Abstract
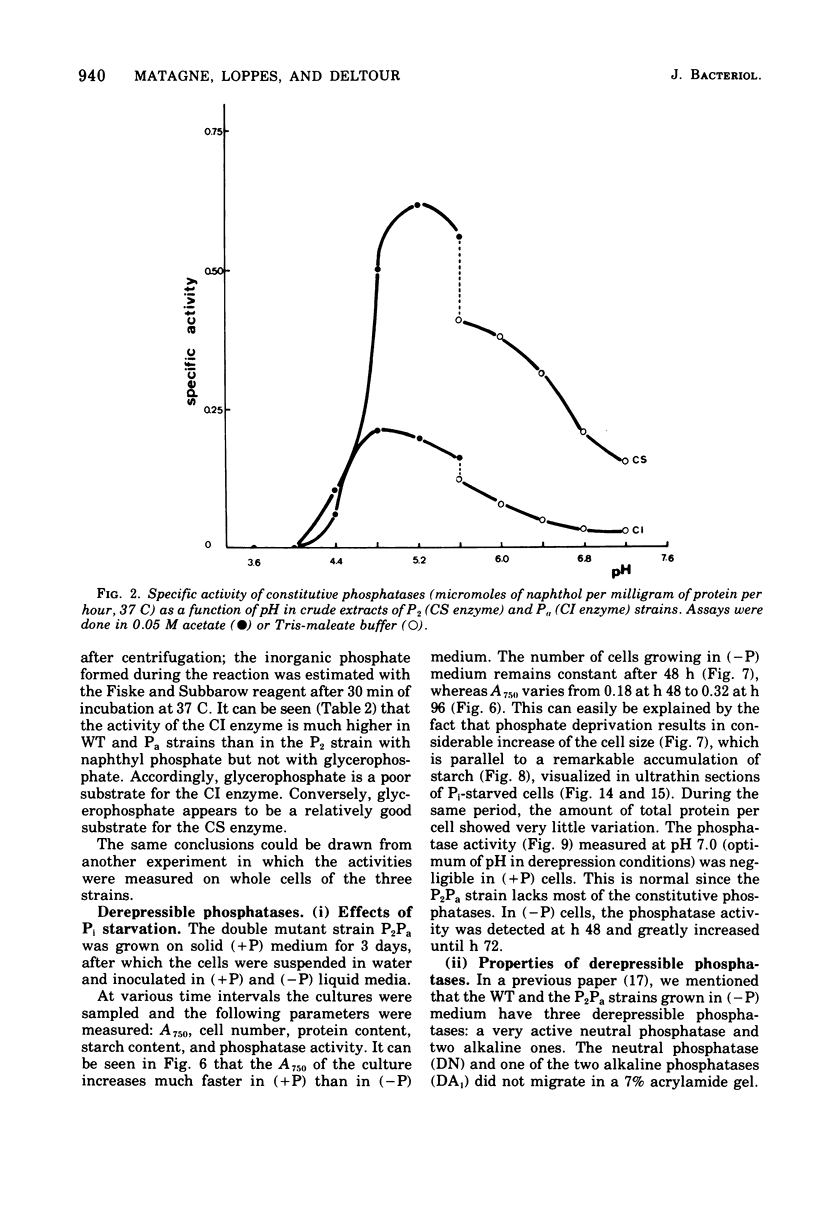
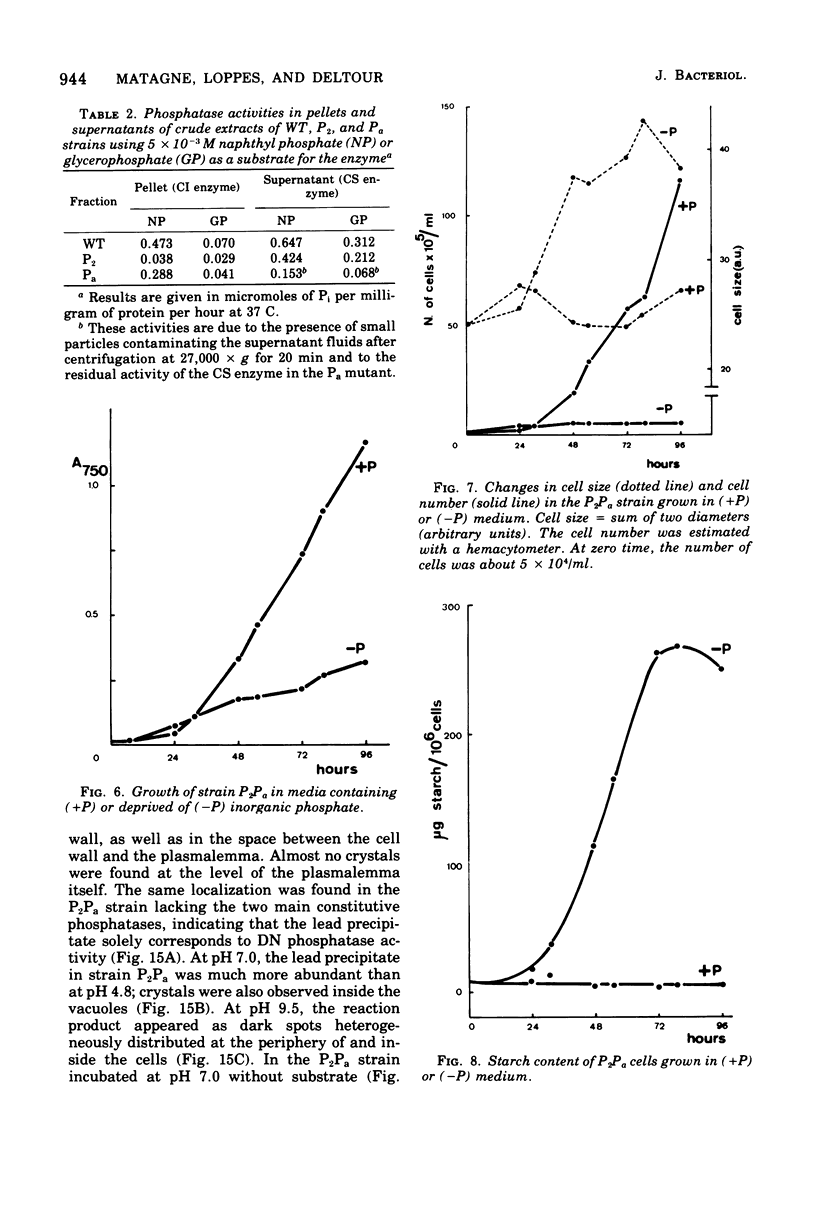
The unicellular alga Chlamydomonas reinhardi produces two constitutive acid phosphatases and three depressible phosphatases (a neutral and two alkaline ones) that can utilize napthyl phosphate as a substrate. Specific mutants depressible phosphatase were used to investigate biochemical properties and the cytochemical localization of these enzymes. The two constitutive phosphatases show similar pH optima (about 5.0) and Km values (2 x 10(-3) to 3.3 x 10(-3) M) but differ in their heat sensitivity and affinity for glycerophosphate.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnold W. N. Location of acid phosphatase and -fructofuranosidase within yeast cell envelopes. J Bacteriol. 1972 Dec;112(3):1346–1352. doi: 10.1128/jb.112.3.1346-1352.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennun A., Blum J. J. Properties of the induced acid phosphatase and of the constitutive acid phosphatase of Euglena. Biochim Biophys Acta. 1966 Oct 17;128(1):106–123. doi: 10.1016/0926-6593(66)90147-0. [DOI] [PubMed] [Google Scholar]
- Cooper R. A., Bowen I. D., Lloyd D. The properties and subcellular localization of acid phosphatases in the colourless alga, Polytomella caeca. J Cell Sci. 1974 Aug;15(3):605–618. doi: 10.1242/jcs.15.3.605. [DOI] [PubMed] [Google Scholar]
- Goff C. W. Localization of nucleoside diphosphatase in the onion root tip. Protoplasma. 1973;78(4):397–416. doi: 10.1007/BF01275775. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Loppes R., Deltour R. Changes in phosphatase activity associated with cell wall defects in Chlamydomonas reinhardi. Arch Microbiol. 1975 May 5;103(3):247–250. doi: 10.1007/BF00436357. [DOI] [PubMed] [Google Scholar]
- Loppes R., Matagne R. F. Acid phosphatase mutants in Chlamydomonas: isolation and characterization by biochemical, electrophoretic and genetic analysis. Genetics. 1973 Dec;75(4):593–604. doi: 10.1093/genetics/75.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlister T. J., Costerton J. W., Thompson L., Thompson J., Ingram J. M. Distribution of alkaline phosphatase within the periplasmic space of gram-negative bacteria. J Bacteriol. 1972 Sep;111(3):827–832. doi: 10.1128/jb.111.3.827-832.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre R. J. A method for measuring activities of acid phosphatases separated by acrylamide gel electrophoresis. Biochem Genet. 1971 Feb;5(1):45–56. doi: 10.1007/BF00485729. [DOI] [PubMed] [Google Scholar]
- Matagne R. F., Loppes R. Isolation and study of mutants lacking a derepressible phosphatase in Chlamydomonas reinhardi. Genetics. 1975 Jun;80(2):239–250. doi: 10.1093/genetics/80.2.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts K., Gurney-Smith M., Hills G. J. Structure, composition and morphogenesis of the cell wall of Chlamydomonas reinhardi. I. Ultrastructure and preliminary chemical analysis. J Ultrastruct Res. 1972 Sep;40(5):599–613. doi: 10.1016/s0022-5320(72)80046-7. [DOI] [PubMed] [Google Scholar]
- SOMMER J. R., BLUM J. J. CYTOCHEMICAL LOCALIZATION OF ACID PHOSPHATASES IN EUGLENA GRACILIS. J Cell Biol. 1965 Feb;24:235–251. doi: 10.1083/jcb.24.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L. M., MacLeod R. A. Biochemical localization of alkaline phosphatase in the cell wall of a marine pseudomonad. J Bacteriol. 1974 Feb;117(2):819–825. doi: 10.1128/jb.117.2.819-825.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]