explain this large difference it can be argued that whereas tRNA may be under selection pressure to fold quickly (17), the 5′-untranslated region has evolved to acquire a delay. Our experiment on the evolution of the nonfolding mutant IN to the retarded folding of pseudorevertant P.2 supports this explanation.
The absolute values of the measured folding times may not be very accurate because we do not know how much time it takes our samples inside the Eppendorf tubes to reach 37°C after insertion in the water bath. Also, had we used other ionic conditions during renaturation the kinetics might have been somewhat different. Nevertheless, it is clear that we are dealing with renaturation times that exceed those of tRNA by many orders of magnitude. There is also little doubt that the relative folding times are reliable and that the ΔSA construct lacking the three arms folds much quicker than the wild-type structure.
The cause of a folding delay is usually attributed to temporal trapping in metastable intermediates. Examples are found in the control of Col E1 replication (9), the Qβ replicase template SV-11 (18), and the IS10 mRNA (14). Our finding that replacement of the three arms by a single 5 nt loop abolishes this delay is in agreement with this temporal trapping. These putative intermediates have not been identified yet, but experiments are underway to clarify this point.
From the renaturation kinetics for several different RNAs shown in Fig. 4, one other important conclusion can be drawn. It is not a general property of a RBS to change its accessibility with renaturation time. One example is ΔSA 1–222. Here, the RBS is part of a single hairpin and its folding cannot be resolved by the assay used. The other example is mutant IN, where the insertion apparently leads to an open RBS. Therefore, the delay observed for the wild-type sequence and for P.2 is the result of a special design that can easily be upset.
Comparison of the renaturation kinetics of full-length and short MS2 RNA (Fig. 4) shows that repression is more complete for full-length RNA. We believe this to be due to an additional pairing involving the start codon and neighboring nucleotides with a sequence around position 365 and therefore not present in the truncated transcript (H. Groeneveld, personal communication). The fact that the slopes reflecting the folding in full-length and in the truncated version of MS2 RNA run parallel suggests that folding of such a large molecule is an ordered process and that the extra 3,300 nt present in the full-sized MS2 RNA do not interfere with folding of the cloverleaf by providing extra pairing alternatives. Presumably, the presence of structure domains in large RNA molecules prevents the chain from becoming entangled in random pairing (4).
Zarrinkar and Williams (15) have studied the folding pathway of the Tetrahymena L-21 ScaI ribozyme, an RNA of 378 nt that folds into a complex secondary structure. Similar to our results, the LDIs were formed in a slow, minute-scale process, whereas hairpin-like structure folded much faster.
Although we feel that our results provide substantial support for our model on control by folding kinetics, we acknowledge that the experiments do not yet completely simulate the in vivo situation. There, the growing chain is folded while synthesized, whereas in our analysis we studied the folding of a prefabricated denatured RNA molecule. We have tried to mimic the in vivo situation by performing ribosome binding and toeprinting in T7 transcription mixes, but these attempts have met with a number of practical problems.
A last point concerns the biological importance of the folding delay. Our experiment in which we evolve the uncontrolled IN mutant clearly shows that there is an advantage in having a transiently accessible maturation gene. The purpose of the design is most likely to create a ribosome-free template when replication is due (19).
Acknowledgments
We thank Dr. Gultyaev for many stimulating discussions. This work was supported by North Atlantic Treaty Organization Grant LG940481 and by the European Union Human Capital and Mobility Program CHRX-CT93–0169. N.V.T. wishes to thank the RNA Research Foundation for support.
Footnotes
This paper was submitted directly (Track II) to the Proceedings office. Abbreviations: LDI, long distance interaction; RBS, ribosome binding site.
References
- 1.de Smit M H, van Duin J. Prog Nucleic Acid Res Mol Biol. 1990;38:1–35. doi: 10.1016/s0079-6603(08)60707-2. [DOI] [PubMed] [Google Scholar]
- 2.Groeneveld H, Thimon K, van Duin J. RNA. 1995;1:79–88. [PMC free article] [PubMed] [Google Scholar]
- 3.de Smit M H, van Duin J. Proc Natl Acad Sci USA. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beekwilder M J, Nieuwenhuizen R, van Duin J. J Mol Biol. 1995;247:903–917. doi: 10.1006/jmbi.1995.0189. [DOI] [PubMed] [Google Scholar]
- 5.Adhin M R, Avots A, Berzin V, Overbeek G P, van Duin J. Biochim Biophys Acta. 1990;1050:104–109. doi: 10.1016/0167-4781(90)90149-v. [DOI] [PubMed] [Google Scholar]
- 6.Hartz D, McPheeters D S, Traut R, Gold L. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 7.Poot R A, Brink M F, Pleij C W A, de Boer H A, van Duin J. Nucleic Acids Res. 1993;21:5398–5402. doi: 10.1093/nar/21.23.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartz D, McPheeters D S, Gold L. J Mol Biol. 1991;218:83–97. doi: 10.1016/0022-2836(91)90875-7. [DOI] [PubMed] [Google Scholar]
- 9.Gultyaev A P, van Batenburg F H D, Pleij C W A. Nucleic Acids Res. 1995;23:3718–3725. doi: 10.1093/nar/23.18.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsthoorn R C L, van Duin J. J Virol. 1996;70:729–736. doi: 10.1128/jvi.70.2.729-736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lodish, H. F. (1975) in RNA Phages, Cold Spring Harbor Monograph Series, ed. Zinder, N. D. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 301–318.
- 12.Steitz, J. A. (1975) in RNA Phages, Cold Spring Harbor Monograph Series, ed. Zinder, N. D. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 319–352.
- 13.Celander D W, Cech T R. Science. 1991;251:401–407. doi: 10.1126/science.1989074. [DOI] [PubMed] [Google Scholar]
- 14.Ma C K, Kolesnikowa T, Rayner J C, Simons E L, Yim H, Simons R W. Mol Microbiol. 1994;14:1033–1047. doi: 10.1111/j.1365-2958.1994.tb01337.x. [DOI] [PubMed] [Google Scholar]
- 15.Zarrinkar P P, Williamson J R. Science. 1994;265:918–923. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 16.Crothers D M, Cole P E, Hilbers C W, Shulman R G. J Mol Biol. 1974;87:63–88. doi: 10.1016/0022-2836(74)90560-9. [DOI] [PubMed] [Google Scholar]
- 17.Higgs P G. J Phys Inst France. 1993;3:43–59. [Google Scholar]
- 18.Biebricher C K, Luce R. EMBO J. 1992;11:5129–5135. doi: 10.1002/j.1460-2075.1992.tb05620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolakofsky D, Weissmann C. Biochim Biophys Acta. 1971;246:596–599. doi: 10.1016/0005-2787(71)90799-4. [DOI] [PubMed] [Google Scholar]