Abstract
The gene encoding the mouse vitamin D receptor has been cloned. A new exon 1 has been found that changes the numbering established for the human VDR gene. Exons 2 and 3 in the human VDR gene (coding for the zinc fingers 1 and 2, respectively) are named exons 3 and 4 in the mouse vitamin D receptor. The 1.5-kb 5′-flanking region of the new exon 1 was analyzed and revealed the presence of putative cis-acting elements. Despite the absence of a TATA box, this 5′-flanking region contains several characteristics of a GC-rich promoter including four Sp1 sites present in tandem and two CCAAT boxes. Interestingly, the Sp1 site that is the most proximal to the new exon 1 overlaps a perfect site for Krox-20/24. Krox-20 is a transcription factor involved in brain development, and also in bone remodeling. In luciferase reporter gene expression assays, we showed that sequences from this 5′-flanking region elicit high transactivation activity. Furthermore, in the NIH 3T3 cell line, a 3- to 5-fold increase in response to forskolin treatment (an activator of adenylate cyclase and in turn of protein kinase A pathway) was observed.
Keywords: 1,25-dihydroxyvitamin D; gene structure; calcium metabolism; steroid receptor
The biological actions of the 1,25-dihydroxyvitamin D3 [1,25-(OH)2D3], the active form of the vitamin D, are mediated through its receptor, the vitamin D receptor (VDR) (1). The VDR belongs to the steroid/thyroid hormone receptor superfamily. Members of this family possess a highly conserved N-terminal DNA binding domain containing two zinc finger structures and a less conserved C-terminal domain containing the ligand binding domain. These receptors are ligand-inducible nuclear transcription factors that recognize cis-acting sequences named hormone responsive elements, located in the promoter of target genes. The DNA consensus sequence bound by the VDR consists of two hexameric half sites spaced by three nucleotides. The VDR binds cooperatively to these vitamin D response elements (VDREs) as a heterodimer with another member of the family, the retinoid X receptor (2–4). VDREs have been identified in several genes regulated by 1,25-(OH)2D3 (for review see ref. 5) involved in calcium and phosphorus homeostasis (e.g., calbindin 28 k and 9 k, parathyroid hormone) or bone metabolism (e.g., osteocalcin and osteopontin).
An unresolved question regarding VDR-mediated responses that is basic to our understanding of a number of metabolic bone diseases is the control of receptor levels in target cells (1, 6). Several regulators of VDR expression have already been identified. However, the fact that the VDR promoter is so far unknown and that the level of VDR mRNA is low in most target tissues have hampered studies on the transcriptional regulation of the VDR gene. Levels of VDR protein have been first determined in binding studies using tritiated 1,25-(OH)2D3. Highly purified VDR from pigs or chicken has been used to generate monoclonal antibodies (7, 8), leading to more precise measurement of VDR levels. Antibodies directed against the VDR DNA binding domain were used to clone different VDR cDNAs. Effectively, the first clone isolated was a partial cDNA sequence coding for the DNA binding domain of the chicken receptor (9). Then, larger VDR cDNA sequences were obtained from rat and human cDNA libraries (10–12), leading to the determination of the primary structure of the full VDR protein (11, 12). Other VDR cDNAs from different species have been cloned recently, including quail (13), chicken (14), mouse (15), and Xenopus (16).
Using these different available tools, the most extensive study done concerned the autoregulation by the ligand itself, 1,25-(OH)2D3 (9, 17). It is now well established that 1,25-(OH)2D3 increases the VDR content in vivo or in cell culture by stabilizing the receptor (18, 19). What is less evident is the role of 1,25-(OH)2D3 in the transcriptional regulation of the VDR gene. If an increase of the level of VDR mRNA content has been shown in some cell lines treated by 1,25-(OH)2D3 (i.e., osteosarcoma Ros 17/2.8 cells), it doesn’t reflect the results found in vivo, where the level of VDR mRNA barely changes after 1,25-(OH)2D3 treatment (18). Other regulators have been shown to increase VDR protein content in vivo, i.e., the glucocorticoids that strongly increase VDR mRNA levels in the intestine of young rats at weaning (20). Calcium plays an important role in the regulation of VDR levels in kidney and parathyroid glands because under conditions of severe hypocalcemia, little or no VDR is present in these tissues (21, 22). In cell culture, various enhancers of VDR protein and mRNA content have been described including estrogen (23), all trans-retinoic acid (24), parathyroid hormone (25), or pharmacological activators of the protein kinase A pathway such as forskolin or cAMP analogs (26, 27). The most dramatic effect on VDR mRNA levels, a 30-fold increase, has been reported after treatment of mouse NIH 3T3 fibroblasts with forskolin (27).
To improve our knowledge of the transcriptional regulation of the VDR expression, the next step would consist of the characterization of responsive elements found in the promoter region. The human (h)VDR gene has been partially characterized and nine exons have been described (28). However, the promoter has not yet been found. Its isolation will permit the study of the transcriptional regulation of the VDR gene. This paper reports the cloning and characterization of the mouse (m)VDR promoter.
MATERIALS AND METHODS
Screening of the P1 Library.
PCR screening of a P1 mouse 129 genomic library was performed by Genome Systems (St. Louis) using the following PCR conditions: forward and reverse primers were designed in the mVDR cDNA sequence (15) to amplify a region corresponding to the first zinc finger of the protein: forward primer, 5′-CAGCCAGCACCTCCCTGCCTG-3′; reverse primer, 5′-AGAAACCCTTGCAGCCTTCAC-3′.
The PCR conditions were as described: the cycles were performed on a thermocycler (Perkin–Elmer model N 801-0150). An initial denaturation of 6 min at 94°C was followed by 40 cycles of 1 min of denaturation at 94°C, 30 sec of annealing at 50°C, and 30 sec of extension at 72°C using Taq polymerase.
The PCR amplification generated a 130 bp product when either reverse transcription product of mouse kidney RNA or mouse genomic DNA from embryonic stem cells (strain 129) were used as template. The P1 library is constituted by linear phage DNA (30 kb) containing genomic inserts in the size range of 75–100 kb. Two positive P1 clones were received in a bacterial host.
P1 Plasmid Purification.
P1 plasmid purification from P1 clones was performed according to the protocol of Genome Systems. Briefly, plasmid extraction was done by alkaline lysis followed by potassium acetate precipitation. The supernatant was then treated by RNase A and further purified by phenol-chloroform extraction and multiple ethanol precipitations. The plasmid was resuspended in water and quantitated on an agarose gel.
Subcloning of mVDR Gene Fragments.
P1 plasmids were cut with BglII restriction enzyme and ligated into the BamHI site of the pBluescript II KS+ vector (Stratagene). Transformant Escherichia coli clones (strain DH5α) were screened using a mVDR probe prepared by reverse transcription–PCR and labeled by random priming. Positive clones were analyzed for mVDR exon/intron content and partially sequenced with an automated sequencer (Pharmacia Biotech). Computer analysis of the sequence was made with the Genetics Computer Group Sequence Analysis Software Package (Madison, WI) using the findpatterns program linked to the transcription factor-listed binding site “tfsites.dat” file.
Cell Culture.
Osteosarcoma Ros 17/2.8 were grown in F-12 Ham’s medium supplemented with 10% fetal calf serum. NIH 3T3 mouse fibroblasts were grown in DMEM with 10% fetal calf serum. When cell cultures reached 50–60% of confluence they were used for transfection experiments.
Functional Analysis of the mVDR Promoter.
Fragments (1.5 and 0.5 kb) of the 5′-flanking region of the new exon 1 were inserted into a pGL2basic vector (Promega) in front of the luciferase gene reporter and transfected into NIH 3T3 or ROS 17/2.8 cells using lipofectin (GIBCO/BRL). An unmodified pGL2basic vector was used as control of no promoter activity in these experiments. Increased VDR expression was induced by treatment of a cell culture with 10 μM of forskolin (Sigma) for 24 hr. Luciferase activity was assayed in each sample and standardized to protein content measured by the Bio-Rad protein assay (Bio-Rad). All experiments have been performed in triplicate.
RESULTS
The 5′-Noncoding Region of the Mammalian VDR cDNAs.
It is well known that the amino acid sequence of the VDR is highly conserved among species. Furthermore, comparison of 5′-noncoding regions of mammalian VDR cDNAs revealed about 90% identity between rat and mouse and more than 80% between either rat or mouse, and human (Fig. 1). The size of the 5′-noncoding regions are 103, 94, and 115 bp, respectively, for the mouse, rat, and human (Fig. 1). Previous work in our laboratory revealed that nucleotides 1–15 from the rat VDR (rVDR) cDNA were not present in a genomic clone containing nucleotides 16–92 corresponding to the human exon 1 (T. K. Ross and H.F.D., unpublished results). Therefore, nucleotides 1–15 likely represent a portion of a new exon in the rVDR gene. In the mVDR cDNA, the presence of 12 nucleotides upstream of the untranslated 5′ end of the rVDR cDNA strengthens the belief in the existence of a new exon containing at least 27 bp in the mVDR gene.
Figure 1.
Comparison of the 5′-noncoding region of mammalian VDR cDNAs. Sequences of hVDR, mVDR, and rVDR cDNAs have been aligned in the 5′-end noncoding region. (|) represents identity between sequences and (−) gaps generated to fit the alignment. The ATG codons initiating translation are in boldface. (><) localizes the intron found in the rVDR gene.
Cloning of the mVDR Gene.
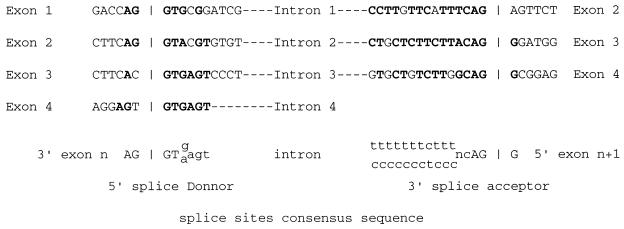
The mVDR gene was cloned by PCR screening of a P1 library performed by Genome Systems. This screening has been chosen because P1 clones contain genomic inserts of between 75 and 100 kb in size, increasing our chances of localizing this putative new exon and the promoter. PCR primers were designed in the region encoding the first zinc finger generating a 130 bp fragment suitable for the screening. Further upstream, exons are too small for that purpose. We received from Genome Systems two distinct clones from a P1 mouse 129 genomic library. Further investigation of these two clones by Southern blot analysis revealed that both clones hybridized to a mVDR oligonucleotide corresponding to the putative new exon 1. P1 clones were digested with BglII and genomic inserts subcloned into the BamHI site of the pBluescript II KS+ vector. One subclone containing the sequence corresponding to exon 1 of the hVDR gene was further analyzed. Similar to the rVDR gene, the 27 bp of the untranslated 5′ end of the mVDR cDNA were not present in this clone. Using an oligonucleotide designed in this putative new 5′-located exon, we were able to isolate another subclone. Partial sequencing of this clone revealed, as expected, the presence of the 5′ end of the untranslated mVDR cDNA, confirming the presence of a new exon–intron couple in the mVDR gene (Fig. 2). Furthermore, the DNA sequences at the intron–exon junctions, as shown in Fig. 3, follow the consensus sequences found in mammalian genes (35). The length of the new exon 1 is in fact 30 bp and not 27 bp as believed from the sequence of the rat genomic clone (T. K. Ross and H.F.D., unpublished results). This is due to the presence of three nucleotides (CAG) identical in the 3′ end of the new exon 1 and in the 3′ end of the intron 1. The exon/intron boundaries analysis led us to conclude that the CAG triplet is part of the first exon (Fig. 3). By isolation of other subclones from the initial P1 clones, we were able to identify the exons 3 and 4 corresponding to the two zinc fingers (Fig. 2) and to characterize their exon/intron borders (Fig. 3). In the hVDR and the mVDR, the two zinc fingers are linked by the identical amino acid sequence G-F-F-R-R-S-M-K-R-K-A-L-F (K, lysine; G, glycine; F, phenylalanine; R, arginine; S, serine; M, methionine; A, alanine; L, leucine; and F, phenylalanine). We found that the intron intervening exon 3 and 4 in the mouse VDR gene interrupted the codon of the first arginine. In the case of the hVDR gene, the intron is reported between the second arginine and the serine (28).
Figure 2.
Partial structure of the mVDR gene. The proposed promoter and four exons have been depicted here. Numbering has been changed to accommodate the new exon 1, and what was known as exon 1 in previous structures becomes exon 2. The zinc finger 1 and 2 of the mVDR are encoded in exons 3 and 4, respectively. The exact starting point of the mVDR RNA is not yet characterized. The 5′-flanking region of exon 1 shown here is 1.5 kb long and contains several sequences characteristic of a TATA-less promoter including four Sp1 sites, two CAAT boxes, and one site for Krox-20/24. All AP-1 sites described here differ in one base from the consensus sequence TGA G/C TCA. The AP-1 site at nucleotide 1123 is similar to the AP-1 site described in the human osteocalcin gene (29). The CREB/ETF site corresponds to the minimal binding site for factors of the CREB and ETF family (30, 31). The PU site binds PU.1, a transcription factor specific to gene expression in B cells and macrophages (32). NMP-2 is a osteoblast-specific factor that binds a site present in the osteocalcin promoter (33, 34).
Figure 3.
Exon–intron splice organization in the 5′-noncoding region of the mVDR gene. The characteristics of the mVDR exon–intron–exon junctions are represented. Nucleotides identical to the consensus sequence for intron exon boundaries are in boldface. The CAG sequences present in the 3′ end of both intron 1 and exon 1 are underlined.
Analysis of the Sequence Upstream of the New Exon 1.
We have sequenced about 1.5 kb upstream of the new exon 1 in an attempt to locate the mVDR promoter. A computer search was performed to systematically characterize putative cis elements for transcriptional regulation. Various protein binding motifs have been identified and are represented in Fig. 3. No TATA box was found in the 1.5 kb fragment. However, GC-rich regions are present and four Sp1 sites in tandem have been located in the immediate 5′-flanking region of the new exon 1. Two of these Sp1 sites, Sp1 #1 and Sp1 #2 (Fig. 4A) match perfectly the canonical sequence GGGGCGGGGC that is the optimal binding site for the nuclear transcription factor Sp1 (36). The Sp1 #3 differed by one base (Fig. 4A) from the consensus sequence G/T GGGCGG G/A G/A C/T, and such an Sp1 site has been reported to weaken the binding affinity for Sp1 factor (36). The Sp1 #4 also differed by one base from the consensus sequence, but such a site has been characterized to be functional in the adenovirus-2 EII late promoter (37). Sp1 site #1 is overlapping a GCGGGGGCGG sequence (Fig. 4 B and C), identical to the consensus sequence for a Krox-20/24 site (38). Similar overlapping sequences have been characterized in the promoter of the human retinoic acid receptor-α (39) and mouse Hox-1.4 genes (40). Two CCAAT boxes oriented in opposite directions have been located upstream of the Sp1 sites. Several AP-1 sites have been identified by a computer search, but none of them match perfectly the consensus sequence TGA G/C TCA. However, at nucleotide 1123 of the mVDR promoter region (Fig. 2), the sequence GGTGACACACC differs by one nucleotide from the AP-1 site GGTGACTCACC (29) found in the osteocalcin promoter. We also found a CTGTGGCTGG sequence that is similar to the CTGTGGTTGG motif from the osteocalcin promoter and containing a binding site for the osteoblast-specific transcription factor NMP-2 (nuclear matrix protein 2) related to the PEBP2/AML1 family (33, 34). No evident VDRE, cAMP-responsive element, glucocorticoid-responsive element, or estrogen-responsive element was identified by a computer search in the 1.5 kb of the 5′-flanking region analyzed. However, the minimum core ACGTGA for the binding of members of the CREB/ETF factor (30, 31) is present at position 935 of the sequence and might correspond to a cAMP-responsive element.
Figure 4.
Analysis of putative Sp1 and Krox-20/24 sites in the 5′-flanking region of the mVDR gene. Sp1 and Krox-20/24 binding sites were identified by computer analysis. (A) Sequence of the four Sp1 binding motifs are indicated in boldface. Nucleotides different from the consensus sequence are underlined. (B) Sequence of the Krox-20/24 binding motif and comparison to the consensus sequence. (C) Comparison between the overlapping sequence of Sp1 and Krox-20/24 binding site identified in mVDR gene and the sequence in the human retinoic acid receptor-α and mHox-1.4 promoters. Sp1 binding sites are underlined and Krox-20/24 binding sites are in boldface.
Functional Analysis of the 5′-Flanking Region.
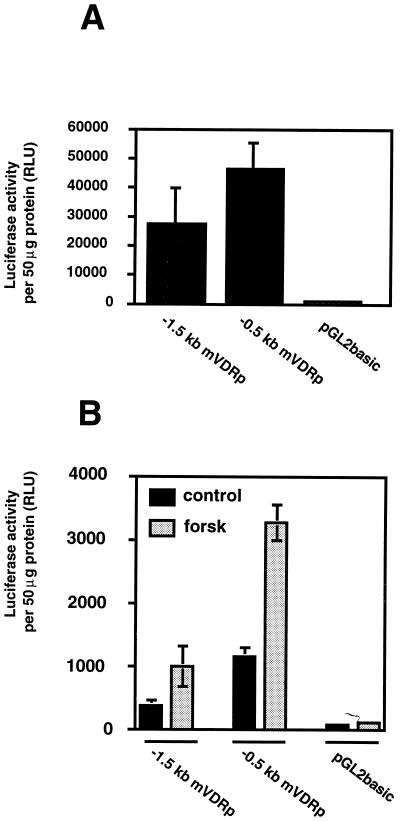
To analyze the transactivation function of the sequence upstream to the new exon 1, we decided to subclone it into a promoterless reporter vector. To be functional, this construct requires the presence of the start site of the promoter. The restriction site HaeII was used to subclone 494 bp from nucleotides 1014–1509 (Fig. 2) into the reporter plasmid pGL2b in front of the luciferase gene (−0.5 kb mVDRp). To obtain more promoter sequence upstream of the HaeII fragment, a PstI fragment (nucleotides 1–1020 in Fig. 2) was inserted in the PstI site adjacent to the HaeII site at 1014 (Fig. 2). The resulting construct possesses 1,509 bp of genomic sequence in front of the luciferase reporter gene (−1.5 kb mVDRp). Using these constructs, transient expression assays were performed in rat osteosarcoma ROS 17/2.8 cells and mouse fibroblast NIH 3T3 cells (Fig. 5). In these experiments, the vector pGL2b was used as control for no promoter activity. In ROS 17/2.8 cells, high levels of activity were observed with both constructs (Fig. 5A). An increase of luciferase activity has also been found when plasmids −1.5 kb mVDRp or −0.5 kb mVDRp were transfected into NIH 3T3 cells. Mouse NIH 3T3 cells have been reported to respond to forskolin treatment by increasing their VDR protein and mRNA content (27). We have found by Northern blot analysis a 25-fold induction of mVDR mRNA content peaking at 2 hr of induction by forskolin (data not shown). When NIH 3T3 cells are transfected with mVDRp reporter plasmids, the luciferase activity is induced 3- to 5-fold (depending on the experiment) by a 24-hr treatment with forskolin (Fig. 5B).
Figure 5.
In vivo transcription analysis of the sequence upstream of the new exon 1. The −1.5 kb mVDRp and −0.5 kb mVDRp constructs were transfected into ROS 17/2.8 cells (A) or NIH 3T3 cells (B) using lipofectin. NIH 3T3 cells were treated with 10 mM forskolin (forsk) or ethanol (control) for 24 hr. Luciferase activity is expressed in relative luciferase units (RLU). Luciferase activity was normalized to protein content of each sample. Values are average (± SD) of experiments in triplicate. Basal level of luciferase activity obtained with the pGL2b construct in ROS 17/2.8 cells and NIH 3T3 cells are 935 RLU ± 182 and 112 RLU ± 28, respectively.
DISCUSSION
An unresolved question regarding VDR-mediated responses is the control of receptor levels in target cells. Different regulation mechanisms have been involved. Glucocorticoids (20), estrogen (23), retinoic acid (24), parathyroid hormone (25), and calcium (21, 22) have each been shown to increase VDR levels. 1,25(OH)2D3 control of VDR levels by transcriptional activation has been reported by two groups (9, 17). However, two studies performed by our group demonstrated that the increase of VDR is the result of receptor stabilization in the presence of the ligand (18, 19). Another regulatory mechanism involves the activation of the cAMP signal pathway. cAMP agonists, like forskolin, have been shown to induce levels of VDR mRNA in mouse NIH 3T3 (27) and UMR 106 sarcoma cell lines (25). To address the issue of these regulation mechanisms, the identification and characterization of the VDR gene promoter will allow the determination of the presence or absence of response elements characteristic to these control mechanisms.
The identification of the promoter region of the VDR gene has been hampered by the insufficient knowledge of the precise 5′ end of the VDR gene. Nucleotides 1–15 from our published sequence of the rVDR cDNA (7) do not match the nucleotide sequence of the corresponding region in a genomic clone (T. K. Ross, unpublished results). Furthermore, the recent publication of the 5′ end of the mVDR cDNA (15), using a rapid amplification of cDNA ends technique, revealed a high identity in nucleotide sequence (only 1 base difference from the rat sequence) and an additional 12 bp upstream from the rat 5′ end (Fig. 1). The first 27 bp of the mVDR cDNA likely represent a portion of a previously unknown 5′-located exon. Indeed, using two mVDR clones from a P1 genomic library, we have confirmed the existence of this new 5′-located exon. This new exon might also be present in the hVDR gene. The sequence homology between rat, mouse, and human species leads us to expect a similar exon–intron boundary in the human gene.
The partial gene structure of the mVDR gene reveals that the intron placed between the two exons encoding the zinc fingers (exons 2 and 3 in the hVDR gene; exons 3 and 4 in the mVDR gene) is located in a different position compared with the hVDR gene (see Results and ref. 28). This might reflect some evolutionary divergence between the hVDR and the mVDR.
The 5′-flanking region of the new exon 1 was sequenced. The 1.5 kb sequence obtained was analyzed by computer using a pattern search to identify sequences homologous to those characterized in transcription regulatory elements. Interestingly, no TATA box but four Sp1 sites in tandem were found in this region (Figs. 2 and 3). Similar TATA-less promoters have been described among the members of the steroid/thyroid hormone receptor superfamily (41). Indeed, with the exception of the estrogen-receptor promoter, promoters of members of this family are TATA-less promoters driven by Sp1 sites. Upstream of the four Sp1 sites in tandem, two CCAAT boxes are present in opposite orientation. An interesting observation is the presence of a perfect Krox-20 site overlapping the Sp1 #1 (Fig. 4 B and C), also reported in the human retinoic acid receptor-α promoter (39) and mouse Hox-1.4 promoter (40). The transcription factor Krox-20 is involved in several brain functions. Alterations of hind brain development and of myelinization of the peripheral nervous system have been observed after disruption of the krox-20 gene in transgenic mice (42, 43). Also in these mice, a defect in bone formation involving chondrocyte–osteoblast interactions, which leads to endosteal bone formation, has been reported (44). This could be due to a defect of the VDR expression in cells known to be important targets of the 1,25-(OH)2D3.
Analysis of the 1.5 kb sequence of the promoter did not allow the finding of any sequences matching the consensus sequence of a VDRE (two direct repeats of six bp following the consensus sequence A/G G G/T T C/G A, spaced by three nucleotides). However, such a sequence might be present farther upstream in the promoter or might be slightly different from the classical consensus sequence.
To test the activity of the putative mVDR promoter, DNA fragments of this region have been placed in front of a luciferase reporter gene and transfected in ROS 17/2.8 osteosarcoma cells or NIH 3T3 fibroblasts. Results of the transfection experiments revealed that either 0.5 kb or 1.5 kb of the 5′-flanking region of the new exon 1 of the mVDR gene increased luciferase activity by 30- to 50-fold (Fig. 5A). This enhancement is higher than that elicited by a simian virus 40 constitutive promoter in Ros 17/2.8 cells (data not shown) confirming the high activity of the putative mVDR promoter. Because the VDR is expressed at very low levels in tissues and cell culture, this means that some silencer might be present upstream of the promoter region that we have cloned. In support of this belief, the −0.5 kb mVDRp construct has 2- to 3-fold higher activity in the transfection assays than does the −1.5 kb mVDRp construct (Fig. 5 A and B). Thus, a silencer may bind the promoter somewhere in the 1.0 kb fragment upstream of the −0.5 kb 5′ fragment of the mVDR promoter.
Treatment of mouse NIH 3T3 cells with forskolin, an activator of adenylate cyclase, has been reported to strongly increase VDR protein and mRNA levels in a biphasic manner (27). Indeed, by Northern blot analysis, the authors showed a first increase in mVDR mRNA with a maximum after 2 hr of treatment and a second increase reaching a plateau after 15 hr of treatment (27). However, we observed a 25-fold increase in mVDR mRNA after 2 hr of treatment with forskolin but we did not find the second increase after 15 hr of treatment (data not shown). In transactivation experiments in these cell lines using the same expression plasmid constructs we have demonstrated a 3- to 5-fold increase in luciferase activity after 24 hr of forskolin treatment (Fig. 5B). This low induction might represent the remnant of the peak of mVDR mRNA induction seen at 2 hr of treatment with forskolin.
Taken together, the following results support the hypothesis that the 1.5 kb sequence immediately upstream of the new exon 1 is the mVDR promoter. (i) Several consensus sites characteristic of a TATA-less promoter have been located in this region by computer search. (ii) A high promoter activity has been demonstrated by transfection experiments. (iii) The 1.5 kb and 0.5 kb constructs are responsive to forskolin and this finding is consistent with the regulation of the mVDR by forskolin previously reported in NIH 3T3 cells.
Acknowledgments
We thank Christine Kimmel-Jehan for her critical reading of the manuscript. This work was supported in part by program project grant DK14881 from the National Institutes of Health, a fund from the National Foundation for Cancer Research, and a fund from the Wisconsin Alumni Research Foundation.
ABBREVIATIONS
- VDR
vitamin D receptor
- mVDR
mouse VDR
- hVDR
human VDR
- 1
25-(OH)2D3, 1,25-dihydroxyvitamin D3
- VDRE
vitamin D response element
- rVDR
rat VDR
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF017779).
References
- 1.DeLuca H F. FASEB J. 1990;2:224–236. [PubMed] [Google Scholar]
- 2.Munder M, Herzberg I M, Zierold C, Hanson K, Clagett-Dame M, DeLuca H F. Proc Natl Acad Sci USA. 1995;92:2795–2799. doi: 10.1073/pnas.92.7.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacDonald P N, Dowd D R, Nakajima S, Galligan M A, Reeder M C, Haussler C A, Ozato K, Haussler M R. Mol Cell Biol. 1993;13:5907–5917. doi: 10.1128/mcb.13.9.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kliewer S A, Umesono K, Mangelsdorf D J, Evans R M. Nature (London) 1992;255:446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darwish H M, DeLuca H F. Crit Rev Eukaryotic Gene Expression. 1993;3:89–116. [PubMed] [Google Scholar]
- 6.Ross T K, Darwish H M, DeLuca H F. Vitam Horm. 1994;49:281–326. doi: 10.1016/s0083-6729(08)61149-8. [DOI] [PubMed] [Google Scholar]
- 7.Dame M C, Pierce E A, Prahl J M, Hayes C E, DeLuca H F. Biochemistry. 1986;25:4523–4534. doi: 10.1021/bi00364a011. [DOI] [PubMed] [Google Scholar]
- 8.Pike J W, Marion S L, Donaldson C A, Haussler M R. J Biol Chem. 1983;258:1289–1296. [PubMed] [Google Scholar]
- 9.McDonnell D P, Mangelsdorf D J, Pike J W, Haussler M R, O’Malley B W. Science. 1987;235:1214–1217. doi: 10.1126/science.3029866. [DOI] [PubMed] [Google Scholar]
- 10.Burmester J K, Maeda N, DeLuca H F. Proc Natl Acad Sci USA. 1988;85:1005–1009. doi: 10.1073/pnas.85.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burmester J K, Wiese R J, Maeda N, DeLuca H F. Proc Natl Acad Sci USA. 1988;85:9499–9502. doi: 10.1073/pnas.85.24.9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker A R, McDonnell D P, Hughes M, Crisp T M, Mangelsdorf D J, Haussler M R, Pike J W, Shine J, O’Malley B W. Proc Natl Acad Sci USA. 1988;85:3294–3298. doi: 10.1073/pnas.85.10.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elaroussi M A, Prahl J M, DeLuca H F. Proc Natl Acad Sci USA. 1994;91:11596–11600. doi: 10.1073/pnas.91.24.11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu Z, Hanson K, DeLuca H F. Arch Biochem Biophys. 1997;339:99–106. doi: 10.1006/abbi.1996.9864. [DOI] [PubMed] [Google Scholar]
- 15.Kamei Y, Kawada T, Fukuwatari T, Ono T, Kato S, Sugimoto E. Gene. 1995;152:281–282. doi: 10.1016/0378-1119(94)00735-b. [DOI] [PubMed] [Google Scholar]
- 16.Li Y C, Bergwitz C, Jüppner H, Demay M B. Endocrinology. 1997;138:2347–2353. doi: 10.1210/endo.138.6.5210. [DOI] [PubMed] [Google Scholar]
- 17.Costa E M, Feldman D. Biochem Biophys Res Commun. 1986;137:742–747. doi: 10.1016/0006-291x(86)91141-1. [DOI] [PubMed] [Google Scholar]
- 18.Wiese R J, Uhland-Smith A, Ross T K, Prahl J M, DeLuca H F. J Biol Chem. 1992;267:20082–20086. [PubMed] [Google Scholar]
- 19.Arbour C, Prahl J M, DeLuca H F. Mol Endocrinol. 1993;7:1307–1312. doi: 10.1210/mend.7.10.8264662. [DOI] [PubMed] [Google Scholar]
- 20.Massaro E R, Simpson R U, DeLuca H F. Am J Physiol. 1983;244:E230–E235. doi: 10.1152/ajpendo.1983.244.3.E230. [DOI] [PubMed] [Google Scholar]
- 21.Sandgren M E, DeLuca H F. Proc Natl Acad Sci USA. 1990;87:4312–4314. doi: 10.1073/pnas.87.11.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhland-Smith A, DeLuca H F. Biochim Biophys Acta. 1993;1176:321–326. doi: 10.1016/0167-4889(93)90061-s. [DOI] [PubMed] [Google Scholar]
- 23.Walters M R. Biochem Biophys Res Commun. 1981;103:721–726. doi: 10.1016/0006-291x(81)90509-x. [DOI] [PubMed] [Google Scholar]
- 24.Petkovitch M, Heershe J N M, Tinker D O, Jones G. J Biol Chem. 1984;259:8274–8290. [PubMed] [Google Scholar]
- 25.Pols H A P, van Leeuwen J P T M, Schilte J P, Visser T J, Birkenhager J C. Biochem Biophys Res Commun. 1988;156:588–594. doi: 10.1016/s0006-291x(88)80883-0. [DOI] [PubMed] [Google Scholar]
- 26.van Leeuwen J P T M, Birjenhager J C, Wijngaarden T V V, van den Bemd G J C M, Pols H A P. Biochem Biophys Res Commun. 1992;185:881–886. doi: 10.1016/0006-291x(92)91709-y. [DOI] [PubMed] [Google Scholar]
- 27.Krishnan A V, Feldman D. Mol Endocrinol. 1992;6:198–206. doi: 10.1210/mend.6.2.1314957. [DOI] [PubMed] [Google Scholar]
- 28.Hughes M R, Malloy P J, Kieback D J, Kesterson R A, Pike J W, Feldman D, O’Malley B W. Science. 1988;242:1702–1705. doi: 10.1126/science.2849209. [DOI] [PubMed] [Google Scholar]
- 29.Schüle R, Umesono K, Mangelsdorf D J, Bolado J, Pike J W, Evans M R. Cell. 1990;61:497–504. doi: 10.1016/0092-8674(90)90531-i. [DOI] [PubMed] [Google Scholar]
- 30.Sassone-Corsi P. Proc Natl Acad Sci USA. 1988;85:7192–7196. doi: 10.1073/pnas.85.19.7192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y-S, Green M R. Proc Natl Acad Sci USA. 1988;85:3396–3400. doi: 10.1073/pnas.85.10.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klemsz M J, McKercher S R, Celada A, Van Beveren C, Maki R A. Cell. 1990;61:113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- 33.Geoffroy V, Ducy P, Karsenty G. J Biol Chem. 1995;270:30973–30979. doi: 10.1074/jbc.270.52.30973. [DOI] [PubMed] [Google Scholar]
- 34.Merriman H L, van Wijnen A J, Bidwell J P, Fey E, Lian J, Stein J, Stein G S. Biochemistry. 1995;34:13125–13132. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro M B, Senapathy P. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadonaga J T, Jones K A, Tjian R. Trends Biochem Sci. 1986;11:20–23. [Google Scholar]
- 37.Bhat G, SivaRaman L, Murthy S, Domer P, Thimmappaya B. EMBO J. 1987;6:2045–2052. doi: 10.1002/j.1460-2075.1987.tb02469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemaire P, Vesque C, Schmitt J, Stunnenberg H, Frank R, Charnay P. Mol Cell Biol. 1990;10:3456–3467. doi: 10.1128/mcb.10.7.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brand N J, Petkovitch M, Chambon P. Nucleic Acids Res. 1990;18:6799–6806. doi: 10.1093/nar/18.23.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chavrier P, Vesque C, Galliot B, Vigneron M, Dolle P, Duboule D, Charnay P. EMBO J. 1990;9:1209–1218. doi: 10.1002/j.1460-2075.1990.tb08228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gronemeyer H, Laudet V. Nuclear Receptors Protein Profile. 1995;2:1173–1235. [PubMed] [Google Scholar]
- 42.Schneider-Maunoury S, Topilko P, Seitanidou T, Levi G, Cohen-Tanoudji M, Pournin S, Babinet C, Charnay P. Cell. 1993;75:1199–1214. doi: 10.1016/0092-8674(93)90329-o. [DOI] [PubMed] [Google Scholar]
- 43.Topilko P, Schneider-Maunoury S, Levi G, Baron-Van Evercooren A, Ben Younes Chennoufi A, Seitanidou T, Babinet C, Charnay P. Nature (London) 1994;371:796–799. doi: 10.1038/371796a0. [DOI] [PubMed] [Google Scholar]
- 44.Levi G, Topilko P, Schneider-Maunoury S, Lasagna M, Mantero S, Cancedda R, Charnay P. Development (Cambridge, UK) 1996;122:113–120. doi: 10.1242/dev.122.1.113. [DOI] [PubMed] [Google Scholar]