Abstract
The intracellular distribution of RNAs depends on interactions of cis-acting nuclear export elements or nuclear retention elements with trans-acting nuclear transport or retention factors. To learn about the relationship between export and retention, we isolated RNAs that are exported from nuclei of Xenopus laevis oocytes even when most RNA export is blocked by an inhibitor of Ran-dependent nucleocytoplasmic transport, the Matrix protein of vesicular stomatitis virus. Export of the selected RNAs is saturable and specific. When present in chimeric RNAs, the selected sequences acted like nuclear export elements in promoting efficient export of RNAs that otherwise are not exported; the pathway used for export of these chimeric RNAs is that used for the selected RNAs alone. However, these chimeric RNAs, unlike the selected RNAs, were not exported in the presence of Matrix protein; thus, the nonselected sequences can cause retention of the selected RNA sequences under conditions of impaired nucleocytoplasmic transport. We propose that most RNAs are transiently immobilized in the nucleus and that release of these RNAs is an essential and early step in export. Release correlates with functional Ran-dependent transport, and the lack of export of chimeric RNAs may result from interference with the Ran system.
Keywords: RNA export, RNA immobilization, selection of ET-RNAs in vivo, vesicular stomatitis virus Matrix protein, Ran-GTPase
The intracellular distributions of RNAs, essential for their correct processing and function, depend on interactions between regions in the RNA and specific RNA binding factors. We refer to the cis-acting elements in the RNAs that promote retention or export as nuclear retention elements (NREs) or nuclear export elements (NEEs), respectively. Localization of an RNA in the nucleus or cytoplasm of a cell may be due either to the activity, inactivity, or saturation of one or both types of these cis-acting elements. Thus, the presence of an RNA in the nucleus could result either from binding of the RNA to nuclear structures, from the absence of an NEE in the RNA, or from saturation of an essential export factor that interacts (directly or indirectly) with an NEE.
Nuclear retention of RNA may be specific (1–3) or it may occur by default. For example, the abundant nuclear antigen La promotes nuclear retention of several RNAs including hY1 RNA (4) and NL-15 RNA, a molecule selected for its localization in nuclei (5). RNAs lacking an NEE, such as small RNAs without m7G caps or certain variants of pre-small nuclear (sn)RNAs (3, 6, 7) are exported very slowly, if at all.
Specific factors have been identified, that exit the nucleus with RNAs during export through the nuclear pore complexes (NPCs). These factors include the cap binding complex, which recognizes the m7G caps of RNA polymerase II transcripts (8), the Rev protein from HIV-1, which recognizes the Rev responsive element in HIV pre-mRNAs (9, 10), and heterogeneous nuclear ribonucleoprotein A1 (11), which binds mRNAs; all of these factors are subsequently imported back into the nucleus. Also, saturation of required factors by large numbers of RNAs of one class, such as pre-snRNAs or tRNAs, can inhibit export of other members of that class (6).
Export of many RNAs is dependent on Ran-GTPase and its associated binding, exchange, and activation factors (12, 13). Inactivation of this system by mutation or inhibition leads to blockage of export of most RNAs, with the exception of tRNA (14) and stress-related mRNAs (15). The precise role of Ran and its associated factors in RNA export remains to be established. The Ran system also is required for protein import (16), so loss of Ran function may result in nuclear depletion of RNA export factors that must shuttle between the nucleus and cytoplasm (17). However, the GTP-bound form of Ran is required for nuclear protein export (18), and the same type of intranuclear signal may also function in nuclear RNA export. We have proposed that movement of RNAs within the nucleoplasm may be controlled by interactions of RNA-protein complexes with the Ran system (14). Thus, retention of RNAs in the nucleus could result from regulation of Ran-mediated access of export substrates to the NPCs.
We recently showed that the Matrix (M) protein of vesicular stomatitis virus very effectively blocks Ran-dependent nucleocytoplasmic transport in Xenopus laevis oocytes (19). Here, we used this inhibitor to select novel RNAs that are exported efficiently, even when Ran-dependent transport and related processes are disrupted. These selected sequences acted like NEEs when present in chimeric molecules containing RNA sequences that are otherwise not exported, or only inefficiently so. However, unlike the RNAs comprised solely of the selected sequences, the chimeric RNAs were very poorly exported in the presence of M protein. Thus, the nonselected sequences in the chimeric molecules imposed on the selected sequences an extra requirement for export, which is most apparent when normal nucleocytoplasmic transport is impaired. We propose that this additional requirement reveals nuclear events that normally happen to most cellular RNAs, resulting in their transient immobilization within the nucleus; export of an RNA would require its release from immobilization, an event that is blocked by M protein and possibly other inhibitors of nucleocytoplasmic transport.
MATERIALS AND METHODS
DNA Templates.
DNA templates for in vitro transcription were generated by PCR amplification of RNA coding regions using appropriate primer pairs. Templates used to transcribe U1, U1124, U2, U3, U6, and hY1 RNAs were described previously (2, 7, 20). The U6Xho DNA template was constructed by amplifying the U6 RNA coding region with a 5′ primer containing the T7 promoter and a 3′ primer containing an XhoI site adjacent to the U6 coding region. The DNA was cut with XhoI prior to transcription. This DNA also was used to construct the DNA templates for the chimeric U6 RNAs (see below). In vitro transcription and purification of RNAs were done as described (5).
To construct the ET-202 dimer (ET-202/di) template, DNA encoding ET-202 RNA under the control of a T7 promoter was cloned into pGEM4Z (Promega), using a HindIII linker 5′ of the T7 promoter and a EcoRI linker 3′ of the ET-202 coding sequence. A second ET-202 coding region (flanked by EcoRI linkers) was then inserted into the EcoRI site of this plasmid. The orientation of the insert was verified by PCR amplification using primer pairs sensitive to the sense or antisense orientation of the second ET-202 DNA. The plasmid containing two ET-202 coding regions in sense orientation and an appropriate pair of primers were used to amplify a DNA template containing the T7 promoter and the coding sequence for ET-202/di with the ET-202 3′ end.
To construct the chimeric adenovirus (Ad)/ET-202 and Ad/αET-202 DNA templates, ET-202 DNA containing ScaI and EcoRV linkers on the 5′ and 3′ side, respectively, was cloned into the SmaI site of pSP64-Ad1 (21), which contains exon one, a shortened form of intron one and 45 nucleotides of exon two of the adeno major late (AdML) coding region (22). Orientation of the ET-202 insert in individual clones was determined by PCR amplification using primer pairs that were sensitive for sense or antisense orientation of the insert. Plasmids containing ET-202 sequences in sense or antisense orientation were used together with appropriate primer pairs to amplify DNA containing an SP6 promoter and the coding region of the chimeric RNAs. Ad/ET-202 RNA had the precise ET-202 3′ end, but Ad/αET-202 RNA had an additional trinucleotide (AGU) at the end of the antisense sequences of ET-202 RNA.
The structure of the DNA template used to prepare the RNA for the first round of in vivo selection has been described (5).
DNA templates for other chimeric RNAs were constructed by ligating DNA of the appropriate RNA coding regions via linker sequences. The ligated products were amplified by PCR using primer pairs selective for the chimeric DNA. The linker sequences separating the two coding regions in the chimeric RNAs were 5′-CTCGAGTACT-3′ (for U2Sm−/ET-202, U1124/ET-202, U1124/ET-208, U6/ET-202, and U6/SLX) and 5′-GAATTCGATTTAGGTGACACTATA-3′ (for ET-202/U2Sm− and SLX/U2Sm−).
Oocyte Injections and in Vivo Selection.
The general scheme for selection was as described (5). To select for RNAs that are exported in the presence of the M protein of vesicular stomatitis virus (M protein; rounds 1–12 and 17 and 18), mRNA for M protein was pre-injected into oocyte cytoplasms (19). Sixteen to 20 hr later, when the M protein was expressed at levels sufficient to inhibit most nucleocytoplasmic transport, the pool of uncapped RNAs containing the randomized sequence (n = 20) was injected into nuclei along with several control RNAs for nuclear retention (e.g., U3 or U6) and export (e.g., U1Sm−). After 2 hr of incubation at 18°C, oocytes were dissected and RNAs were prepared from both nuclear and cytoplasmic fractions. The exported RNAs (in the cytoplasms) were purified by size selection in a denaturing polyacrylamide gel containing 7 M urea and the selected RNAs were amplified by reverse transcription coupled to PCR (RT-PCR). The resulting DNA templates were used to prepare the RNA for the next round of selection. The counterselection (rounds 13–16) to ensure active transport was done by injecting RNAs into oocytes kept at 0°C. After 24 hr of incubation on ice, the not-exported RNAs (in the nucleus) were isolated and amplified as above. The final RT-PCR products of exported RNAs in presence of M protein (round 18; ET-RNAs, see Fig. 1) were cloned and sequenced as described (5).
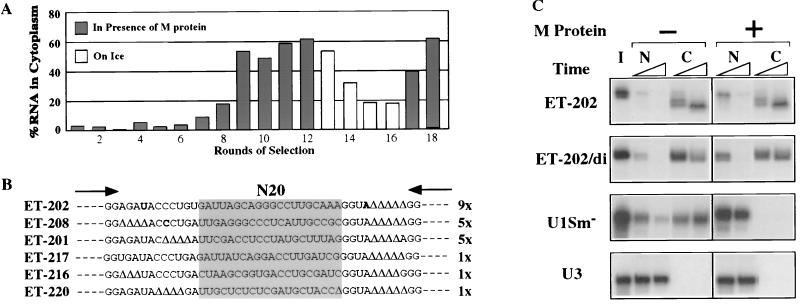
Figure 1.
Selection of RNAs containing NEEs. (A) Enrichment of injected RNA pools for RNAs containing NEEs. The percentage of exported RNAs at 2 hr (rounds 1–12 and 17 and 18) or 24 hr (rounds 13–16) after nuclear injection was calculated as [C/(N+C)] × 100. Rounds of selection are indicated at the bottom. Solid bars, selection in presence of M protein; open bars, counterselection on ice. (B) Sequences of ET-RNAs (Exceptional Transport RNAs) after 18 rounds of selection. Shaded box represents the randomized region (N20). Δ indicates nucleotides that were deleted from the fixed sequence during the selection procedure and arrows indicate the 3′ ends of the primers used for reverse transcription and PCR. (C) Export of the monomeric (ET-202) and dimeric (ET-202/di) forms of the winner RNA ET-202 (Top). Both forms of ET-202 RNA were uncapped, whereas the control RNAs (U1Sm− and U3; Bottom) were m7GpppG capped. RNAs were injected into nuclei of oocytes that did (+) or did not (−) contain M protein and prepared from nuclei (N) and cytoplasms (C) after 1 and 3 hr. Nucleocytoplasmic distribution of the RNAs was determined by denaturing 8% PAGE containing 7 M urea. I, input RNA.
Oocyte injections and dissections were done as described (2); for export in the presence of mAb414, RNAs were mixed with 3 mM DTT, RNasin, and the antibody (at a final concentration of 5 mg/ml) prior to injection. tRNAPhe from yeast (Sigma) was used as unlabeled competitor tRNA.
Error-Prone PCR.
The DNA template encoding ET-202 RNA was mutagenized by error-prone PCR essentially as described (23). The DNA was amplified using the primer pair indicated in Fig. 1B using either 1 mM each of dGTP, dTTP, and dCTP plus 0.2 mM dATP, or 1 mM dGTP and dTTP and 0.2 mM dCTP and dATP. In both cases, amplification was for 25 cycles of 1 min each at 95°C, 68°C, and 72°C in an otherwise standard PCR reaction containing 7.1 mM MgCl2 and 10% dimethyl sulfoxide. Amplified DNA was transcribed into RNA, which was then injected into nuclei of M-treated oocytes. Individual cDNAs made from RNA isolated from cytoplasm or nuclei were cloned and sequenced as described (5).
RESULTS
Selection of RNAs Exported in the Presence of M Protein.
Using M protein as an inhibitor of Ran-dependent nucleocytoplasmic transport in X. laevis oocytes (19), we selected RNAs that were capable of being exported by an alternative pathway. A combinatorial library of sequences comprising 20 nucleotides inserted into a shortened version of U1 RNA (5) was injected into nuclei of oocytes containing M protein (M oocytes). Exported RNAs were isolated from the cytoplasm and amplified to produce templates for the subsequent round of selection.
After 12 rounds of selection, over 60% of the injected RNAs were in the cytoplasm within 2 hr of nuclear injection (Fig. 1A). This mixture of molecules was then subjected to four rounds of counterselection (no export at 0°C) to remove RNAs whose export was by diffusion rather than by active transport. After two additional rounds of selection for export in the presence of M protein, individual cDNAs were cloned and sequenced (Fig. 1B). Three “winner sequences” predominated among the selected ET-RNAs (Exceptional Transport RNAs). Almost one-half of the isolates, represented by clone ET-202, had the same sequence; two other clones (ET-208 and ET-201) each accounted for about one-quarter of the total.
As expected, individual RNAs transcribed from the cloned cDNAs, such as ET-202 RNA, were exported very efficiently in both control and M oocytes (Fig. 1C). Likewise, export of the selected ET-RNAs was not greatly affected by other inhibitors of RNA export such as mAb414 (cf. Fig. 5A; data not shown), which effectively blocks export of most RNAs (17) or wheat germ agglutinin, an inhibitor of tRNA export (ref. 24; unpublished results). The lack of inhibition by mAb414 was surprising since the antibody was not used in the selection of the ET-RNAs. Thus, these RNAs must use an export pathway that does not depend on the XFXFG amino acid repeats in the proteins of the NPC, to which this antibody binds. The lack of inhibition by wheat germ agglutinin indicates that ET-202 RNA uses a different export pathway from that of tRNAs, despite the fact that export of both RNAs is largely unaffected by M protein (see below).
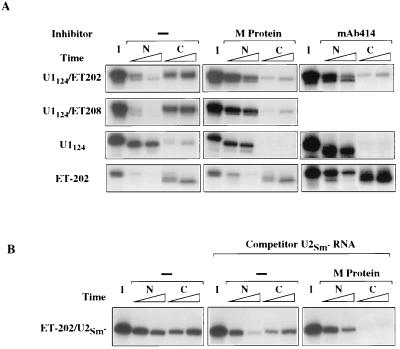
Figure 5.
Nuclear retention of chimeric RNAs. (A) The ability of the selected NEEs in chimeric RNAs to support export in the absence or presence of M protein or mAb414 was tested by injection of the RNAs into nuclei of control oocytes (−) or oocytes containing M protein or mAb414. Export of chimeric U1124/ET-202 (Top) and U1124/ET-208 (Upper Middle) RNAs was tested in oocytes containing M protein or mAb414, as indicated; export of U1124 RNA and ET-202 RNA is shown for comparison. All RNAs were ApppG capped (except for ET-202 RNA, which was uncapped). RNAs were isolated from nuclear (N) and cytoplasmic (C) fractions 1 and 3 hr after nuclear injection and analyzed as in Fig. 1C. (B) Specificity of export and retention of the chimeric RNAs was tested by coinjection of a mixture of unlabeled RNAs containing 1.0 pmol of ApppG-capped U2 RNA and 0.5 pmol of NL-15 RNA; the former RNA served as competitor for nonselected RNA sequences present in the chimeric ET-202/U2Sm− RNA and the latter RNA was a competitor for nonspecific binding to nuclear La protein. Intracellular distribution of the chimeric RNA was assayed as in Fig. 5A.
Passive diffusion of ET-202 RNA from the nucleus was ruled out by the efficient export of a >50 kDa dimeric version of ET-202 (ET-202/di, >160 nucleotides long), both in the presence and absence of M protein (Fig. 1C). Furthermore, the appearance of ET-202 RNA in the cytoplasm was sensitive to low temperatures (0°C; Fig. 1A and data not shown), indicating that the process requires energy. Addition of an m7G cap to the normally uncapped RNAs was without effect on export (data not shown), indicating an export mechanism that is not dependent on interaction of the RNA with the cap binding complex, which is involved in export of m7G-capped U1 RNA (8). As expected (19), export of m7G-capped U1 snRNA was blocked by M protein (Fig. 1C). Coinjected U3 or U6 RNAs served as controls for the accuracy of nuclear injection and nucleocytoplasmic fractionation in this and all other experiments (ref. 2; data not shown).
Database searches failed to detect natural RNAs with sequences strongly homologous to those in the selected RNAs. Probing of the ET-202 RNA structure by enzymatic digestions and chemical modifications suggested the existence of extensive stem-loops and of several tertiary interactions stabilized by Mg2+ (A. E. Pasquinelli and C.G., data not shown); however, no common secondary structure among the selected RNAs was predicted by computer analysis. Several variants of ET-202 were generated by error-prone PCR and tested for export efficiency, but no specific nucleotide or sequence could be identified as being essential for export (Fig. 2). These results suggest that the efficient transport of ET-202 RNA is mediated by the structure rather than the sequence of the RNA.
Figure 2.
Intracellular distributions of sequence variants of ET-202 RNAs. Mutated templates of ET-202, generated by error-prone PCR, were transcribed to make a pool of variant ET-202 RNAs that were injected into nuclei of oocytes containing M protein. RNAs isolated from the cytoplasm (C) or nucleus (N) after 2 hr of incubation were reverse transcribed, cloned, and sequenced. Underlined nucleotides in the ET-202 sequence represent the ends of the primers used in the PCR amplification. ∗ indicates individual RNAs that were tested further for their abilities to be exported to the presence of M protein. Nucleotides are shown that differ from the original ET-202 RNA; an outlined letter represents inserted nucleotides and Δ represents a deleted nucleotide. The average numbers of changes in the RNAs were 2.4 and 5.5 per molecule in the C and N clones, respectively.
Factor-Mediated Export of the Selected RNAs.
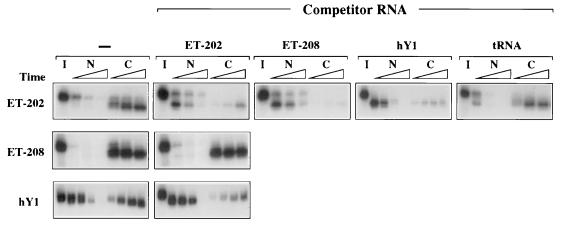
To determine if export of ET-202 RNA required factors used by other RNAs, we injected nuclei of control oocytes with mixtures containing labeled RNAs and unlabeled competitors. When present in high amounts, unlabeled ET-202 RNA competed for its own export, demonstrating that export of this RNA requires a saturable factor(s) (Fig. 3). Both ET-208 RNA and the small cytoplasmic hY1 RNA inhibited export of ET-202 RNA (Fig. 3 Top), indicating that these three RNAs interact with a common factor(s) and that the selected RNAs access the export pathway normally used by hY1 RNA; this is supported by the observation that ET-202 RNA can inhibit export of hY1 RNA (Bottom). The destabilization of ET-202 RNA upon blockage of its export by competitor RNA may result either from removal of stabilizing proteins or from an inherent instability of the RNA in the nucleus. Unlabeled ET-202 RNA did not block export of ET-208 RNA (Middle). The reason for the lack of reciprocity in the saturation of export by these two molecules is unclear. Perhaps ET-208 binds but does not require a factor needed for export of ET-202. We have not yet probed the other selected RNAs to determine the number of independent export pathways accessed by them.
Figure 3.
Common factors among exported RNAs. Competition for export of other RNAs by ET-202 RNA. One to 10 fmol of 32P-labeled ET-202 (Top), ET-208 RNA (Middle), or hY1 RNA (Bottom) were injected into nuclei of control oocytes in the absence (−) or presence of 0.5 pmol unlabeled competitor RNAs as indicated across the top, except 5.0 pmol of tRNA were used in the right-most panel; the preparation of labeled hY1 RNA also contained 0.5 pmol NL-15 RNA, to bind the nuclear La protein, thereby allowing efficient export of the hY1 RNA (5). Most RNAs were prepared from nuclear (N) and cytoplasmic (C) fractions 1, 2, and 4 hr after nuclear injection, but samples containing hY1 RNA (either labeled or as competitor) were isolated at 0.5, 1, 2, and 6 hr and samples with tRNA were isolated at 0.5, 2, and 4 hr. All RNAs were uncapped and were analyzed as in Fig. 1C.
The export pathways of both the selected ET-RNAs and tRNAs are resistant to inhibition by M protein and mAb414. However, neither ET-202 nor tRNA interfered with export of the other (Fig. 3 Top and data not shown), indicating that their export pathways do not use a common, limiting factor(s); also, the lectin wheat germ agglutinin inhibits export of tRNA (ref. 24; E.L. and J.E.D., unpublished work) but not of ET-202 RNA (data not shown). Thus, several export pathways exist that are insensitive to inhibition by M protein.
Efficient Export of Chimeric ET-RNAs in Normal Oocytes.
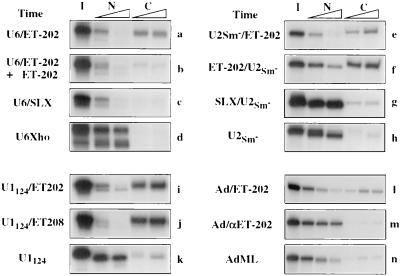
Because of the efficiency of export of ET-202 and ET-208 (Fig. 3), we asked if these sequences could act as NEEs for other RNAs that ordinarily are not exported, or only poorly so. We made several chimeric RNAs containing both a selected ET sequence and normal cellular RNA sequences. All of these chimeras were exported very efficiently in the absence of M protein (Fig. 4). For example, U6 RNA was retained in nuclei as expected (d), but most of the chimeric U6/ET-202 RNA appeared in the cytoplasm within 1 hr of injection into oocyte nuclei (a). Likewise, ApppG-capped versions of U2 snRNA, a truncated variant of U1 snRNA (U1124 RNA), and AdML pre-mRNA were exported inefficiently on their own (h, k, and n, respectively) but all of these RNAs were exported very rapidly when they were present in chimeric molecules containing ET-202 or ET-208 sequences (e, f, i, j, and l).
Figure 4.
Transposable NEEs in the selected RNAs. ET-202 RNA in the sense (a, b, e, f, i, and l) or antisense (m) orientation, SLX RNA (unselected RNAs from round 2; c and g), or ET-208 RNA (j) were fused to the 5′ or 3′ end of U6 RNA (a–c), U2Sm− RNA (e–g), U1124 RNA (i and j), or AdML pre-mRNA (l and m) as indicated. Export of these chimeric RNAs and the control RNAs U6Xho (d), U2Sm− (h), U1124 (k), and AdML pre-mRNA (n) was tested in control oocytes. Export of intron-containing AdML pre-mRNAs (l–n) was analyzed in the presence of 300 fmol of unlabeled, m7G-capped AdML pre-mRNA serving as competitor for splicing. RNAs containing U6 snRNA (a–d) were γpppG capped, whereas all other labeled RNAs (e–n) were ApppG capped. RNAs were isolated from nuclear (N) and cytoplasmic (C) fractions 1 and 3 hr (a–k) or 1, 3, and 5.5 hr (l–m) after nuclear injection and analyzed as in Fig. 1C.
The enhanced export of chimeric molecules probably uses the pathways of the selected sequences, since it can be competed by the homologous selected ET-RNA (b) and does not occur if the chimeric RNA contains either an antisense version of the ET-RNA (m) or sequences from an early round (round 2) of selection (c and g). In RNAs containing the U2 RNA sequences, export was efficient regardless of the location of the ET-RNA within the molecule (e and f). Thus, the selected RNA sequences confer on the poorly exported, nonselected RNAs the ability to use alternate, efficient export pathways.
Nuclear Retention of Chimeric ET-RNAs in Oocytes Containing M Protein.
Because the ET-RNA sequences act as NEEs for their own export in the presence of M protein, one might expect that they could also direct export of the chimeric RNAs under these conditions. Surprisingly, export of the chimeric RNAs was strongly inhibited in the presence of M protein (Fig. 5A Middle) or mAb414 which, as noted above, does not inhibit export of the ET-RNAs alone. Thus, the nonselected sequences contain elements that lead to nuclear retention in the presence of inhibitors like M protein or mAb414, and thereby override the export function of the attached ET-RNA sequences. This retention is not due to the presence of a specific NRE (2) in the nonselected RNAs, since it is even observed with ET-RNA chimeras that contain normally exported sequences, such as m7G-capped U1 or U2 snRNAs.
To test if the blockage of export is due to binding of the chimeric RNAs to saturable nuclear structures, we coinjected competitor RNAs that might release the chimeras from sites specific for the nonselected RNAs. Injection of 1.0 pmol of competitor U1 or U2 RNA did not cause export of an ET-202 RNA chimera containing U1 or U2 RNA sequences (Fig. 5B; data not shown) indicating that nuclear retention sites, if they exist (see Discussion), must have a very high capacity. Moreover, such sites must be nonspecific, in that an ET-RNA chimera containing a random RNA sequence from the pGEM4Z vector also was retained in the presence of M protein (not shown). The high capacity of retention raises the possibility that nuclear retention of the chimeric RNAs is due to lack of access to RNA export factors, perhaps as a result of improper folding of the nonselected RNAs in the presence of M protein or mAb414.
These experiments indicate that prior to export, RNAs must be mobilized, either by release from a nuclear structure or by folding into an alternative tertiary structure. We refer to these events as immobilization and release.
DISCUSSION
Transport of most RNAs between the nucleus and cytoplasm requires Ran-GTPase and associated activating, exchange, and binding factors (12, 13). To date, only the export of tRNAs and stress-related mRNAs has been reported to be exempt from this requirement. Because the Ran system is needed for protein import, export of these RNAs may not require shuttling transport factors (17). The data presented here make it likely that export of the ET-RNAs also does not require protein factors that shuttle between the nucleus and cytoplasm, since these RNAs were exported in the presence of M protein, a potent inhibitor of protein import (19).
Our results also reveal an additional step in RNA export, the transient immobilization of RNAs within the nucleus. Because immobilization prevents export, even RNAs that contain functional NEEs must be released before they can exit the nucleus. We propose that the immobilization and release of RNA is normally a transient but ongoing process, which is revealed only when release is blocked by, for example, M protein. RNAs containing NEEs that function in the presence of M protein were retained in the nucleus in the presence of this inhibitor, if the RNAs were chimeras that contained additional, nonselected, sequences. Thus, extra sequences, even those derived from normally exported molecules like U1 or U2 RNAs, made the chimeras susceptible to immobilization.
Because the sequences responsible for retention of the chimeric RNAs need not be specific NREs like those of U3, U6, or U8 RNAs (2, 3, 25), we conclude that the nuclear retention of the chimeras results from blockage of their release from immobilization. M protein inhibits both RNA export and protein import, so we do not know if its effect on release is direct or secondary to the inhibition of import of one or more proteins needed for that process. Thus, we are unable to say if the Ran system is directly involved in release.
ET-202 RNA and hY1 RNAs compete with each other for export under normal conditions, indicating that the two RNAs share at least some export factors that can be saturated by the other RNA (Fig. 2). However, the RNAs differ in that export of hY1 RNA is sensitive to inhibition by M protein and mAb414. Perhaps the two RNAs use the same export system, but certain sequences in hY1 RNA make it sensitive to retention, as is the case with the chimeric RNAs.
The ET-RNAs, which were selected under conditions where release was blocked, apparently were able to avoid immobilization on their own. It is unclear what features of RNAs lead to their immobilization or what makes the ET-RNAs resistant to immobilization; no secondary structure motifs seem to be shared by the various ET-RNAs, but chemical and enzymatic probing indicates that the RNAs are highly structured. As expected from their 5′ and 3′ ends (derived from U1 RNA), these RNAs do not fold into the cloverleaf structure of tRNAs, which also are exported in the presence of M protein (19). It is striking that almost all of the selected RNAs fall into one of three sequences, implying a high degree of selective pressure for specific sequences or structures. We note, however, that the size of the random region in the selected RNAs was rather small (20 nucleotides) so additional motifs may emerge if larger libraries are screened. Also, possible roles of the flanking regions in the transport of these RNAs may be revealed by learning if the same sequences are selected from the combinatorial library when the vector RNAs have different flanking sequences.
It was surprising that the ET-RNAs could be exported even in the presence of mAb414, since the antibody was not used in the selection of these RNAs. This result implies that ET-RNAs do not require functional XFXFG nucleoporin repeats for their export. However, the chimeric RNAs, which appear to use the ET-RNA pathway for translocation through the NPC (Fig. 4), are retained in the presence of mAb414 (Fig. 5A), as they are in the presence of M protein. It is possible that M protein and mAb414 inhibit export of immobilized RNAs by different mechanisms. For example, the release of an RNA may direct it into an export pathway that is sensitive to inhibition by the antibody, and RNAs that are not subject to immobilization and release could use an alternate pathway.
Even in the absence of M protein, extra sequences in chimeric RNAs retarded the export of the ET-RNA sequences (Fig. 5 and data not shown). This slowing of export may well reflect the time required for release of the RNAs, a process that the selected RNAs do not have to undergo. Transient immobilization may also account for the relatively slow export of injected pre-snRNAs and the wide range in the rates at which fully processed mRNAs are exported (E.L. and J.E.D., unpublished).
Immobilization itself could be caused either by direct association of the RNA with a nuclear structure, or by the folding of the RNA into a structure that prevents its association with export factors. Possible candidates for release factors include the putative RNA helicases that have been implicated in the release of spliced mRNAs from spliceosomes (26–28) or proteins such as the Rev protein of HIV-1 that facilitate export of incompletely spliced viral pre-mRNAs. One function of cis-acting elements such as the Rev response element of HIV-1 and the constitutive transport element of Mason–Pfizer Monkey Virus may be to recruit the postulated release factors to the immobilized RNAs (A. E. Pasquinelli, personal communication; refs. 9, 29, and 30).
Acknowledgments
We thank Amy Pasquinelli, Lu-Shiun Her, and Virgil Varvel for useful comments, Ariane Grandjean and Michelle Barr for excellent technical assistance, and George Q. Pennabble. This work was supported by National Institutes of Health Grant GM30220 and a fellowship (823A-042935) from the Swiss National Research Foundation.
ABBREVIATIONS
- NREs
nuclear retention elements
- NEEs
nuclear export elements
- snRNA
small nuclear RNA
- M
Matrix
- Ad
adenovirus
- AdML
adeno major late
References
- 1.Chang D D, Sharp P A. Cell. 1989;59:789–795. doi: 10.1016/0092-8674(89)90602-8. [DOI] [PubMed] [Google Scholar]
- 2.Terns M P, Grimm C, Lund E, Dahlberg J E. EMBO J. 1995;14:4860–4871. doi: 10.1002/j.1460-2075.1995.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boelens W C, Palacios I, Mattaj I W. RNA. 1995;1:273–283. [PMC free article] [PubMed] [Google Scholar]
- 4.Simons F H M, Rutjes S A, Van Venroij W J, Pruijn G J M. RNA. 1996;2:264–273. [PMC free article] [PubMed] [Google Scholar]
- 5.Grimm C, Lund E, Dahlberg J E. EMBO J. 1997;16:793–806. doi: 10.1093/emboj/16.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I M. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terns M P, Dahlberg J E, Lund E. Genes Dev. 1993;7:1898–1908. doi: 10.1101/gad.7.10.1898. [DOI] [PubMed] [Google Scholar]
- 8.Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj I W. Nature (London) 1995;376:709–712. doi: 10.1038/376709a0. [DOI] [PubMed] [Google Scholar]
- 9.Malim M H, Hauber J, Le S, Maizel J, Cullen B R. Nature (London) 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 10.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 11.Michael W M, Choi M, Dreyfuss G. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 12.Izaurralde E, Mattaj I W. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 13.Goerlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 14.Cheng Y, Dahlberg J E, Lund E. Science. 1995;267:1807–1810. doi: 10.1126/science.7534442. [DOI] [PubMed] [Google Scholar]
- 15.Saavedra C, Tung K S, Amberg D C, Hopper A K, Cole C N. Genes Dev. 1996;10:1608–1620. doi: 10.1101/gad.10.13.1608. [DOI] [PubMed] [Google Scholar]
- 16.Moore M S, Blobel G. Nature (London) 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 17.Dahlberg J E, Lund E. Semin Cell Dev Biol. 1997;8:65–70. doi: 10.1006/scdb.1996.0123. [DOI] [PubMed] [Google Scholar]
- 18.Richards S A, Carey K L, Macara I G. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- 19.Her L-S, Lund E, Dahlberg J E. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 20.Simons F H M, Pruijn G J M, Van Venroij W J. J Cell Biol. 1994;125:981–988. doi: 10.1083/jcb.125.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powers M A, Forbes D J, Dahlberg J E, Lund E. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Konarska M M, Sharp P A. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 23.Cadwell R C, Joyce G F. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 24.Neuman deVegvar H E, Dahlberg J E. Mol Cell Biol. 1990;10:3365–3375. doi: 10.1128/mcb.10.7.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terns M P, Dahlberg J E. Science. 1994;264:959–961. doi: 10.1126/science.8178154. [DOI] [PubMed] [Google Scholar]
- 26.Company M, Arenas J, Abelson J. Nature (London) 1991;349:487–493. doi: 10.1038/349487a0. [DOI] [PubMed] [Google Scholar]
- 27.Ohno M, Shimura Y. Genes Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- 28.Liang S, Hitomi M, Hu Y, Liu Y, Tartakoff A M. EMBO J. 1996;16:5139–5146. doi: 10.1128/mcb.16.9.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ernst R K, Bray M, Rekosh D, Hammarskjold M-L. Mol Cell Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang H, Gaietta G M, Fischer W H, Ellisman M H, Wong-Staal F. Science. 1997;276:1412–1415. doi: 10.1126/science.276.5317.1412. [DOI] [PubMed] [Google Scholar]