Abstract
Glutamic acid 286 (E286; Escherichia coli cytochrome bo3 numbering) in subunit I of the respiratory heme-copper oxidases is highly conserved and has been suggested to be involved in proton translocation. We report a technique of enzyme reconstitution that yields essentially unidirectionally oriented cytochrome bo3 vesicles in which proton translocation can be measured. Such experiments are not feasible in the E286Q mutant due to strong inhibition of respiration, but this is not the case for the mutants E286D and E286C. The reconstituted E286D mutant enzyme readily translocates protons whereas E286C does not. Loss of proton translocation in the D135N mutant, but not in D135E or D407N, also is verified using proteoliposomes. Stopped-flow experiments show that the peroxy intermediate accumulates in the reaction of the E286Q and E286C mutant enzymes with O2. We conclude that an acidic function of the 286 locus is essential for the mechanism of proton translocation.
The heme-copper oxidases are responsible for cell respiration in animals, plants, and aerobic bacteria. They contribute to primary energy conservation by catalyzing proton translocation that is linked to the redox chemistry of reduction of O2 (1). The mechanism of proton translocation is presently under intensive study. Site-directed mutagenesis combined with measurements of proton translocation under multiturnover conditions is a powerful approach to determining a functional role of individual amino acids. However, this method also is beset with problems. To measure proton translocation reliably, the mutant enzyme must have sufficient catalytic activity (2). The method becomes unreliable when the natural back-leak of protons across the membrane is significantly faster than enzyme turnover. Moreover, if proton translocation remains tightly coupled to the redox chemistry, the coupling efficiency may be unaffected by a mutation even if it has primarily inhibited a proton translocation event.
To date three residues (N124, D135, and N142)‡ have been found in the heme-copper oxidases where nonconservative mutations inhibit proton translocation relative to electron transfer in multiturnover experiments (2–4). All of these reside in the transmembrane helix II-III loop domain of subunit I, near the proton input side of the membrane, as recently confirmed by the crystal structures (5, 6). Accordingly, this domain was suggested to form an input site for a proton translocation pathway (2–4). X-ray work supports this proposal, showing that this domain may be part of a proton-conducting structure (the D channel), which ends near a cavity almost halfway across the membrane. Iwata et al. (5) proposed that the end of this channel may connect to a well conserved glutamic acid residue (E286 in the Escherichia coli enzyme) by hydrogen bonding through solvent molecules, and that the glutamic acid may be a key residue in the proton translocation mechanism. In agreement with this, recent statistical-mechanical calculations on the crystal structure of the bovine enzyme based on the “potential of mean force” formalism (7, 8) predict that three bound water molecules link the end of the D channel with E286 by hydrogen bonding (9).
In this work we have set out to test the role of E286 in proton translocation. Early reports on the mutant E286Q indicated that it was fully active (2, 10, 11). However, this was due to reversion back to wild-type enzyme, as revealed by both sequencing and a more recent construction of a new E286Q mutant (12).
Proton translocation by the cytochrome bo3 enzyme mainly has been measured in cells rather than in proteoliposomes, because the vesicle-reconstituted quinol oxidase would have to be almost completely unidirectionally oriented to allow measurements of proton translocation due to the membrane permeability of ubihydroquinone. This was not previously achieved with cytochrome bo3 using conventional dialysis techniques of reconstitution (13). Here we report a method of reconstituting cytochrome bo3 into proteoliposomes that fulfills this requirement. We show that the reconstituted E286D enzyme translocates protons with the same efficiency as the wild type, whereas proton pumping is abolished in the E286C mutant. In E286C and E286Q the “peroxy” intermediate accumulates when the reduced mutant enzyme reacts with dioxygen. The implications of these findings for the mechanism of proton translocation are discussed.
MATERIALS AND METHODS
Enzyme Isolation and Mutagenesis.
Bacterial growth conditions and purification of cytochrome bo3 enzymes were as described previously (14, 15). Site-directed mutagenesis was performed according to published methods (16, 17) and confirmed by DNA sequencing. This also was done for the double-stranded expressing plasmid from the host strain. E286D is in the pJT40 plasmid and is expressed in the host strain GO105 (cyo Δcyd recA; ref. 18). E286C is in the plasmid pHTC8, a derivative of pL1 (18), from which all cysteines of subunits I, III, and IV were mutated (12). These cysteines are not conserved and were mutated to residues commonly found in the respective locus among the heme-copper oxidases. The phenotype of pHTC8 expressed in GO105 cells exhibits turnover and proton translocating activities similar to wild type (A.P., unpublished work). The E286C mutant plasmid was transformed into the host strain GL101 (cyo sdh recA; ref. 16), because it does not complement aerobic growth of E. coli well enough. All used plasmids contained a construct for a “histidine tag” to facilitate isolation of the enzyme in a single step by a nickel affinity column (14, 19).
Enzyme Reconstitution into Proteoliposomes.
This procedure of reconstituting cytochrome bo3 is based on the methodology developed by Rigaud et al. (20). Asolectin (8 mg/ml) was solubilized in 200 mM Hepes⋅KOH, pH 7.4/55 mM octyl glucoside by brief ultrasonic treatment. Purified cytochrome bo3 (40–160 μM) was added to a final concentration of 0.18 μM, and the mixture was stirred for 15 min. SM2 Bio-Beads (80 mg wet beads/ml) were added directly to the mixture, which was stirred. After a 30-min incubation a second portion of 80 mg/ml Bio-Beads was added. A third portion (160 mg/ml) was added after 1 hr and an equivalent amount after 2 hr. The mixture was stirred for 2 more hr to complete the removal of detergent. All incubations were performed at room temperature. The liquid phase with the proteoliposomes was carefully removed by suction with a thin pipette tip that did not pass the Bio-Beads. The proteoliposomes were diluted 10 times with 200 mM KCl and sedimented at 144,000 × g (Ti50 rotor) for 90 min. The pellet was carefully rinsed with 200 mM KCl to remove as much buffer as possible, suspended in the same medium, and used within 24 hr. No loss in oxidase activity or respiratory control was found after overnight storage on ice. Freezing caused a drastic decrease in respiratory control, but no loss of enzymatic activity.
Respiration, Proton Translocation, and Kinetics.
Respiration of reconstituted enzyme was measured in 200 mM KCl/10 mM Hepes⋅KOH/2 mM DTT/0.1 mM ubiquinone-1. Respiratory control was determined by the increase in respiratory rate upon addition of 1 μM valinomycin plus 1 μM carbonylcyanide p-chloromethoxy phenylhydrazone. Proton translocation was determined by the O2 pulse method (2, 3). Stopped-flow experiments were performed as described by Puustinen et al. (15).
RESULTS AND DISCUSSION
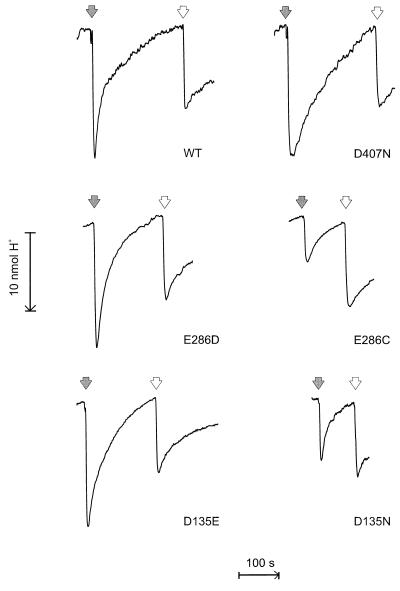
Wild-type cytochrome bo3 exhibits respiratory control when reconstituted into proteoliposomes by the present methodology (see Materials and Methods). The rate of O2 consumption with ubiquinone-1 plus DTT is accelerated 3- to 5-fold upon uncoupling with valinomycin plus carbonylcyanide p-chloromethoxy phenylhydrazone (Table 1). This suggests that the enzyme is, in the main, incorporated unidirectionally into the liposomal membrane, and that the proteoliposomes are well sealed with a reasonably low proton conductance. These conclusions are supported by the results from proton translocation measurements using the O2 pulse technique (Fig. 1; Table 1). The H+/e− ratio of proton release was found to be 1.75 for the reconstituted wild-type enzyme, which is in good agreement with the previous finding that this ratio is near 2.0 in E. coli cells or sphaeroplasts (2, 3, 21). In the presence of carbonylcyanide p-chloromethoxy phenylhydrazone the H+/e− ratio fell to zero with no net proton ejection or consumption, in agreement with the fact that the overall reaction involves reduction of O2 by ubihydroquinone. It should be recalled that with the quinol oxidases the oxidation of ubihydroquinone by the enzyme results in scalar release of 1 H+/e− to the outside of the membrane, whereas the second released proton is due to proton translocation across the membrane (2, 3, 21). Although the H+/e− ratio of proton release is twice that found for the cytochrome c oxidases, both types of enzyme translocate two equivalents of electrical charge across the membrane per electron transferred (3).
Table 1.
Proton translocation by liposome-reconstituted mutants of cytochrome bo3
| Enzyme | Activity, e−/s | Respiratory control | H+/e− ± SD (no. expts) |
|---|---|---|---|
| Wild type | 850 | 3–5 | 1.75 ± 0.08 (11) |
| Glu286gln | ≈5 | ND | ND |
| Glu286asp | 680 | 3.0 | 1.69 ± 0.06 (3) |
| Glu286cys | 125 | 3.5 | 0.74 ± 0.10 (7) |
| Asp135glu | 1,200 | 3.0–4.2 | 1.73 ± 0.09 (6) |
| Asp135asn | 350 | 3.1 | 0.90 ± 0.07 (7) |
| Asp407asn | 690 | 4.7 | 1.69 ± 0.06 (6) |
Catalytic activities are mean values for the isolated, unreconstituted enzymes. Respiratory control denotes the ratio of O2 consumption rates in proteoliposomes in the presence and absence of carbonylcyanide p-chloromethoxy phenylhydrazone and valinomycin (see Materials and Methods). ND, not determined.
Figure 1.
Proton translocation with reconstituted cytochrome bo3. Proteoliposomes containing wild-type or mutant cytochrome bo3 (0.14–0.18 μM) were incubated anaerobically in a medium containing 200 mM KCl, 2 mM DTT, 0.1 mM ubiquinone-1, and 1 μM valinomycin, and in the pH range 6.4–6.8. The filled arrows indicate the addition of O2 in the form of 10 μl of air-saturated pure water at 25o (2.58 nmol O2, or 10.32 nmol electron acceptor). Empty arrows show H+ calibrations by addition of 10 μl anaerobic 1 mM HCl (10 nmol H+). In the presence of 1 μM carbonylcyanide p-chloromethoxy phenylhydrazone no proton ejection or consumption upon addition of O2 was experienced.
Table I and Fig. 1 report proton translocation experiments in proteoliposomes for several mutants of cytochrome bo3. The respiratory control ratios were in the 3–5 range in all cases, as it was with wild-type enzyme. D407 is a well conserved residue that lies on the periplasmic (proton output) side of heme a3, only ca. 3 Å from one of the heme’s propionate carboxyl oxygens (5, 6). Mutation of this residue to asparagine slows down turnover, but the efficiency of proton translocation is unaffected. A similar result recently was reported for the corresponding mutation in cytochrome aa3 of Rhodobacter sphaeroides, and D407 was concluded not to be a critical residue for proton pumping (22). This does not exclude the possibility that D407 still may be involved in optimal proton transfer, but that alternative proton pathways are possible with slightly less favorable kinetics.
E286 is the only highly conserved acidic residue deep within the enzyme’s membrane domain and has been proposed to be involved directly in the mechanism of proton translocation (5). Experimental assessment of this possibility has been hampered by the fact that mutations at this locus have led to almost complete inactivation of the enzyme (cf. refs. 10 and 23). For example, we find that the turnover of the E286Q mutant bo3 enzyme is only ca. 5 s−1, or <1% of the wild-type value. However, the mutants E286D and E286C are much more active (Table 1), and hence amenable to testing by the O2 pulse method. With aspartic acid in the 286 locus the proton-pumping efficiency was found to be normal, but proton translocation is absent with cysteine in this site (Fig. 1; Table 1).
Fourier transform infrared studies have shown that little perturbation of the binuclear site in the E286D and E286C mutant enzymes occurs, except for subtle changes in the C≡O stretching mode of CuB-CO (12), and optical spectroscopy does not reveal any significant effects on the hemes (not shown). Hence the inhibition of proton translocation in the E286C mutant indicates that E286 is a key residue in the mechanism of proton translocation. This is a similar finding to that previously reported at the cellular level for three mutations in the loop domain between transmembrane helices II and III of subunit I (2). The D135N mutant is one of these, and we show here that it also fails to translocate protons in proteoliposomes (Fig. 1; Table 1). As for the E286 locus, the D135 site also can accommodate a side chain of different length (D135E, Table 1).
It also may be noted that the reconstituted D135N cytochrome bo3 exhibits respiratory control, which was reversed in the corresponding mutant of cytochrome aa3 from R. sphaeroides (4). The latter may have been caused by the very low catalytic activity in combination with inhibition of proton transfer.
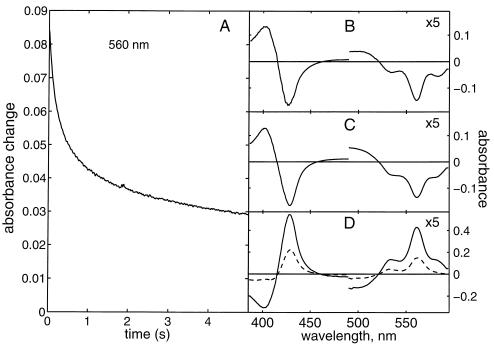
Svensson-Ek et al. (23) showed that the E286A mutation blocks oxygen reduction and net proton uptake in the bo3 enzyme. Ädelroth et al. (24) recently reported similar data for the E286Q mutant of cytochrome aa3 from R. sphaeroides. Fig. 2 shows a stopped-flow experiment with the E286Q mutant of the bo3 enzyme isolated in dodecyl maltoside. After reduction, this preparation contains one bound ubiquinol molecule, so that any low-spin heme that may become oxidized during mixing will be quickly re-reduced (15). In the wild-type enzyme, the oxidation of the low-spin heme has a slower component with a time constant of ca. 3–4 ms (not shown; see ref. 15). In contrast, oxidation of the low-spin heme is a biphasic process with time constants of 0.17 and 1.6 s (Fig. 2A) in the E286Q mutant, i.e., two orders of magnitude slower than in the wild-type enzyme. Spectral analysis of this event (Fig. 2 B-D) shows a disappearing absorption peak at 560 nm due to oxidation of the low-spin heme, but also the disappearance of another absorption feature near 582 with similar kinetics. We previously have assigned the 582-nm band in the bo3 enzyme to the P (peroxy) species (14). Very similar results were obtained with the E286C mutant, except that the rate of oxidation of the low-spin heme was faster than in E286Q by a factor of 2–3 (not shown).
Figure 2.
The reaction of the reduced E286Q mutant enzyme with O2. (A) Stopped-flow experiment monitoring a biphasic absorption decrease at 560 nm (τ = 0.17 and 1.6 s, respectively). (B and C) Difference spectra of these two phases. (D) Difference absorption spectra vs. oxidized enzyme after mixing (i.e., before the events shown in A; solid curve), and after the biphasic event shown in A (dashed curve), respectively. The α-band in B-D is expanded 5-fold.
Accumulation of the P intermediate indicates that its conversion to the ferryl state is strongly inhibited in both of these mutants. This is of interest because the conversion of the P state to F is known to be coupled to proton translocation (25, 26). Whether the further conversion of F to the oxidized enzyme also is inhibited in these mutants is difficult to judge from the present experiments, in part because the optical spectrum of F resembles that of the reduced low-spin heme. The biphasicity of low-spin heme oxidation (Fig. 2A), and the slight difference in shape of the spectra of the two phases (Fig. 2 B and C), suggest that the decay of F also may be slowed down. However, because no clear accumulation of F occurs and the decay of P is seen in both phases, the decay of F cannot be significantly more inhibited than the P→F transition. Because the depressed H+/e− ratio in the E286C mutant proteoliposomes implicates this glutamic acid residue in proton transfer, it is reasonable to assume that inhibition of proton transfer is the cause of inhibition of the catalytic oxygen chemistry as well.
It is of interest that the turnover in D135N is inhibited by only ca. 60% although proton translocation is abolished (ref. 2; Table 1). It should be recalled, however, that it is the reductive phase of the catalytic cycle that normally limits turnover in the heme-copper oxidases. Proton transfer coupled to the oxidative phase must be much faster. Hence, the fact that turnover is inhibited by 60% in the D135N mutant means that proton transfer through the D channel has, in fact, been severely inhibited. The proton pump mechanism presumably requires proton input at a fast rate because proton translocation may compete kinetically with the highly exergonic proton consumption of the oxygen reduction chemistry. If proton delivery is significantly retarded the oxygen reduction chemistry will dominate, and the enzyme will turn over without proton translocation. In the histidine cycle model (9, 27), for example, a proton taken up via the D channel will protonate a histidine CuB ligand through a pathway that involves E286. This causes the histidine to dissociate from CuB, which, in turn, opens up protonic connectivity from the D channel, via E286, to the iron-bound oxygen ligand. If proton replenishment via this pathway is slowed down sufficiently, then the reduced oxygen ligand will instead capture the proton from the histidine, whereby proton translocation is abolished or short-circuited. The turnover in D135N thus may reflect the rate of this leak. In cytochrome bo3 the lowered efficiency of proton translocation above pH 8 also suggests a relatively high intrinsic leak of the proton pump mechanism in this enzyme (28). This leak may be much smaller in the aa3-type enzyme from Rhodobacter as judged from the very low turnover of the corresponding mutant (4). It must be emphasized, however, that in wild-type cytochrome bo3 this leak of some 300–400 s−1 competes with proton transfer reactions that must have rates of at least 5,000 s−1 to match the kinetics of dioxygen reduction (15).
Acknowledgments
We thank Hoffman-LaRoche for the kind gift of ubiquinone-1. This work was supported by grants from the University of Helsinki, the Academy of Finland, the Sigrid Jusèlius Foundation, and Biocentrum Helsinki.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
The amino acid numbering throughout is for the E. coli cytochrome bo3.
References
- 1.Babcock G T, Wikström M. Nature (London) 1992;356:301–309. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- 2.Thomas J W, Puustinen A, Alben J O, Gennis R B, Wikström M. Biochemistry. 1993;32:10923–10928. doi: 10.1021/bi00091a048. [DOI] [PubMed] [Google Scholar]
- 3.Garcìa-Horsman J A, Puustinen A, Gennis R B, Wikström M. Biochemistry. 1995;34:4428–4422. doi: 10.1021/bi00013a035. [DOI] [PubMed] [Google Scholar]
- 4.Fetter J R, Qian J, Shapleigh J, Thomas J W, Garcìa-Horsman A, Schmidt E, Hosler J, Babcock G T, Gennis R B, Ferguson-Miller S. Proc Natl Acad Sci USA. 1995;92:1604–1608. doi: 10.1073/pnas.92.5.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwata S, Ostermeier C, Ludwig B, Michel H. Nature (London) 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 6.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Tamaguchi H, Shinzawa-Itoh K, Nakashina R, Yaono R, Yoshikawa S. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 7.Hummer G, García A E, Soumpasis D M. Biophys J. 1995;68:1639–1652. doi: 10.1016/S0006-3495(95)80381-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hummer G, García A E, Soumpasis D M. Faraday Discuss Chem Soc. 1997;103:175–189. doi: 10.1039/fd9960300175. [DOI] [PubMed] [Google Scholar]
- 9.Wikström M, Morgan J E, Hummer G, Woodruff W H, Verkhovsky M I. In: Frontiers of Cellular Bioenergetics. Papa S, Guerrieri F, Tager J M, editors. New York: Plenum; 1997. , in press. [Google Scholar]
- 10.Hosler J P, Ferguson-Miller S, Calhoun M W, Thomas J W, Hill J, Lemieux L, Ma J, Georgiou C, Fetter J, Shapleigh J, Tecklenburg M M J, Babcock G T, Gennis R B. J Bioenerg Biomembr. 1993;25:121–136. doi: 10.1007/BF00762854. [DOI] [PubMed] [Google Scholar]
- 11.Puustinen A, Morgan J E, Verkhovsky M, Thomas J W, Gennis R B, Wikström M. Biochemistry. 1992;31:10363–10369. doi: 10.1021/bi00157a026. [DOI] [PubMed] [Google Scholar]
- 12.Puustinen, A., Bailey, J. A., Dyer, R. B., Mecklenburg, S. L., Wikström, M. & Woodruff, W. H. (1997) Biochemistry, in press. [DOI] [PubMed]
- 13.Puustinen A, Finel M, Haltia T, Gennis R B, Wikström M. Biochemistry. 1991;30:3936–3942. doi: 10.1021/bi00230a019. [DOI] [PubMed] [Google Scholar]
- 14.Morgan J E, Verkhovsky M I, Puustinen A, Wikström M. Biochemistry. 1995;34:15633–15637. doi: 10.1021/bi00048a005. [DOI] [PubMed] [Google Scholar]
- 15.Puustinen A, Verkhovsky M I, Morgan J E, Belevich N P, Wikström M. Proc Natl Acad Sci USA. 1996;93:1545–1548. doi: 10.1073/pnas.93.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemieux L J, Calhoun M W, Thomas J W, Ingledew W J, Gennis R B. J Biol Chem. 1992;267:2105–2113. [PubMed] [Google Scholar]
- 17.Thomas J W, Calhoun M W, Alben J O, Gennis R B. Biochemistry. 1993;32:11173–11180. doi: 10.1021/bi00092a029. [DOI] [PubMed] [Google Scholar]
- 18.Calhoun M W, Lemieux L J, Thomas J W, Hill J J, Goswitz V C, Alben J O, Gennis R B. Biochemistry. 1993;32:13254–13261. doi: 10.1021/bi00211a038. [DOI] [PubMed] [Google Scholar]
- 19.Rumbley, J. N., Furlong Nickels, E. & Gennis, R. B. (1997) Biochim. Biophys. Acta, in press. [DOI] [PubMed]
- 20.Rigaud J-L, Pitard B, Levy D. Biochim Biophys Acta. 1995;1232:223–246. doi: 10.1016/0005-2728(95)00091-v. [DOI] [PubMed] [Google Scholar]
- 21.Puustinen A, Finel M, Virkki M, Wikström M. FEBS Lett. 1989;249:163–169. doi: 10.1016/0014-5793(89)80616-7. [DOI] [PubMed] [Google Scholar]
- 22.Qian J, Shi W, Pressler M, Hoganson C, Mills D, Babcock G T, Ferguson-Miller S. Biochemistry. 1997;36:2539–2543. doi: 10.1021/bi962721+. [DOI] [PubMed] [Google Scholar]
- 23.Svensson-Ek M, Thomas J W, Gennis R B, Nilsson T, Brzezinski P. Biochemistry. 1996;35:13673–13680. doi: 10.1021/bi961466q. [DOI] [PubMed] [Google Scholar]
- 24.Ädelroth, P., Svensson-Ek, M., Mitchell, D. M., Gennis, R. B. & Brzezinski, P. (1997) Biochemistry, in press. [DOI] [PubMed]
- 25.Verkhovsky M I, Morgan J E, Verkhovskaya M L, Wikström M. Biochim Biophys Acta. 1996;1318:6–10. doi: 10.1016/0005-2728(95)00142-5. [DOI] [PubMed] [Google Scholar]
- 26.Wikström M. Nature (London) 1989;338:776–778. doi: 10.1038/338776a0. [DOI] [PubMed] [Google Scholar]
- 27.Morgan J E, Verkhovsky M I, Wikström M. J Bioenerg Biomembr. 1994;26:599–608. doi: 10.1007/BF00831534. [DOI] [PubMed] [Google Scholar]
- 28.Verkhovskaya M, Verkhovsky M, Wikström M. J Biol Chem. 1992;267:14559–14562. [PubMed] [Google Scholar]