Abstract
Recolonization of Europe by forest tree species after the last glaciation is well documented in the fossil pollen record. This spread may have been achieved at low densities by rare events of long-distance dispersal, rather than by a compact wave of advance, generating a patchy genetic structure through founder effects. In long-lived oak species, this structure could still be discernible by using maternally transmitted genetic markers. To test this hypothesis, a fine-scale study of chloroplast DNA (cpDNA) variability of two sympatric oak species was carried out in western France. The distributions of six cpDNA length variants were analyzed at 188 localities over a 200 × 300 km area. A cpDNA map was obtained by applying geostatistics methods to the complete data set. Patches of several hundred square kilometers exist which are virtually fixed for a single haplotype for both oak species. This local systematic interspecific sharing of the maternal genome strongly suggests that long-distance seed dispersal events followed by interspecific exchanges were involved at the time of colonization, about 10,000 years ago.
Keywords: founder effect, geostatistics, introgression, long-distance dispersal
Oak species are major constituents of the deciduous forests of Europe. The timing and speed of the postglacial spread by oak forests has been described by workers using pollen analytical methods (1). However, there are limitations to the paleoecological approach. For instance, initial colonization may have occurred at a density too low to be recorded in the fossil pollen counts (2), and the spatial resolution is limited in lowland regions, due to the scarcity of peat bogs and mires. Moreover, the pollen grains of the different oak species are extremely difficult to distinguish (3). The study of the genetic structure of such long-lived species, if compared with fossil pollen data, may help reconstruct the recent Quaternary history of these trees.
Since the pioneering work of Skellam (4), who in 1951 modeled the postglacial colonization by oaks, there has been a growing interest in the study of the mechanisms of biological invasions. Recent developments emphasize the “stratified” diffusion of many organisms—i.e., the occurrence side by side of neighborhood diffusion, considered by Skellam, and of long-distance dispersal (5). It appears from these theoretical studies that very rare events of long-distance dispersal, at frequencies 10−5 times lower than the short-distance dispersal events, can be of great importance in the pattern and speed of colonization. Recently, Dyer (6) studied migration of oaks at a regional scale in the eastern United States and used such a stratified dispersal process.
Rangewide surveys of chloroplast DNA (cpDNA) variability in European oaks (7, 8) had revealed a clear pattern of genetic structure, which was interpreted as reflecting largely the routes of postglacial recolonization by these species. The study of the genetic structure of oak forests at a regional scale could deepen our knowledge of the mode of colonization of these species. Indeed, historical events, such as the founding of new populations, can have durable genetic consequences (9). Hence, present genetic structure may be used to infer past colonization events. Being maternally inherited in many angiosperm species, including oaks (10), cpDNA is much more susceptible to drift than are nuclear genes (11) and is clearly more appropriate to study recent or past events of seed dispersal.
We therefore decided to survey PCR-based cpDNA variation in oaks at a regional scale, with a sampling intensity high enough to identify the detailed spatial patterns of genetic variation. Furthermore, the inclusion of two sympatric species, Quercus robur L. (pedunculate oak) and Quercus petraea Matt. Liebl. (sessile oak) (Fagaceae), was motivated by the finding of a complete absence of cpDNA differentiation between them at a continental scale (7, 8), similar to what had been described for American white oaks (12). It had been argued that these interspecific gene flows may have arisen after prolonged contact in the glacial refugia (7). The recent estimation by our laboratory of the extent of hybridization in a mixed forest (13) suggested a more pervading process, which could have played an important role during the recolonization process itself.
We show that the spatial distribution of the cpDNA polymorphisms is patchy, with whole forests virtually fixed for a single haplotype, and that exactly the same spatial genetic structure is found for both species. In conjunction with the results of a simulation study based on the same data (14), we discuss the likeliness that these patches indeed reflect cases of rare, wide-ranging, founder events at the time of recolonization, and we further examine the consequences of hybridization and introgression in these species.
MATERIALS AND METHODS
The region studied is located along the Loire river in western France between Nantes and Rennes in the west and Orléans in the east and covers approximately 60,000 km2. Forests, dominated by pedunculate and sessile oaks, represent about 10% of the area. Small branches from two to four oaks were sampled in woods or forests at an interval of 20–25 km except in the region located between the Sarthe and the Mayenne rivers, which was sampled more thoroughly. Wherever possible, one or more individuals of each oak species were taken, with a preference for older trees, which are more likely to be native (7). They were identified according to the morphological characters of their leaves (15). Material was collected from each of 447 morphologically typical individuals from both species, originating from 188 localities. A total of three to five buds per individual were peeled and their total genomic DNA was extracted by using a simplified cetyltrimethylammonium bromide (CTAB)/dichloromethane protocol (10). This DNA was used as a template in the PCR. Two pairs of universal primers (16, 17) were then selected since they were known, from preliminary experiments, to detect several polymorphisms in this part of the range of the two oaks. These primers amplify mostly noncoding DNA sequences. A first fragment of 1.8 kb corresponds to an intergenic region located between the genes trnD (GUC) and trnT (GGU). It was digested by the endonuclease TaqI. A second fragment of 1.9 kb was amplified by using two primers located in the genes trnT (UGU) and trnF (GAA). It was digested with the endonuclease DdeI. Both digestion mixtures were subjected to electrophoresis in an 8% acrylamide gel, before staining with ethidium bromide, as described previously (17). The combined information of all polymorphisms found in both fragments allows the identification of six haplotypes.
Two techniques of analysis of the spatial structure were applied to the cpDNA data. First, spatial autocorrelation (18) was used to test the geographic structure and to study the response surface of the haplotype frequencies in a correlogram. The distance in kilometers between all pairs of populations was calculated, permitting the assignment of all pairs to distance classes with class intervals of 10 km. Then for each class and each of the four more frequent haplotypes, Moran’s index I (analogous to a coefficient of correlation of the haplotype frequencies in the pairs) (19) was computed and its values were plotted for the haplotypes as a function of the distance. Second, geostatistical methods were used to obtain synthetic contour maps of each of the haplotype frequencies through interpolation (20). There are two steps in such a procedure. First, the spatial structure of the variable (each haplotype frequency) is modeled with the help of a variogram. The variogram is similar to the autocorrelogram except that the variance of the haplotype frequency (instead of the correlation) is plotted versus distance, also for class intervals of 10 km. Then the variogram is modeled by two functions, a spherical one first and then a linear one (as soon as the slope of the variogram decreases). Finally, a prediction of the variable everywhere in the studied space is obtained (kriging) and allows a cartography. We used the statistical package GEOSTAT-PC for these purposes (21).
RESULTS
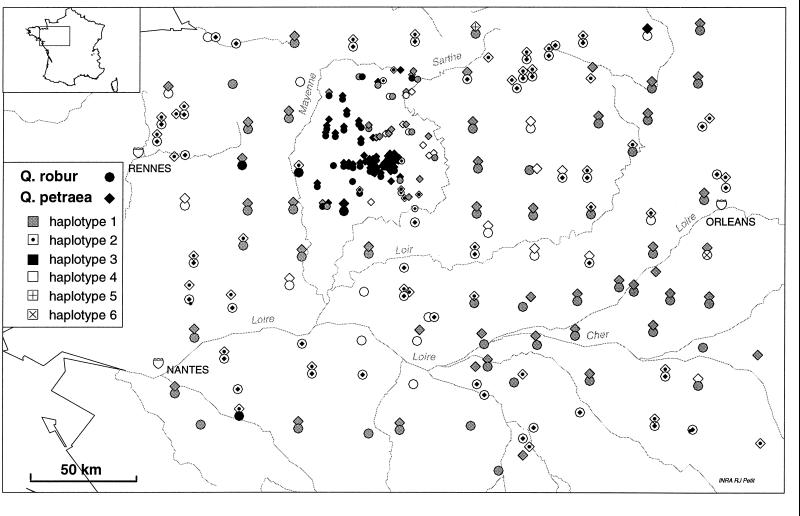
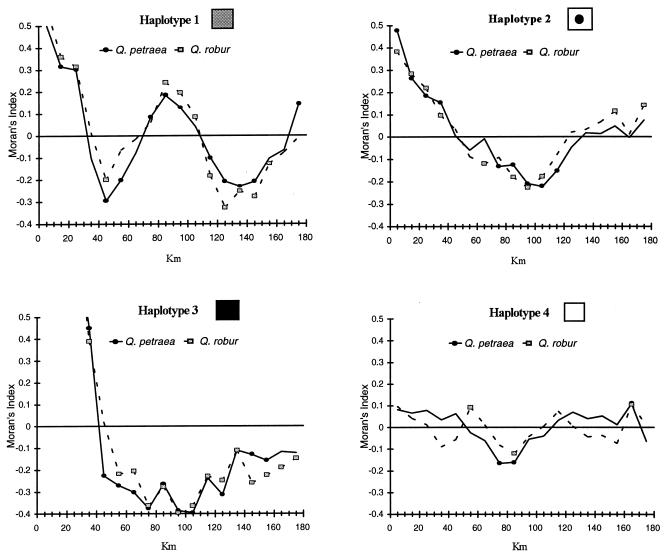
A total of five mutations (three insertions/deletions and two point mutations) were detected within the 1.8-kb fragment [trnD (GUC) to trnT (GGU)] and two insertions/deletions were detected within the 1.9-kb fragment [trnT (UGU) to trnF (GAA)]. Altogether, the seven mutations allow the identification of six haplotypes among the 447 trees studied. The distribution of these haplotypes is represented in Fig. 1. Two haplotypes (nos. 5 and 6) are extremely rare, each being observed in a single individual. Among the four more frequent haplotypes, the patch formed by haplotype 3, since it is located in the region more intensively sampled, is particularly conspicuous, but throughout the studied area the pattern of genetic structure is well marked. Amazingly, in 101 of the 125 forests where both species had been sampled, the same haplotype was shared by the two species. Autocorrelation analyses indicate the presence of a strong genetic structure, significant up to 30–40 km and very similar for both species (Fig. 2). The high positive autocorrelation values in the lowest distance classes, up to 30 km (haplotype 1) or 40 km (haplotypes 2 and 3), indicate that the scale of the pattern is comparable for the three more frequent haplotypes. The response surfaces are remarkably close for the pedunculate and the sessile oak, confirming the impression of systematic sharing of the haplotypes between the two species.
Figure 1.
Map of the distribution of cpDNA haplotypes in the lower Loire region. The six cpDNA haplotypes identified are plotted on this map for both oak species.
Figure 2.
Comparison of the spatial genetic structure for the two oak species. For each of the four more frequent haplotypes, Moran’s correlation indexes were computed for all distance classes of 10 km. Significant autocorrelation coefficients (P ≤ 0.05) in each class are indicated by squares (pedunculate oak) or circles (sessile oak).
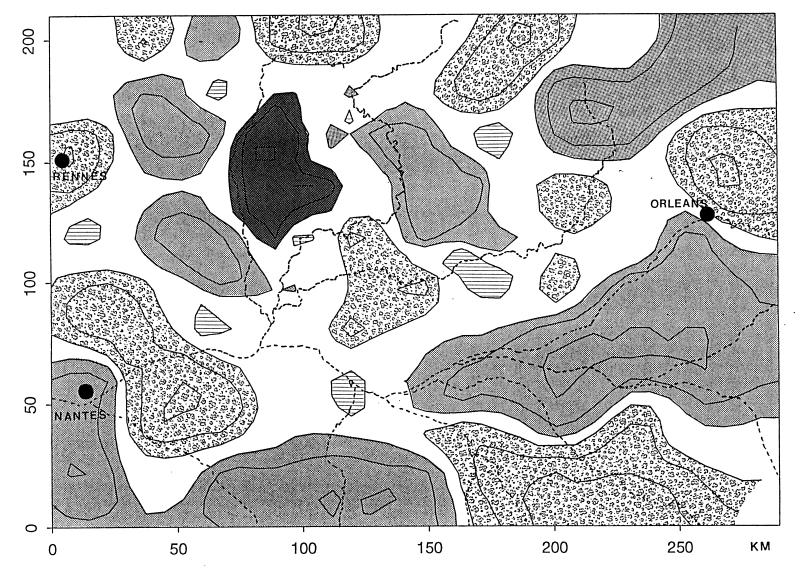
The filtered map of the spatial pattern of haplotype frequencies over all the sampled area for the complete data set (regardless of the taxonomic status of the trees) was obtained by using geostatistical methods. Only the predicted contour frequencies of 0.6, 0.8, and 1.0 are represented to outline the regions where a single haplotype is predominant. This gives a clear representation of the patchy cpDNA genetic pattern in the region studied (Fig. 3).
Figure 3.
Map of the estimated haplotype frequencies obtained by using geostatistical methods applied to the complete data set (both species simultaneously). Predicted frequencies of 0.6, 0.8, and 1.0 are represented for the four more frequent haplotypes: ░⃞, haplotype 1; , haplotype 2; ▪, haplotype 3; and ▤, haplotype 4.
DISCUSSION
The strong patchy cpDNA structure observed in the region is remarkable. In particular, the more intensive sampling in the Sarthe area demonstrates that whole forests can be fixed for a single haplotype, despite considerable diversity at the regional level. It is now clearly established, from theoretical expectations (11) and from empirical results (e.g., refs. 8 and 22), that maternally inherited markers in plants will generally reveal much more genetic structure than will nuclear markers. We decided to take advantage of this phenomenon and to study cpDNA variation in numerous localities within a region. The scale of the present study (a limited geographic region) is very unusual in forest tree population genetic studies, though clearly relevant for conservation and management purposes.
A relatively short span of time (in generations) has elapsed since recolonization by oaks took place in the studied region 10,000 to 9,000 years ago (1). This recent history may have left durable prints in the genetic structure of these long-lived trees. Recent studies have shown that colonization of empty territories offers great opportunities for genetic structure to build up (11, 23). This possibility was examined in a related simulation study of colonization in forest trees (14). The hypothesis tested was that the observed geographic distribution of the haplotypes at a regional scale was reflecting maternal founder effects due to long-distance seed dispersal at the outset of postglacial recolonization, when vacant habitats were available. There is indeed increasing evidence that a dual stratified process involving local diffusion and long-distance dispersal is involved in many cases of range expansion (5). We found that such a stratified dispersal could indeed create patchy patterns concordant with the present experimental investigation (14).
Taking into account the large size of the chloroplast patches, we propose, therefore, that very long (at least 30–40 km, the mean size of the patches) and rare dispersal events were involved at the time of recolonization. Note that rather long dispersal distances are necessary to account for the observed postglacial rate of colonization. Indeed, as already noted by Reid as early as 1899 (24), it will take several seasons until a tree which has colonized a new territory forms a fresh starting point. In particular, oaks are unlikely to reproduce before an age of 20–30 years in conditions similar to those of the early Postglacial (25). Since the apparent rate of spread for oaks reached 500 m per year in Europe (1), minimal dispersal distances varying from 10 to 15 km are implied each generation. However, it is very unlikely that a single isolated tree will be at the origin of a new leap the first time it produces acorns. Moreover, Bennett (2, 3), studying pollen accumulation rates of several tree species in England, concluded that initial doubling times for oak populations were between 60 and 73 years. This suggests that the previous figures of 10–15 km could be underestimates. Very long dispersal events (25–130 km) were also inferred to explain the spatial pattern of postglacial beech forest expansion in North America, using only paleoecological methods (26). Finally, dispersal distances, rather than number of dispersal events per generation, are the variable to which migration rates are most sensitive, as shown by simulations (6, 14). Even if frequent “medium” distance dispersal events could account for the rate of oak dispersal, coalescence of the newly founded populations would then occur too rapidly to generate the patchy pattern observed.
The initial genetic structure created by these founder events is expected to have “frozen” when dense forests became established (8, 14), because an increase in density will reduce genetic drift (9, 27). First, the local production of acorns will quickly saturate the few remaining favorable germinating sites. Second, seed dispersal distances may be lower in established forests: Carter Johnson and Webb, studying the possible role of jays in the postglacial dispersal of fagaceous trees, suggested that isolated “nut” patches should promote longer seed dispersal by jays, a consequence of the behavior of these birds (28). Dyer found, in his model of oak dispersal, that preexisting forests were actually slowing the rate of colonization as compared with more open environments (6).
It seems difficult to imagine that local selection of haplotypes could be responsible for the observed cpDNA pattern. Obviously, the environmental heterogeneity within the chloroplast patches for factors such as soil pH, humidity, etc. far outweigh possible differences among patches. Moreover, the genetic pattern is similar for the two oak species, which have quite different ecological requirements (29).
There is no suggestion in the fossil pollen data that large extinction/recolonization processes occurred (due, for instance, to fire) at the scale of interest after postglacial recolonization (1). The large mixed oak forest which became established after the last ice age remained in place until deforestation by humans started, at the Neolithic revolution. In France, peaks of deforestation occurred in Roman times and during the Middle Ages until the beginning of the 18th century (30). Since then, the proportion of forested land has doubled, either by natural means or through planting. But at a scale of tens of kilometers, human activities are more likely to blur a preexisting genetic structure than to generate it. In a study of cpDNA variation in pedunculate oak in East Anglia, Ferris et al. (31) also arrived at the same conclusion. In our study, the consistency of the geographic pattern obtained with two different oak species is one of the best indications of its ancestral character. Indeed, recent artificial transfers are more likely to involve a single oak species and to result in local intra- and interspecific differences.
During the glacial maximum, the studied region was a tundra with, at most, scattered populations of Betula and Pinus, so the presence of near-glacial refugia of oaks that could have given rise to the strong clustering of haplotypes via fragmentation and drift seems highly unlikely (J.-L. de Beaulieu, personal communication). But the early presence of “islands” of oak forests much before the general increase of oak populations is possible. Pollen records from a site located 100 km east of our studied area show that oak pollen reached 2% as early as 12,000 years ago (32). An earlier (and hence more rapid) spread would actually support our model of long-distance colonization.
The European jay Garrulus glandarius L., the most likely long-distance animal vector (33), has been observed caching viable acorns in the ground at maximal distances of 8 km from the foraging sites (34). In Great Britain, Jones (35) cites a seedling found in the island of Hoy which must have come from an acorn carried at least 10 miles (16 km). Moreover, occasional long-distance transport of acorns in rivers is possible, especially for the water-tolerant pedunculate oak. In South Africa, where this species was introduced in the 18th century, dispersal downstream in rivers has played an important role (36). Given the rarity of the inferred long-distance dispersal events (14), it is unlikely that they can be documented.
Although previous reports indicate that there is very little species differentiation with cpDNA markers at the European scale (7, 8), the finding of a nearly identical cpDNA spatial structure at a regional scale requires careful examination. Pedunculate and sessile oaks are the two most widespread and certainly the best studied oak species in Europe (37). They are largely sympatric: the sessile oak range lies nearly completely within the pedunculate oak range. Darwin, in his famous Origin of Species, used these oaks as a paradigm to define his species concept. Although it is still a controversial issue, most modern taxonomic treatments consider these forms as distinct species, and the discovery of a partial (asymmetric) incompatibility between the two forms in controlled crosses supports this view (38). In natural conditions, the study of the mating system has shown that the contribution of the sessile oak to the pedunculate oak progenies can vary from 17% to 48%, but ovules of sessile oak trees are preferentially fertilized by “extreme” sessile genotypes, resulting in progenies more different from pedunculate oaks across generations (13). This could indicate that backcrosses are occurring and that they are also asymmetric. Furthermore, numerous studies indicate that when the two oaks form mixed stands, the shade-tolerant sessile oak has a higher competitive ability and replaces the more pioneer pedunculate oak (13, 29). The striking conclusion is that gene exchanges between these species reinforce succession. In the stand studied, sessile oak represented 47% of the adult trees but at least 63% of the seedlings (13). More than half of this increase was due to directional hybridization or backcrossing, the rest being attributed to the ecological dynamics.
Contrasting the pollen and seed flow in oaks is also useful to understand how congruent cpDNA spatial patterns could arise for the two species at a regional scale. Recent analyses of parentage in oaks, using hypervariable microsatellite nuclear markers, indicate extremely high levels of long-distance pollination and more limited acorn dispersal (39). This is consistent with indirect methods based on measures of genetic differentiation for nuclear versus cpDNA markers in oaks, which suggest that pollen flow is much higher (by two orders of magnitude) than seed flow (11, 40).
When the pioneer pedunculate oak colonized new territories during postglacial recolonization, the newly established populations produced flowers receptive to sessile oak pollen. The overwhelming importance of pollen versus seed flow implies that the establishment of sessile oak through pollen flow, by hybridization and repeated backcrossing, should greatly exceed the colonization through seed dispersal. Interestingly, in this hypothesis, the initial cpDNA spatial structure formed during colonization would remain intact, while pedunculate oak’s nuclear genes are progressively replaced by sessile oak’s genes. In other words, the late successional species would spread largely through pollen, by “swamping out” the nuclear genome of the pioneer species. Recent studies of the nuclear genome of these species further support this hypothesis. First, an extremely low level of nucleotide divergence between the two species was estimated (0.5% on average, but up to 3.3% in the most discriminating regions) (41). Second, this interspecific divergence was found to vary throughout the range (42). Altogether, given the magnitude of asymmetric gene flow between the two species, the prevalence of pollen flow in oaks, and the necessity to account for the identical regional distribution of cpDNA, we consider that the scenario described above is the most parsimonious.
Ironically, if these oak species are considered distinct, the proposed mechanism constitutes an exception to Harper’s rule, which states that in plants “the haploid phase of environmental exploration cannot act further than has already been colonized by the diploid phase of the species” (43). This mechanism also differs diametrically from the interspecific “cytoplasmic captures” discussed by other authors, where native cytoplasms are replaced by alien ones (44).
Acknowledgments
We are grateful to J.-L. de Beaulieu, P. Taberlet, and two anonymous reviewers for critical reading of this paper. The study was supported by grants from the Ministère de l’Agriculture et de la Pêche (Direction de l’Espace rural et des Forêts) and from the European Union Research Program in Biotechnology (BIO2-CT93–0373).
ABBREVIATION
- cpDNA
chloroplast DNA
References
- 1.Huntley B, Birks H J B. An Atlas of Past and Present Pollen Maps for Europe 0–13000 Years Ago. Cambridge, U.K.: Cambridge Univ. Press; 1983. [Google Scholar]
- 2.Bennett K D. Phil Trans R Soc Lond B. 1986;314:523–531. [Google Scholar]
- 3.Bennett K D. Nature (London) 1983;303:164–167. [Google Scholar]
- 4.Skellam J G. Biometrika. 1951;38:196–218. [PubMed] [Google Scholar]
- 5.Shigesada N, Kawasaki K, Takeda Y. Am Nat. 1995;146:229–251. [Google Scholar]
- 6.Dyer J M. Ecol Modell. 1995;79:199–219. [Google Scholar]
- 7.Ferris C, Olivier R P, Davy A J, Hewitt G M. Mol Ecol. 1993;2:334–337. doi: 10.1111/j.1365-294x.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 8.Petit R J, Kremer A, Wagner D B. Theor Appl Genet. 1993;87:122–128. doi: 10.1007/BF00223755. [DOI] [PubMed] [Google Scholar]
- 9.Boileau M G, Hebert P D N, Schwartz S S. J Evol Biol. 1992;5:25–39. [Google Scholar]
- 10.Dumolin S, Demesure B, Petit R J. Theor Appl Genet. 1995;91:1253–1256. doi: 10.1007/BF00220937. [DOI] [PubMed] [Google Scholar]
- 11.Petit R J, Kremer A, Wagner D B. Heredity. 1993;71:630–641. [Google Scholar]
- 12.Whittemore A T, Schaal B A. Proc Natl Acad Sci USA. 1991;88:2540–2544. doi: 10.1073/pnas.88.6.2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bacilieri R, Ducousso A, Petit R J, Kremer A. Evolution. 1996;50:900–908. doi: 10.1111/j.1558-5646.1996.tb03898.x. [DOI] [PubMed] [Google Scholar]
- 14.Le Corre V, Machon N, Petit R J, Kremer A. Genet Res Camb. 1997;69:117–125. [Google Scholar]
- 15.Dupouey J L. Ann Sci For. 1983;40:265–282. [Google Scholar]
- 16.Taberlet P, Gielly L, Pautou G, Bouvet J. Plant Mol Biol. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- 17.Demesure B, Sodzi N, Petit R J. Mol Ecol. 1995;4:129–131. doi: 10.1111/j.1365-294x.1995.tb00201.x. [DOI] [PubMed] [Google Scholar]
- 18.Sokal R R, Oden N L. Biol J Linn Soc. 1978;10:199–228. [Google Scholar]
- 19.Moran P A P. Biometrika. 1950;37:17–23. [PubMed] [Google Scholar]
- 20.Monestier P, Goulard M, Charmet G. Theor Appl Genet. 1994;88:33–41. doi: 10.1007/BF00222391. [DOI] [PubMed] [Google Scholar]
- 21.Boivin P. GEOSTAT-PC. Paris: ORSTOM; 1990. [Google Scholar]
- 22.McCauley D E. Proc Natl Acad Sci USA. 1994;91:8127–8131. doi: 10.1073/pnas.91.17.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wade M J, McKnight M L, Shaffer H B. Evolution. 1994;48:1114–1120. doi: 10.1111/j.1558-5646.1994.tb05298.x. [DOI] [PubMed] [Google Scholar]
- 24.Reid C. The Origin of the British Flora. London: Dulau; 1899. [Google Scholar]
- 25.Jensen T S, Nielsen O F. Oecologia (Berlin) 1986;70:214–221. doi: 10.1007/BF00379242. [DOI] [PubMed] [Google Scholar]
- 26.Webb S L. Ecology. 1987;68:1993–2005. doi: 10.2307/1939890. [DOI] [PubMed] [Google Scholar]
- 27.Handel S N. In: Pollination Biology. Real L, editor. New York: Academic; 1983. pp. 163–211. [Google Scholar]
- 28.Carter Johnson W, Webb T., III J Biogeo. 1989;16:561–571. [Google Scholar]
- 29.Lévy G, Becker M, Duhamel D. Forest Ecol Manag. 1992;55:51–63. [Google Scholar]
- 30.Gadant J. L’Atlas des Forêts de France. Paris: de Monza; 1994. p. 14. [Google Scholar]
- 31.Ferris C, Oliver R P, Davy A J, Hewitt G M. Mol Ecol. 1993;2:337–344. doi: 10.1111/j.1365-294x.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 32.Planchais N. Pollen et Spores. 1970;12:381–428. [Google Scholar]
- 33.Bossema I. Behavior. 1979;70:1–117. [Google Scholar]
- 34.Schuster L. Vogelwelt. 1950;71:9–17. [Google Scholar]
- 35.Jones E W. J Ecol. 1959;47:169–222. [Google Scholar]
- 36.Knight R S. S Afr J Bot. 1985;51:265–269. [Google Scholar]
- 37.Kleinschmit J. Ann Sci For. 1993;50:166–186. [Google Scholar]
- 38.Steinhoff S. Ann Sci For. 1993;50:137–143. [Google Scholar]
- 39.Dow B D, Ashley M V. Mol Ecol. 1996;5:615–627. [Google Scholar]
- 40.Ennos R A. Heredity. 1994;72:250–259. [Google Scholar]
- 41.Bodénès C, Laigret F, Kremer A. Heredity. 1997;78:433–444. [Google Scholar]
- 42.Bodénès, C., Labbé, T., Pradère, S. & Kremer, A. (1997) Mol. Ecol. 6, in press.
- 43.Harper J L. Population Biology of Plants. London: Academic; 1977. p. 775. [Google Scholar]
- 44.Rieseberg L H, Soltis D E. Evol Trends Plants. 1991;5:65–84. [Google Scholar]