Abstract
Spores harboring an ACC1 deletion derived from a diploid Saccharomyces cerevisiae strain, in which one copy of the entire ACC1 gene is replaced with a LEU2 cassette, fail to grow. A chimeric gene consisting of the yeast GAL10 promoter, yeast ACC1 leader, wheat cytosolic acetyl-CoA carboxylase (ACCase) cDNA, and yeast ACC1 3′ tail was used to complement a yeast ACC1 mutation. The complementation demonstrates that active wheat ACCase can be produced in yeast. At low concentrations of galactose, the activity of the “wheat gene” driven by the GAL10 promoter is low and ACCase becomes limiting for growth, a condition expected to enhance transgenic yeast sensitivity to wheat ACCase-specific inhibitors. An aryloxyphenoxypropionate and two cyclohexanediones do not inhibit growth of haploid yeast strains containing the yeast ACC1 gene, but one cyclohexanedione inhibits growth of the gene-replacement strains at concentrations below 0.2 mM. In vitro, the activity of wheat cytosolic ACCase produced by the gene-replacement yeast strain is inhibited by haloxyfop and cethoxydim at concentrations above 0.02 mM. The activity of yeast ACCase is less affected. The wheat plastid ACCase in wheat germ extract is inhibited by all three herbicides at concentrations below 0.02 mM. Yeast gene-replacement strains will provide a convenient system for the study of plant ACCases.
Acetyl-CoA carboxylase (ACCase) catalyzes the first committed step in de novo fatty acid biosynthesis and provides malonyl-CoA for the synthesis of very-long-chain fatty acids and a variety of important secondary metabolites, and for malonylation. In plants, these primary and secondary metabolic pathways are located in plastids and cytoplasm, respectively. Plants have two forms of ACCase (reviewed in ref. 1). The one located in plastids, the site of plant fatty acid synthesis, can be either a eukaryotic-type high molecular weight multifunctional enzyme (e.g., wheat) or a prokaryotic-type multisubunit enzyme (e.g., pea, soybean, tobacco, and Arabidopsis). The cytosolic plant ACCase is of the eukaryotic type.
In yeast, a single ACC1 gene encodes an ACCase that provides malonyl-CoA for both de novo fatty acid synthesis as well as for subsequent fatty acid elongation. A null mutation of ACC1 is not rescued by fatty acid supplementation, suggesting an additional essential function for ACCase in yeast (2). This function has recently been identified as providing malonyl-CoA for the biosynthesis of very-long-chain fatty acids, which are required to maintain a functional nuclear envelope (3).
Interaction with aryloxyphenoxypropionate and cyclohexanedione inhibitors is an important aspect of plant ACCase biochemistry (4–6). Some of these compounds are used as powerful graminicides. The herbicide action is caused by the inhibition of the eukaryotic-type plastid ACCase and, as a result, inhibition of fatty acid biosynthesis in sensitive plants. Plants containing prokaryotic-type plastid ACCase are resistant to these compounds, as are other eukaryotes and prokaryotes. The molecular mechanism of inhibition/resistance of the enzyme is not known.
We have shown previously that wheat plastid ACCase is highly sensitive to an aryloxyphenoxypropionate inhibitor (haloxyfop) (7), but the corresponding cytosolic enzyme could not be studied effectively. We have recently obtained full-length cDNA and genomic sequences for both cytosolic and plastid ACCases from wheat (refs. 8 and 9, and P.G. and R.H., unpublished work). In this paper we report the establishment of yeast expression systems that allow investigation of the structure and function, including interaction with inhibitors, of individual wheat ACCases.
MATERIALS AND METHODS
Assembling a Chimeric Gene.
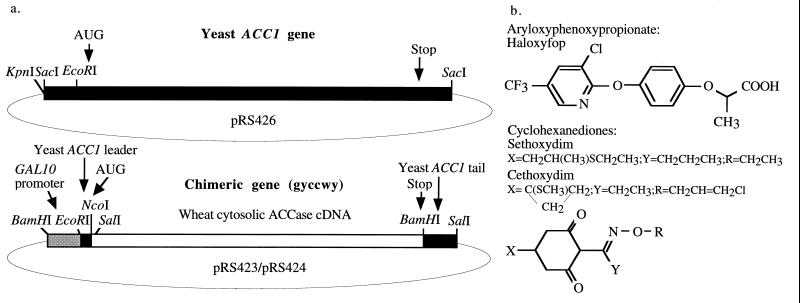
Full-length cDNA encoding wheat cytosolic ACCase was assembled in a multistep “cut and paste” cloning experiment. The assembly process was monitored by sequencing selected regions of the construct. Restriction fragments from cDNA clones described before (8), as well as PCR-generated fragments with new restriction sites, were used for the construction. The chimeric gene (gyccwy, Fig. 1a) consists of the following eight fragments inserted in this order between BamHI and SalI sites of pRS423 or pRS424 (10): (i) BamHI–EcoRI fragment containing GAL10 promoter from pBM150 (11); (ii) EcoRI–NcoI fragment containing the leader sequence of the ACC1 gene from Saccharomyces cerevisiae (2) prepared by PCR to introduce a NcoI site; (iii) NcoI–XbaI fragment of clone 39–1 for which a subfragment was prepared by PCR to introduce the NcoI site at the ACCase translation start codon; (iv) XbaI–SphI fragment of clone 20–1; (v) SphI–BamHI fragment of clone 45–1; (vi) BamHI–EcoRI fragment of clone 24–3; (vii) EcoRI–BamHI fragment of clone 01–4; (viii) BamHI–SalI fragment consisting of 17 nucleotides of wheat ACCase cDNA [sequence identical to clone 01–4 (8)], 277 nucleotides of the yeast ACC1 gene containing the 3′ tail, and 24 nucleotides of the pRS426 vector followed by a SalI site. This fragment was prepared by PCR using ACC1-pRS426 (described below) as template. A 7,969-bp SacI fragment of yeast genomic DNA from plasmid YCp50-ACC1 (2), kindly provided by S. D. Kohlwein (Technical University, Graz, Austria), containing the ACC1 gene, was cloned into the SacI site of vector pRS426 to create ACC1-pRS426. The 5′ end of the ACC1 gene in this construct is adjacent to the KpnI site of the vector.
Figure 1.
(a) Structure of yeast expression plasmids encoding yeast and wheat cytosolic ACCase. (b) Structure of ACCase inhibitors used in this study.
Yeast Transformation, Growth, and Tetrad Analysis.
The yeast complex medium for vegetative growth contained 1% bacto-yeast extract, 2% bacto-peptone, 0.1% adenine sulfate, and was supplemented with the following carbon sources: 2% dextrose in yeast extract/peptone/dextrose (YPD), 2% raffinose in yeast extract/peptone/raffinose (YPR), 2% galactose, and 2% raffinose in yeast extract/peptone/raffinose/galactose (YPRG). The yeast synthetic minimal medium (SD) contained 0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% dextrose, and was supplemented with various additives required for growth: adenine sulfate (10 mg/liter), uracil (20 mg/liter), histidine (20 mg/liter), leucine (200 mg/liter), and/or tryptophan (50 mg/liter). The SRG medium is identical to SD, with dextrose replaced by 2% raffinose and 2% galactose. Two percent bacto-agar was used in solid media. SPII (2% potassium acetate, supplemented with essential amino acids at 7.5 mg/liter) and SPIII (0.1% dextrose, 0.25% bacto-yeast extract and 2% bacto-agar added to SPII) media for sporulation are as described (12, 13). S. cerevisiae strain W303D-ACC1ΔLEU2 was also provided by S. D. Kohlwein. Yeast cells were transformed as described (14) using Frozen-EZ kit (Zymo Research, Orange, CA) according to the manufacturer’s protocol, and transformants were selected using SD plates lacking a marker amino acid. Sporulation was induced in SPII or SPIII medium at 30°C for 2–3 days. Dissection of asci was performed as described (15). YPD or YPRG plates were used for vegetative growth of ascospore clones following dissection. Galactose-dependent strains (MJ 6.8, MJ 6.9, MJ 1.12, and MJ 1.13) were grown in YPRG medium and other strains in YPD medium, all at 30°C. To measure galactose induction, the strains were grown for ≈24 hr in YPR medium until all galactose was exhausted (growth of galactose-dependent strains ceased), and then diluted ≈100-fold with YPR medium containing varying amounts of galactose.
In Vivo and in Vitro Herbicide Inhibition.
Sethoxydim and cethoxydim (our name, CGA215684) were provided by CIBA–GEIGY (now Novartis, Research Triangle Park, NC) and haloxyfop was provided by DowElanco (Indianapolis). The inhibitor structures are shown in Fig. 1b. To measure inhibition in vivo, galactose-dependent strains were grown in YPR containing 0.01% galactose (to A600 of ≈0.5) and then diluted (≈100-fold) to the same initial density with fresh YPR medium containing 0.01% galactose and varying amounts of herbicide added as 100-fold concentrated stock solutions in dimethyl sulfoxide. ACCase activity in vitro was measured using protein extracts prepared as described (16) from 25–50 ml cultures of yeast strains MJ 1.12 and MJ 9.11 grown for 24–40 hr. Cells were suspended in one volume of extraction buffer (0.3 M sorbitol/0.1 M NaCl/5 mM MgCl2/10 mM Tris, pH 7.4) supplemented with protease inhibitors: aprotinin (6.6 μg/ml), leupeptin (1.5 μg/ml), pepstatin A (3 μg/ml), and chymostatin (1 μg/ml), and vortexed with 2 vol of glass beads for 5 min at 4°C. A 1/200 vol of 100 mM ethanol solution of phenylmethylsulfonyl fluoride was added twice during extraction. The cell debris were removed by centrifugation and the supernatant (≈0.5 ml) was loaded immediately on a 20-ml Sephadex G50 column equilibrated with 100 mM KCl, 20 mM Tris⋅HCl (pH 7.5), 20% glycerol, 7 mM 2-mercaptoethanol. Protein was eluted with the same buffer and stored at −70°C. ACCase activity was measured as described previously (7) using aliquots of the column fractions. Herbicides were added to the reaction mixtures (1/10 or 1/20 vol) as a solution in 0.25–0.75 M Tris⋅HCl (pH 8.0) containing 25% dimethyl sulfoxide. Wheat germ extract was purchased from Promega.
DNA and Protein Analysis.
DNA and protein manipulations and analyses were performed as described (17) or following the manufacturer’s protocols. DNA was isolated from yeast as described (18). Biotinylated peptides present in protein extracts prepared as described above but without gel filtration were analyzed on Western blots using [35S]streptavidin as described (7).
RESULTS
Complementation of a Yeast ACC1 Null Mutation with Wheat Cytosolic ACCase.
We assembled a chimeric gene consisting of the yeast GAL10 promoter, yeast ACC1 leader, wheat cytosolic ACCase cDNA, and yeast ACC1 3′ tail in high-copy-number yeast expression vectors of the pRS series (10). The coding sequence was assembled from fragments of different cDNA clones described earlier, all encoding the cytosolic isozyme but representing transcripts of different genes (8). Some fragments were generated by PCR to introduce new restriction sites to simplify the cloning process and to enable future manipulations. The GAL10 promoter was selected for its strength and tight regulation. Leader and tail sequences from the ACC1 gene were fused to the wheat ACCase coding sequence to provide proper yeast transcription and translation signals as well as mRNA processing and stability elements. This design, in addition to the high-copy-number of the vectors, was selected in an attempt to ensure a high steady-state level of the transcript that would overcome potential problems with efficient translation of the foreign coding sequences, protein degradation, and inefficient modification (biotinylation) of a foreign protein of the ACCase size (250 kDa). In addition, down-regulation of the expression of the chimeric gene was desired for in vivo inhibition experiments described below. The overall structure of the plasmids containing the chimeric gene as well as the control plasmid containing the yeast ACC1 gene are shown in Fig. 1a.
The chimeric gene was introduced into heterozygous S. cerevisiae strain W303D-ACC1ΔLeu2 (Table 1), in which one allele of the ACC1 gene was replaced with a LEU2 cassette. Sporulation was induced and 14, 39, and 16 asci were dissected in a standard tetrad analysis for strains complemented with gyccwy-pRS423, gyccwy-pRS424, and ACC1-pRS426, respectively. About 45% of the resulting spores were viable. Eight sets of three or four viable spores derived from a single tetrad were obtained for strains containing the chimeric wheat gene. This result indicates that the gene can complement the ACC1 mutation, since haploid ACC1 disruptants do not grow beyond a few cell divisions (2). Marker analysis of a set of 28 haploid strains derived from 10 tetrads followed by Southern blot analysis of 16 of these haploid strains confirmed this conclusion (data not shown). Low spore viability has other causes than lack of complementation. Complementation was the expected outcome for the ACC1-containing plasmid as the same SacI fragment of yeast genomic DNA was shown to be sufficient for complementation (2).
Table 1.
Yeast strains
| No. strain | Construct/gene | Parent vector | MAT | ACC1 | URA3‡ | LEU2§ | HIS3‡ | TRP1‡ | Galactose dependence | ACCase content* |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 MJ 1.12 | gyccwy† | pRS424 | a | − | − | + | − | + | + | 0.80 |
| 2 MJ 1.13 | gyccwy† | pRS424 | α | − | − | + | − | + | + | 0.88 |
| 3 MJ 6.8 | gyccwy† | pRS423 | a | − | − | + | + | − | + | 0.69 |
| 4 MJ 6.9 | gyccwy† | pRS423 | α | − | − | + | + | − | + | 0.78 |
| 5 MJ 9.11 | Yeast ACC1‡ | pRS426 | a | + | + | + | − | − | − | 1.12 |
| 6 MJ 9.12 | Yeast ACC1§ | None | α | + | − | (+)¶ | − | − | − | 3.76 |
| 7 MJ 9.13 | Yeast ACC1§ | None | α | + | − | − | − | − | − | 1.27 |
| 8 MJ 9.14 | Yeast ACC1‡ | pRS426 | a | + | + | + | − | − | − | 1.23 |
| 9 W303D | Yeast ACC1§ | None | a/α | + | − | + | − | − | − | 1.00 |
| 10 W303D-ACC1ΔLeu2 | Yeast ACC1§ | None | a/α | + | − | + | − | − | − | 0.59 |
All MJ strains were obtained by sporulation of W303D-ACC1ΔLeu2 transformed with an appropriate plasmid. The related haploid strains (1.12 and 1.13, 6.8 and 6.9, 9.11–9.14) described here were obtained from single tetrads. W303D relevant genotype (3): MATa/MATα, leu2-3,112/leu2-3,112 his3-11,15/his311,15 ade2-1/ade2-1 ura3-1/ura3-1 trp1-1/trp1-1 can1-100/can1-100 ACC1/ACC1; W303D-ACC1ΔLeu2 relevant genotype; MATa/MATα, leu2-3,112/leu2-3,112 his3-11,15/his311,15 ade2-1/ade2-1 ura3-1/ura3-1 trp1-1/trp1-1 can1-100/can1-100 ACC1/acc1::LEU2.
Relative amount of the 250-kDa biotinylated peptide estimated from band intensities on Western blots probed with [35S]streptavidin (e.g., Fig. 2) and normalized with respect to pyruvate carboxylase (PCase) as an internal standard. Averages of three measurements are shown.
GAL10 promoter/yeast ACC1 leader/cytosolic carboxylase wheat cDNA/yeast ACC1 tail (see Fig. 1).
Plasmid born.
Genomic.
Unexplained.
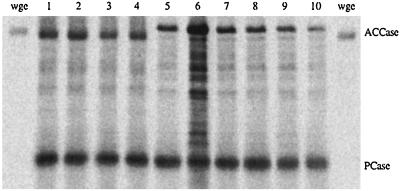
A collection of eight haploid strains was subjected to marker and mating type analysis (Table 1). The DNA content of these strains was verified by Southern blot analysis using yeast and wheat ACCase-specific DNA probes (results not shown). Finally, Western blot analysis with [35S]streptavidin (Fig. 2) and with antibodies specific for wheat ACCase (results not shown) confirmed expression of full-size wheat ACCase in the gene-replacement strains. The amount of biotinylated peptides does not vary greatly between the strains (Fig. 2) except for strain MJ 9.12 (lane 6), which produces a significantly higher amount of ACCase. The reason for this “over-production” is not known. Wheat ACCase migrates in an SDS gel significantly faster than the yeast protein (Fig. 2). This is true for ACCase in wheat germ extract as well. Wheat cytosolic ACCase is 2 kDa larger than yeast ACCase, calculated from the amino acid sequence. The slower migration of yeast ACCase may be due to protein modification.
Figure 2.
Biotinylated proteins in yeast strains. Western blot was probed with [35S]streptavidin to reveal biotinylated peptides. Equal amounts of protein from strains 1–10 listed in Table 1 were loaded (lanes 1–10); wge, wheat germ extract; PCase, pyruvate carboxylase (lanes 1–10).
The level of ACCase activity in Sephadex G50-purified protein is always 10–40 times higher for yeast strains expressing wheat ACCase than in the control strain expressing yeast ACCase from a multicopy plasmid (results not shown). The wheat peptide is biotinylated efficiently, assessed by its functionality and by the ability to bind streptavidin on Western blots. Unexpectedly, however, the level of biotinylated 250-kDa wheat polypeptide expressed under full induction of the GAL10 promoter is comparable to that of the wild-type yeast (Table 1). Comparison of relative signal intensities on Western blots probed with streptavidin and with antibodies against wheat ACCase (data not shown) suggests that biotinylation is not complete. It is lower than biotinylation of ACCase in wheat germ extract used as a reference. Higher activity of the wheat ACCase expressed in yeast may be explained by lack of functional phosphorylation that inactivates yeast ACCase. Higher instability of the yeast protein, inhibition by other mechanisms, or suboptimal conditions used to assay ACCase activity in crude protein extracts might also be responsible.
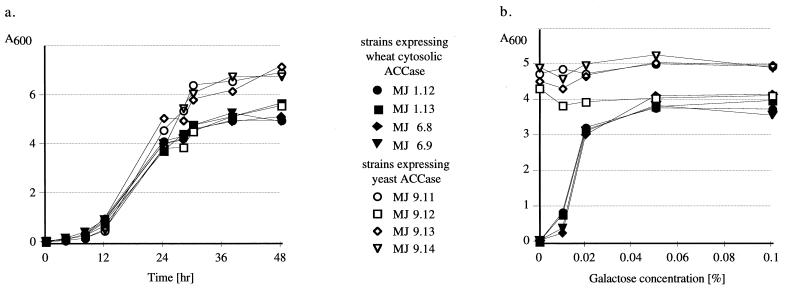
The gene-replacement strains grow in liquid medium almost as fast and to only ≈20% lower density than the haploid strains containing a native yeast ACC1 gene either on the chromosome or on a plasmid (Fig. 3a). No differences in cell morphology were observed between the strains at the level of the light microscope.
Figure 3.
Growth curves (a) and galactose dependence (b) of gene-replacement and wild-type yeast strains. Growth in YPRG medium (a) or in the presence of varying amounts of galactose in YPR medium (b) was monitored at 600 nm. Galactose induction was measured 24 hr after inoculation from cultures grown in YPR medium for about 24 hr.
The activity of the GAL10 promoter can be regulated in medium without glucose by adjusting the galactose concentration. As expected, ACCase becomes limiting for growth in the gene-replacement strains grown at low concentrations of the inducer (Fig. 3b). In medium containing glucose (liquid or plates), the activity of the chimeric gene is effectively repressed and the strains do not grow at all. Cells transferred from YPRG to YPR medium grow for several generations until internal galactose and ACCase and its products are exhausted and then growth ceases. Addition of galactose to a concentration of about 0.05% leads to full induction of the chimeric gene and normal growth (Fig. 3b).
In Vivo and in Vitro Analysis of Wheat ACCase Interaction with Inhibitors Using the Yeast Expression System.
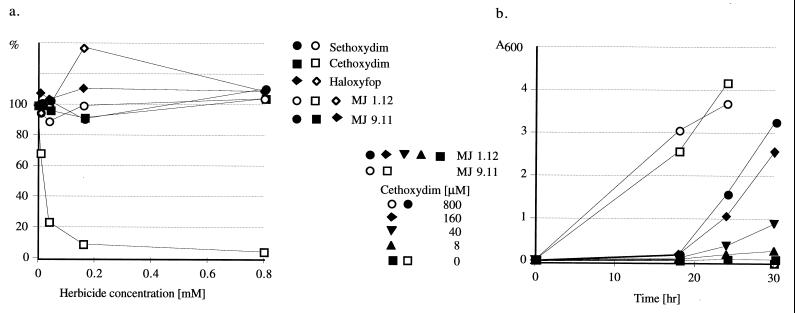
Two strains were selected for a detailed analysis. Haploid strain MJ 9.11 containing one copy of the yeast ACC1 gene (on the chromosome) and expressing ACCase under its own promoter was grown in YPDA medium. Haploid gene-replacement strain MJ 1.12 expressing wheat ACCase from a multi-copy plasmid under the GAL10 promoter was grown in YPR medium supplemented with 0.01% galactose. Under these conditions, ACCase is limiting and the growth rate for MJ 1.12 is 10–20% of the rate observed under full galactose induction or the rate of growth of MJ 9.11 in YPD. The growth of strain MJ 1.12 is strongly inhibited by cethoxydim (50% inhibition at ≈20 μM). Sethoxydim and haloxyfop have no effect on MJ 1.12. MJ 9.11 is not affected by any of the three inhibitors at concentrations up to 800 μM (Fig. 4a). The drastically higher sensitivity of MJ 1.12 to cethoxydim is evident when growth curves are compared in an experiment that also reflects the different growth regimens described above (Fig. 4b). At 800 μM cethoxydim, MJ 1.12 does not grow at all and MJ 9.11 is unaffected.
Figure 4.
Growth inhibition of yeast strains expressing wheat cytosolic or yeast ACCase. Growth in YPR medium containing 0.01% galactose and varying amounts of inhibitors was monitored at 600 nm. (a) Relative culture densities 24 hr after inoculation. (b) Growth curves in the presence of cethoxydim.
In vitro sensitivity of yeast ACCase and wheat cytosolic ACCase expressed in yeast to the three inhibitors was measured using yeast crude protein extracts purified by gel filtration. Wheat germ extract was used in parallel to assess inhibition of the wheat plastid ACCase. It was shown previously that the ACCase activity in wheat germ extract is highly sensitive to haloxyfop, resembling the sensitivity of ACCase activity in purified chloroplasts (7). We conclude that the major component of the ACCase activity in wheat germ represents the plastid enzyme. Two peaks of ACCase activity were observed after ion-exchange chromatography of maize protein (19). In that experiment, activity in the first minor peak was not inhibited by fluazifop (50 μM), but activity in the major peak was. Two peaks of activity were also observed during purification of wheat germ ACCase by ion-exchange chromatography. An internal amino acid sequence of an ACCase peptide from the major peak, eluted at higher salt concentration (7), matches the sequence of wheat plastid ACCase deduced from DNA sequence (unpublished results), but differs from the cytosolic ACCase sequence (8). The two activities most likely represent cytosolic and plastid isozymes, respectively.
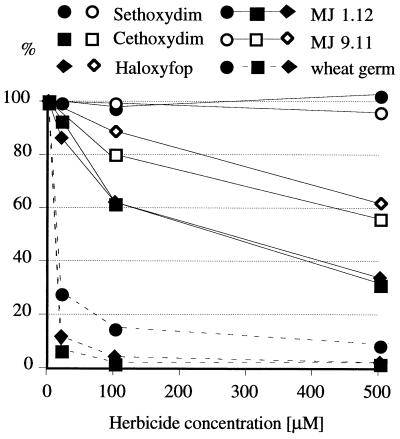
In our in vitro experiments, the yeast ACCase is the least sensitive, wheat cytosolic enzyme expressed in yeast is significantly more sensitive, and the plastid enzyme in wheat germ extract is very sensitive (Fig. 5). Sethoxydim does not inhibit the yeast or wheat cytosolic enzymes and is the weakest inhibitor of the wheat plastid enzyme (Fig. 5). These results show that the wheat cytosolic enzyme is indeed much less sensitive to aryloxyphenoxypropionate and cyclohexadione inhibitors than the wheat plastid enzyme.
Figure 5.
In vitro inhibition of wheat cytosolic ACCase expressed in the yeast gene-replacement strain, along with yeast and wheat germ ACCases. Relative acetyl-CoA-dependent conversion of radioactive bicarbonate into acid-stable malonyl-CoA was measured in the presence of varying amounts of the inhibitors.
The difference in in vitro sensitivity of yeast and wheat cytosolic ACCase to cethoxydim translates to a dramatic difference in the in vivo sensitivity (Fig. 4). This phenomenon can be best explained by the limiting amount of ACCase in strain MJ 1.11, grown at low galactose concentration. This condition is expected to amplify the effect of the increased ACCase sensitivity to inhibitors introduced by the gene replacement. On the other hand, haloxyfop, which shows an almost identical inhibition pattern in vitro (Fig. 5), has no effect in vivo (Fig. 4a). That could result from poor uptake or inactivation of the inhibitor by yeast.
DISCUSSION
We have established an efficient and flexible yeast expression system to produce plant ACCase polypeptides in their active conformation. We can answer the first important questions: our cDNA can produce full-length and active enzyme in yeast, and the “wheat gene” complements the deletion of the ACC1 gene in yeast. The high level of ACCase activity in transgenic yeast strains also reflects efficient modification of the wheat apoprotein by yeast apoprotein:biotin ligase. This occurs despite the fact that amino acid sequences of the two carboxylases are only about 40% identical (8). The yeast expression system was selected over Escherichia coli to avoid potential problems such as protein solubility, folding, or efficient modification (biotinylation). Any ACCase isozyme or variant can be analyzed in vivo (in an appropriate yeast strain) and in vitro (yeast extracts), and purified for biochemical and structural studies. Expression of ACCase fragments, e.g., the two-domain biotin carboxylase plus biotin carboxyl carrier subfragment or inhibitor-binding domain should also be possible. The strong GAL10 promoter and high-copy-number plasmids were chosen to ensure high level and regulated expression. At low concentrations of the inducer, ACCase becomes limiting for growth, which enhances transgenic yeast sensitivity to ACCase specific inhibitors. Under full induction, enough active ACCase can be obtained for biochemical and structural studies.
Plasmids that can drive the expression of active wheat ACCase in yeast will serve as a source of DNA for further manipulations. Because the size of the wheat ACCase cDNA (≈8 kb) makes such manipulations difficult, we introduced new restriction sites that allow us to move almost the entire ORF as one SalI fragment. A short linker encoding the AUG start codon (with engineered NcoI site) was then added to fuse the ORF to a proper yeast leader and promoter (GAL10). This two-step construction scheme is compatible with the pRS series of yeast shuttle vectors; therefore, in addition, we can use different selectable markers present in these vectors (10). This modular design makes many modifications possible, e.g., adding a chloroplast transit peptide for later use in a plant system or a tag to aid peptide purification.
In yeast, a single gene (ACC1) encodes ACCase, which provides malonyl-CoA for both fatty acid biosynthesis as well as for subsequent fatty acid elongation. The product of the same gene is needed to provide malonyl-CoA for the biosynthesis of very-long-chain fatty acids for the nuclear envelope (3). Complementation of the ACC1 null mutation by wheat cytosolic ACCase is consistent with the conclusion that the only essential role of yeast ACCase is to provide malonyl-CoA (3).
In yeast, disruption of one ACC1 allele decreases the level of ACCase activity (to about one-half in the heterozygous diploid disruptant), and the presence of the ACC1 gene on a high copy plasmid increases the enzyme activity in the wild-type diploid strain about 3-fold. The level of ACCase activity and amount of the biotinylated protein correlate with the steady-state level of ACCase-specific mRNA, indicating that the observed changes in ACCase amount and activity result from changes in the gene dosage (2). Expression of ACCase in yeast is correlated with phospholipid biosynthesis. In addition to transcription control, ACCase activity is regulated by phosphorylation/dephosphorylation reminiscent of the well-documented regulation of ACCase activity by AMP-activated protein kinase in vertebrates. None of the four conserved motifs containing serine residues, which correspond to phosphorylation sites in rat, chicken, and human ACCases (20), is present at a similar position in the wheat cytosolic ACCase, including the serine residue critical for regulation of the human enzyme by phosphorylation (21). The high relative activity of the wheat ACCase expressed in yeast may reflect that lack of phosphorylation (22).
Inhibitors are valuable tools for studies of enzyme mechanism. In addition, they find applications, e.g., as herbicides in agriculture. Understanding the molecular basis of inhibitor action is an important goal. Two classes of potent inhibitors specifically target plastid ACCase from graminae (4–6). Inhibitor-sensitive yeast gene-replacement strains, such as the strains described here, can be used to study the mechanisms of these inhibitors. Mutants that confer resistance can be selected and the mutation site(s) identified. These mutations will define inhibitor binding sites studied by other methods.
Plant-specific inhibitors have no effect on the growth of the wild-type yeast. On the other hand, we have identified one compound, cethoxydim, that completely inhibits growth of our gene-replacement yeast strains in liquid cultures at reasonable concentration. It is also an effective inhibitor under conditions used to select for resistant survivors on solid medium (data not shown). This is despite the fact that the wheat cytosolic ACCase expressed in our gene-replacement strain is less sensitive than the plastid enzyme. Indeed, in an in vitro assay, this compound moderately inhibits yeast-expressed wheat cytosolic ACCase. Yeast ACCase, although not completely resistant, is less sensitive. Cethoxydim, as other aryloxyphenoxypropionates and cyclohexanediones, is a much stronger inhibitor of the wheat plastid ACCase, as reflected by the in vitro assay of wheat germ extracts. Chimeric proteins composed of cytosolic (herbicide-resistant or tolerant) or plastid-resistant mutant (isolated as described above) and plastid (herbicide-sensitive) sequences can be analyzed for activity and inhibitor sensitivity/resistance using the yeast strains.
Acknowledgments
We are grateful to E. Ward, S. D. Kohlwein, and A. Ivessa for providing valuable materials and discussion. This work was supported by grants from CIBA–GEIGY (now Novartis) and Monsanto, and by the H. and F. Block Research Fund of the University of Chicago.
ABBREVIATIONS
- ACCase
acetyl-CoA carboxylase
- YPD
yeast extract/peptone/dextrose
- YPR
yeast extract/peptone/raffinose
- YPRG
yeast extract/peptone/raffinose/galactose
References
- 1.Konishi T, Shinohara K, Yamada K, Sasaki Y. Plant Cell Physiol. 1996;37:117–122. doi: 10.1093/oxfordjournals.pcp.a028920. [DOI] [PubMed] [Google Scholar]
- 2.Haslacher M, Ivessa A, Paltauf F, Kohlwein S. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- 3.Schneiter R, Hitomi M, Ivessa A S, Fasch E-V, Kohlwein S D, Tartakoff A M. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golz A, Focke M, Lichtenthaler H K. J Plant Physiol. 1994;143:426–433. [Google Scholar]
- 5.Holt J S, Powles S B, Holtum J A M. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:203–229. [Google Scholar]
- 6.Konishi T, Sasaki Y. Proc Natl Acad Sci USA. 1994;91:3598–3601. doi: 10.1073/pnas.91.9.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gornicki P, Haselkorn R. Plant Mol Biol. 1993;22:547–552. doi: 10.1007/BF00015984. [DOI] [PubMed] [Google Scholar]
- 8.Gornicki P, Podkowinski J, Scappino L A, DiMaio J, Ward E, Haselkorn R. Proc Natl Acad Sci USA. 1994;91:6860–6864. doi: 10.1073/pnas.91.15.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Podkowinski J, Sroga G E, Haselkorn R, Gornicki P. Proc Natl Acad Sci USA. 1996;93:1870–1874. doi: 10.1073/pnas.93.5.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 11.Johnston M, Davis R W. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klapholz S, Esposito R E. Genetics. 1982;100:387–412. doi: 10.1093/genetics/100.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klapholz S, Wadell C S, Esposito R E. Genetics. 1985;110:187–216. doi: 10.1093/genetics/110.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen D C, Yang B C, Kuo T T. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 15.Guthrie C, Fink G R. Methods Enzymol. 1991;194:21–37. [Google Scholar]
- 16.Mitchell D A, Marshall T K, Deschenes R J. Yeast. 1993;9:715–722. doi: 10.1002/yea.320090705. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Hoffman C S, Winston F. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 19.Howard J L, Ridley S M. FEBS Lett. 1990;261:261–264. [Google Scholar]
- 20.Ha J, Daniel S, Kong I-S, Park C-K, Tae H-J, Kim K-H. Eur J Biochem. 1994;219:297–306. doi: 10.1111/j.1432-1033.1994.tb19941.x. [DOI] [PubMed] [Google Scholar]
- 21.Ha J, Daniel S, Broyles S S, Kim K-H. J Biol Chem. 1994;269:22162–22168. [PubMed] [Google Scholar]
- 22.Witters L A, Watts T D. Biochem Biophys Res Commun. 1990;169:369–376. doi: 10.1016/0006-291x(90)90341-j. [DOI] [PubMed] [Google Scholar]