Abstract
Oligonucleotides that recapitulate the acceptor stems of tRNAs are substrates for aminoacylation by many tRNA synthetases in vitro, even though these substrates are missing the anticodon trinucleotides of the genetic code. In the case of tRNAAla a single acceptor stem G⋅U base pair at position 3·70 is essential, based on experiments where the wobble pair has been replaced by alternatives such as I⋅U, G⋅C, and A⋅U, among others. These experiments led to the conclusion that the minor-groove free 2-amino group (of guanosine) of the G⋅U wobble pair is essential for charging. Moreover, alanine-inserting tRNAs (amber suppressors) that replace G⋅U with mismatches such as G⋅A and C⋅A are partially active in vivo and can support growth of an Escherichia coli tRNAAla knockout strain, leading to the hypothesis that a helix irregularity and nucleotide functionalities are important for recognition. Herein we investigate the charging in vitro of oligonucleotide and full-length tRNA substrates that contain mismatches at the position of the G⋅U pair. Although most of these substrates have undetectable activity, G⋅A and C⋅A variants retain some activity, which is, nevertheless, reduced by at least 100-fold. Thus, the in vivo assays are much less sensitive to large changes in aminoacylation kinetic efficiency of 3·70 variants than is the in vitro assay system. Although these functional data do not clarify all of the details, it is now clear that specific atomic groups are substantially more important in determining kinetic efficiency than is a helical distortion. By implication, the activity of mutant tRNAs measured in the in vivo assays appears to be more dependent on factors other than aminoacylation kinetic efficiency.
The genetic code is established in the aminoacylation reactions, where specific amino acids are attached to tRNAs that bear the anticodon trinucleotides. However, in at least some instances, the relationship between the triplet of the code and its corresponding amino acid is indirect. An example is alanyl-tRNA synthetase (AlaRS) where a single G3⋅U70 base pair in the tRNA acceptor stem is essential for aminoacylation (1, 2), and where the synthetase makes no contact with the anticodon trinucleotide (3). Small oligonucleotide substrates that reconstruct the acceptor stem of tRNAAla are efficiently acylated by AlaRS in a G3⋅U70-dependent manner (4). This observation led to the investigation of many other systems and the demonstration that at least 11 synthetases can charge oligonucleotide substrates based on the acceptor stem, with a specificity and efficiency that is highly dependent on the nucleotide sequence of the substrate (5). The relationship between these acceptor stem sequences/structures and specific amino acids constitutes an operational RNA code for amino acids that may have predated the genetic code (6).
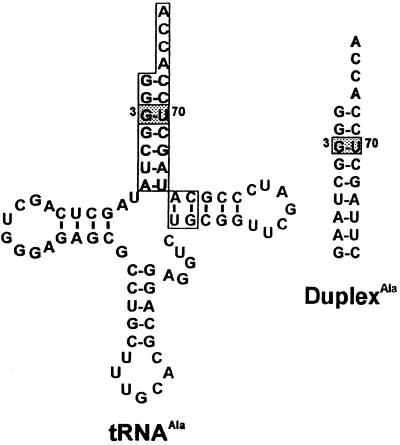
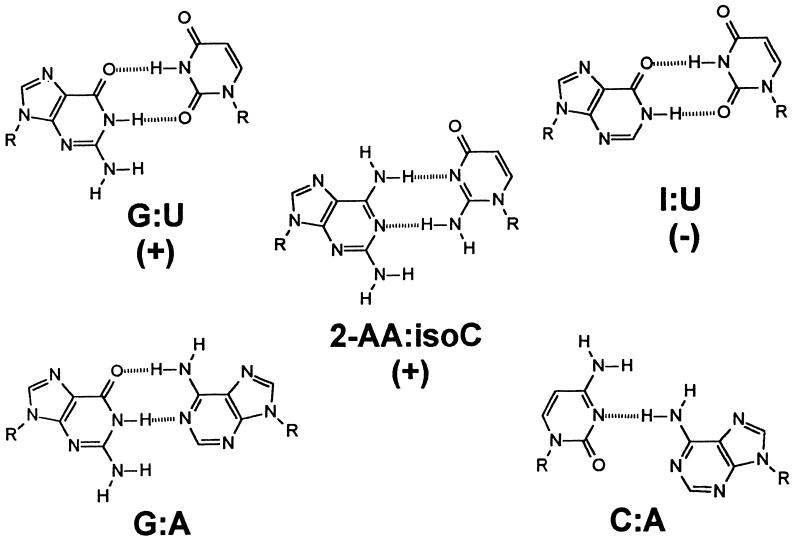
To learn more about the basis of acceptor helix recognition, chemically synthesized duplex substrates that mimic the acceptor stem portion of tRNAAla (Fig. 1) have been used (7). The G3⋅U70 base pair is in the wobble configuration, wherein the 2-amino group of guanosine is unpaired (8). In vitro aminoacylation assays using more than 40 duplexAla variants provide support for the role of specific minor-groove functional groups in AlaRS recognition (9–11). Most significantly, when the exocyclic 2-amino group of G3 was removed by substitution of the modified base inosine (Fig. 2), aminoacylation was abolished (9). Moreover, substitution at 3·70 with a 2-aminoadenosine–isocytidine base pair to place the 2-amino group in the same location in the minor groove as that of a G⋅U pair (Fig. 2) partially restored aminoacylation (11). These results and additional work probing the role of backbone 2′-hydroxyls (10) led to the conclusion that specific minor groove atomic groups are major determinants for charging by AlaRS.
Figure 1.
Sequence of E. coli tRNAAla/UGC and synthetic RNA duplex substrates used in this work. The duplex substrates were designed to mimic the acceptor-TΨC helix of E. coli tRNAAla/GGC (boxed) and consisted of a 5′-ribononamer annealed to a 3′-ribotridecamer (7).
Figure 2.
Proposed structures of base pairs incorporated at the 3⋅70 position of duplexAla and tRNAAla substrates in this work and in previous work. I, inosine; 2-AA, 2-aminoadenosine; isoC, isocytidine. The aminoacylation results for the G⋅U, I⋅U, and 2-AA⋅isoC variants as determined by in vitro experiments with RNA duplexes are indicated in parentheses (9, 11). +, Variant was active; −, activity was not detectable.
To help understand how the G⋅U wobble pair of tRNAAla contributes to the aminoacylation specificity by AlaRS in vivo, 16 tRNAAla mutants that represent all possible base combinations at the 3⋅70 position were previously constructed (12, 13). Expressing these tRNAs from a plasmid in Escherichia coli by using two different in vivo assay systems (amber suppressor tRNA and an E. coli knockout strain) revealed a range of functional responses with respect to the specificity and efficiency of alanine acceptance. Remarkably, particular base combinations gave concordant responses in the two systems. For example, loss-of-function mutant tRNAs with G⋅C and A⋅U accepted some alanine but also other amino acids, they exhibited very low levels (near background) of aminoacyl-tRNA, and they did not support growth of cells lacking chromosomal tRNAAla genes. The active tRNAs with C⋅A and G⋅A accepted only alanine, were substantially aminoacylated (80% of G⋅U tRNA) under steady-state cellular conditions, and they supported growth of cells lacking chromosomal tRNAAla genes nearly as well as did the G⋅U tRNA. Since tRNAAla variants containing other wobble base pairs and mismatches such as C3⋅A70 and G3⋅A70 substantially function as alanine acceptors in vivo (whereas G3⋅C70 and A3⋅U70 do not), it was proposed that a “helical distortion” and functional groups of G⋅U are components of AlaRS recognition (12, 13). Experimentally assessing the separate contributions may be difficult, however. The solution structure of a 22-mer RNA microhelix mimicking the acceptor stem of E. coli tRNAAla was recently solved by NMR spectroscopy (14). The structure reveals that, although the overall geometry of the acceptor stem is close to A-form RNA, the G3⋅U70 pair serves to distort the backbone conformation, resulting in unusual phosphate–phosphate distances and presentation of the functional groups of U70 in an “unusual spatial location.”
Thus, in vitro and in vivo experiments have given a different picture of what is important for RNA acceptor stem recognition by AlaRS. However, different sequence variants were used in the in vivo studies than were used in vitro. With this in mind, we conducted an in vitro analysis of the same variants that were used for the in vivo work. These studies were done to clarify the apparent paradox about the basis for E. coli AlaRS recognition of the acceptor stem of tRNAAla (15, 16).
MATERIALS AND METHODS
Materials.
RNA synthesis chemicals and the controlled pore glass supports were from Glen Research (Sterling, VA). All other RNA phosphoramidite monomers were purchased from Chemgenes (Waltham, MA). Ultra-high purity acetonitrile and dichloroethane were obtained from Baxter Scientific Products (McGaw Park, IL).
Preparation of RNA.
RNA oligonucleotides were synthesized by using the phosphoramidite method on a Gene Assembler Special (Pharmacia), deprotected, gel-purified on denaturing 16% polyacrylamide-TBE gels (where TBE is 89 mM Tris/45 mM boric acid/1 mM EDTA, pH 8.3), eluted, and desalted as described (17, 18). Full-length tRNAAla was prepared by in vitro transcription with T7 RNA polymerase (19). Site-directed mutagenesis of full-length tRNAs was accomplished by using the Kunkel method (20). For the determination of RNA concentrations, the following extinction coefficients were used (units, M−1⋅cm−1): full-length tRNA, 60.4 × 104; 13-mer, 10.7 × 104; 9-mer, 8.9 × 104 (7).
Enzyme Preparation and Assays.
Histidine-tagged E. coli AlaRS (pQE-875) was purified as described (21). For aminoacylation assays, the AlaRS concentration was based on the active-site titration (22). Aminoacylation assays were performed using the published conditions (7). Prior to performing the assays, duplexAla substrates were annealed by heating in 50 mM Hepes (pH 8) at 80°C for 2 min, then cooling to 60°C for 2 min, and finally adding MgCl2 to a final concentration of 10 mM before placing the samples on ice. The final concentration of duplex substrates in the assays was 10 μM. The reactions were initiated by adding purified AlaRS to 0.25 μM. Rates of aminoacylation, which are proportional to kcat/Km under the conditions used, were determined from the slope of a plot of pmol of alanine charged versus time and represent the average of at least two determinations. A 1,000-fold decrease in aminoacylation represents the detection limit of the duplex assays.
Aminoacylation of the full-length tRNAAla transcripts was carried out using 2.5 nM AlaRS in reactions with wild-type and G3⋅A70 tRNAAla and 25 nM AlaRS in reactions with G3⋅C70, A3⋅C70, and C3⋅A70 variants. Transfer RNA concentrations ranged from 0.25 to 4 μM for wild-type and G3⋅A70 tRNAAla and from 2.5 to 60 μM for the G3⋅C70, A3⋅C70, and C3⋅A70 variants. The kcat/Km values for full-length substrates were determined from Lineweaver–Burk plots and represent the average of at least two determinations.
In inhibition experiments, all reaction components were incubated for at least 5 min prior to initiating the reaction with full-length wild-type in vitro transcribed tRNAAla (final concentration, 0.5 μM). The conditions for inhibition assays were essentially the same as for the charging assays. The concentration of AlaRS was 2.5 nM, and the concentrations of inhibitor tRNAAla 3⋅70 variants (A⋅C, C⋅A, and G⋅C) were 0.5 to 50 μM. Under these conditions, the concentration of inhibitor at which 50% inhibition is achieved (IC50) was estimated from the x-axis intercept of a plot of 1/v (where v is the initial steady-state velocity) versus inhibitor concentration. Inhibition results are averages of two determinations.
RESULTS
RNA Duplex Substrates.
In this work, we first tested a number of new 3⋅70 variants, including those found to be active alanine acceptors in vivo, by using in vitro aminoacylation assays with RNA duplex substrates (Fig. 1). As shown in Table 1, all base-pair combinations that we examined either eliminated activity (A⋅A, A⋅C, A⋅G, C⋅A, C⋅C, C⋅G, C⋅U, U⋅A, and U⋅C) or displayed significantly reduced (1% or less) aminoacylation kinetics in vitro (G⋅A, G⋅G, and U⋅U). Interestingly, some substitutions retained more function than others, as in the in vivo system (13).
Table 1.
Aminoacylation efficiencies of RNA duplex variants
| Base pair (3·70) | kcat/Km duplexAla |
|---|---|
| G·U (wild-type) | 1.0 |
| G·C | 0 |
| G·A | 4.5 × 10−3 |
| G·G | 4.4 × 10−3 |
| A·U | 0 |
| A·C | 0 |
| A·A | 0 |
| A·G | 0 |
| U·U | 6.7 × 10−3 |
| U·C | 0 |
| U·A | 0 |
| U·G | 0 |
| C·U | 0 |
| C·C | 0 |
| C·A | 0 |
| C·G | 0 |
| U3·U70, G4·U69 | 4.7 × 10−3 |
| A3·U70, G4·U69 | 3.5 × 10−3 |
| G3·C70, G4·U69 | 0 |
Relative kcat/Km is based on in vitro aminoacylation. Results are relative to the wild-type duplexAla (ribononamer + ribotridecamer), which was set at a value of 1.0. Values reported are averages of at least two determinations, with average standard deviations of ±26%. A value of zero indicates that the aminoacylation level was greater than 1,000-fold decreased relative to the wild-type duplex.
A recent in vivo experiment also showed that shifting the G⋅U pair by one position conferred some alanine acceptance on a A3⋅U70/G4⋅U69-tRNALys variant (23). When we tested this double base-pair mutation and a U3⋅U70/G4⋅U69 variant in our in vitro duplexAla system, we found that alanine acceptance was measurable but reduced 200- to 300-fold (Table 1). This result confirms that, although the positional location of the G⋅U base within the acceptor stem is critical for optimal aminoacylation, shifting the G⋅U to an adjacent site allows weak recognition by AlaRS in vitro, as well as in vivo. As previously reported, a G3⋅C70/G4⋅U69 variant was inactive both in vivo (12, 23) and in vitro (9).
Full-Length tRNA Substrates.
Although the major determinants of the aminoacylation efficiency of tRNAAla are in the acceptor stem (4, 24), a footprint analysis demonstrated contacts of AlaRS with parts of tRNAAla that are outside of the acceptor stem. These included contacts with the dihydrouridine-anticodon stem, but not with the anticodon itself (3). Thus, in the context of the full-length tRNA, interactions with the acceptor helix could be modified by contacts in the dihydrouridine-anticodon stem and thereby account for the difference in in vitro versus in vivo results.
To address this concern, the wild-type and four 3⋅70 variants were prepared in the context of full-length tRNAAla transcripts. These four variants were chosen to span the full range of charging levels seen in vivo. Specifically, these levels were 83% (G3⋅U70 (wild-type)), 66% (C3⋅A70), 65% (G3⋅A70), 42% (A3⋅C70), and 1% (G3⋅C70) (Table 2) (13). Thus, the difference between the most active (G3⋅U70) and least active (G3⋅C70) substrates in vivo is about 83-fold (83% versus 1%).
Table 2.
Comparison of in vitro aminoacylation efficiencies of full-length tRNAAla with in vivo activity of tRNAAla variants
| Base pair (3·70) | Kcat/Km tRNAAla | Ala-tRNAAla (%, in vivo) | Viability of tRNAAla knockout strain |
|---|---|---|---|
| G·U (wild-type) | 1.0 | 83 (100) | + |
| G·A | 1.1 × 10−2 | 65 (78) | + |
| A·C | 2.8 × 10−4 | 42 (51) | − |
| C·A | 1.8 × 10−4 | 66 (80) | + |
| G·C | 2.1 × 10−5 | 1 (1) | − |
Relative kcat/Km is based on in vitro aminoacylation. Results are relative to the wild-type tRNAAla transcript, which was set at a value of 1.0. Values reported are averages of at least two determinations, with average standard deviations of ±18%. In vivo percentage of alanyl-tRNAAla [100 × charged/(charged + uncharged)] has been reported (13) and reproduced for comparison. Numbers in parentheses are percent relative to wild-type tRNAAla, which was set to 100%. The values for A·C and G·C are corrected for non-alanine specificity. Viability is the ability to support growth of a knockout tRNAAla cell line (13). +, Variant supported cell growth; −, variant did not support cell growth.
In contrast, the difference in aminoacylation kinetic efficiencies in vitro between the most and least active substrates is about 50,000-fold (Table 2). Although with some variants there is qualitative correlation of in vivo and in vitro results, there is no systematic numerical correlation in the two sets of data. The G3⋅C70 tRNA is one of the poorest substrates in both systems, with kcat/Km reduced approximately 48,000-fold in in vitro aminoacylation assays and about 1% of the wild-type alanyl-tRNAAla level measured in vivo. The kcat/Km of the G3⋅A70 variant, which displayed approximately 78% of the wild-type alanine acceptance levels in vivo, was only reduced approximately 90-fold in vitro (Table 2). This result is also consistent with previous studies in the duplex system showing that a free 2-amino group is important for AlaRS substrate recognition (Fig. 2) (9).
Two of the full-length mismatched variant tRNAs that were good substrates in vivo were inefficient substrates in vitro. We measured large reductions in kcat/Km for both the A3⋅C70 (about −3,000-fold) and C3⋅A70 (about −5,000-fold) variants (Table 2). In contrast, high levels of alanyl-tRNAAla (51% of the wild-type for A⋅C and 80% of the wild-type for C⋅A) were determined for these variants in vivo (Table 2). In this case, the in vitro result is consistent with previous findings in the duplex system that substrates lacking a minor groove 2-amino group at position 3 are poor alanine acceptors.
For the data presented in Table 2, we have rank-ordered the full-length tRNAAla substrates according to their in vitro kinetic activity and compared this ordering with that seen in vivo. With the full-length transcripts, the C3⋅A70, A3⋅C70, and G3⋅C70 variants were reduced by 3,600- to 48,000-fold in vitro. Note that the C3⋅A70 and A3⋅C70 variants are also better substrates than G3⋅C70, as in vivo (12, 13). These variants had no detectable activity as duplex substrates. (Because the full-length tRNA substrates are more active than the duplex variants, there is an even larger range of activity detectable for the full-length molecules than was possible with the duplexes.) In addition, with the G⋅A variant that was studied both as a duplex and as a full tRNA substrate, the results with the two systems are in reasonably good agreement. For example, the activities of the two G⋅A variants are within about a factor of two relative to their respective wild-type counterparts (Tables 1 and 2). Thus, we have no evidence for context effects with the full-length molecules that would alter any of the conclusions reached with the duplex variants.
Potential for Helical Distortion or Unwinding in the Transition State.
The in vitro data suggest that a helical distortion does not play a major role in the recognition of the G3⋅U70 base pair. If a helical distortion contributes to recognition, then that contribution might reflect a transition state in which local unwinding of the helix occurs. For example, if local unwinding is required for the transition state of catalysis, then the roughly 10-fold higher activity in vitro of the A3⋅C70 and C3⋅A70 variants compared with the G3⋅C70 construct could be due to the easier melting of the acceptor stem helices with mismatched A⋅C or C⋅A pairs. Because transition state analogs are typically good inhibitors of enzymes, we tested the possibility that the A⋅C, C⋅A, and G⋅C (a control) full-length constructs would act as good inhibitors of aminoacylation. We found that G3⋅C70 and A3⋅C70 tRNAAlas exhibited little or no inhibition of aminoacylation. These results are similar to the earlier observation of Park et al. (25) who concluded that the A3⋅U70 variant is also defective for binding to the synthetase. In contrast, C3⋅A70 tRNAAla showed significant inhibition, with an IC50 of 46 ± 12 μM. This compares with a Km for wild-type tRNAAla of about 2.2 μM (25). The in vivo results show that the C3⋅A70 variant was substantially aminoacylated (80% of G3⋅U70) and supported growth of the E. coli tRNAAla knockout strain, whereas the A3⋅C70 variant was a poorer substrate and did not support cell growth (Table 2). Thus, the inhibition studies are qualitatively consistent with the respective in vivo activities of these variants.
DISCUSSION
Although much speculation has centered around why in vitro and in vivo studies of tRNAAla recognition have led to different interpretations for the role of the G3⋅U70 base pair, the present study addresses some of the major questions that arose when considering the two sets of data. In particular, some of the mismatched base-pair combinations at the 3⋅70 position that were studied in vivo have now been investigated in vitro and, in addition, the concern about the use of duplex substrates in the earlier in vitro work has now been addressed.
We estimated earlier that the 2-amino group of the G3⋅U70 base pair contributed at least 3.3 kcal/mol (1 cal = 4.184 J) to lowering the apparent free energy of activation for charging an alanine acceptor stem (9, 10). This estimate was arrived at by simply comparing the activity of a G3⋅U70 to an I3⋅U70 duplex where only the exocyclic 2-amino group of G3 is removed and no “negative” or blocking determinants are introduced. Neither the A3⋅C70 nor the C3⋅A70 duplex variants had an activity that was sufficiently greater than that of the I3⋅U70 construct to make charging detectable. As an alternative, we can make use of the full-length tRNA transcripts to make some estimations.
In particular, if we use the full-length G3⋅C70 tRNAAla as a common reference, then the difference in free energy of activation between G⋅U and G⋅C corresponds to about 6.5 kcal/mol (Table 2). In contrast, the difference between A⋅C and G⋅C is 1.5 kcal/mol and between C⋅A and G⋅C is 1.3 kcal/mol (Table 2). By this measure, the unpaired 2-amino group of G⋅U is substantially more significant for recognition of tRNAAla (by at least 3.5 kcal/mol) than is a helical distortion.
In addition to the possible role of a helical distortion, the weak charging activity of the mismatched variants at the 3⋅70 position is due to the other RNA determinants known to influence aminoacylation efficiency. Both in vitro and in vivo data show that secondary recognition elements, in addition to G3⋅U70 exist (12, 13, 21, 24, 26–29). The A73 “discriminator” base and at least three other positions within the acceptor helix and external to this helix have been identified as important for either positive or negative discrimination by AlaRS.
Among the most interesting and relevant conclusions reached herein is that a severe defect in aminoacylation kinetic efficiency can still result in the steady-state level accumulation of charged tRNAAla in vivo (Table 2). Thus, the high steady-state level of charged mutant tRNAAlas appears to be due to factors other than aminoacylation efficiency. For example, an additional cellular component such as the abundant elongation factor Tu may play a critical role. This factor binds specifically to charged tRNAs and may sequester (and thereby help to accumulate) the charged tRNAAla molecules that are poor substrates for aminoacylation.
Another factor is the in vivo experimental conditions that may compensate for a poor kinetic efficiency. For example, if a defective tRNA or enzyme is provided on a multicopy plasmid, then overexpression can compensate in vivo for a high Km or low kcat of the defective enzyme or tRNA (30–33). Other factors that may also contribute to differences between in vitro and in vivo systems have been discussed (34).
In conclusion, both in vitro and in vivo assays have contributed to our understanding of tRNAAla recognition. We have established that mismatched 3⋅70 variants are poor substrates in vitro and that a helical distortion per se is not as important as specific atomic groups in determining kinetic efficiency. But our data do not exclude a free energy contribution to aminoacylation kinetic efficiency of as much as 1.5 kcal/mol from a helical distortion. This potential contribution, the contribution of nucleotide determinants such as the A73 discriminator base, and the effect of additional factors other than aminoacylation efficiency that operate in vivo (vide supra) are collectively sufficient to reconcile the in vitro and in vivo studies of the recognition of tRNAAla.
Acknowledgments
We thank Dr. Stephen Hale for critically reading the manuscript before submission for publication. This work was supported by Grants GM-49928 from the National Institutes of Health (to K.M.-F.) and GM-15539 (to P.S.). K.M.-F. also acknowledges the donors of The Petroleum Research Fund, administered by the American Chemical Society, for partial support of this research.
ABBREVIATION
- AlaRS
alanyl-tRNA synthetase
References
- 1.Hou Y-M, Schimmel P. Nature (London) 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 2.McClain W H, Foss K. Science. 1988;240:793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- 3.Park S J, Schimmel P. J Biol Chem. 1988;263:16527–16530. [PubMed] [Google Scholar]
- 4.Francklyn C, Schimmel P. Nature (London) 1989;337:478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 5.Martinis S A, Schimmel P. In: tRNA Structure, Biosynthesis, and Function. Söll D, RajBhandary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 349–370. [Google Scholar]
- 6.Schimmel P, Giegé R, Moras D, Yokoyama S. Proc Natl Acad Sci USA. 1993;90:8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musier-Forsyth K, Scaringe S, Usman N, Schimmel P. Proc Natl Acad Sci USA. 1991;88:209–213. doi: 10.1073/pnas.88.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limmer S, Reif B, Ott G, Lubos A, Sprinzl M. FEBS Lett. 1996;385:15–20. doi: 10.1016/0014-5793(96)00339-0. [DOI] [PubMed] [Google Scholar]
- 9.Musier-Forsyth K, Usman N, Scaringe S, Doudna J, Green R, Schimmel P. Science. 1991;253:784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- 10.Musier-Forsyth K, Schimmel P. Nature (London) 1992;357:513–515. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]
- 11.Musier-Forsyth K, Shi J-P, Henderson B, Bald R, Fürste J P, Erdmann V A, Schimmel P. J Am Chem Soc. 1995;117:7253–7254. [Google Scholar]
- 12.McClain W H, Chen Y-M, Foss K, Schneider J. Science. 1988;242:1681–1684. doi: 10.1126/science.2462282. [DOI] [PubMed] [Google Scholar]
- 13.Gabriel K, Schneider J, McClain W H. Science. 1996;271:195–197. doi: 10.1126/science.271.5246.195. [DOI] [PubMed] [Google Scholar]
- 14.Ramos A, Varani G. Nucleic Acids Res. 1997;25:2083–2090. doi: 10.1093/nar/25.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibba M, Söll D. Nature (London) 1996;381:656. doi: 10.1038/381656a0. [DOI] [PubMed] [Google Scholar]
- 16.Schimmel P, Musier-Forsyth K. Nature (London) 1996;384:422. doi: 10.1038/384422a0. [DOI] [PubMed] [Google Scholar]
- 17.Scaringe S A, Francklyn C, Usman N. Nucleic Acids Res. 1990;18:5433–5441. doi: 10.1093/nar/18.18.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sproat B, Colonna F, Mullah B, Tsou D, Andrus A, Hampel A, Vinayak R. Nucleosides Nucleotides. 1995;14:255–273. [Google Scholar]
- 19.Liu H, Musier-Forsyth K. Biochemistry. 1994;33:12708–12714. doi: 10.1021/bi00208a023. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beuning, P. J., Gulotta, M. & Musier-Forsyth, M. (1997) J. Am. Chem. Soc., in press.
- 22.Fersht A R, Ashford J S, Bruton C J, Jakes R, Koch G L E, Hartley B S. Biochemistry. 1975;14:1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]
- 23.McClain W H, Gabriel K, Schneider J. RNA. 1996;2:105–109. [PMC free article] [PubMed] [Google Scholar]
- 24.Francklyn C, Shi J-P, Schimmel P. Science. 1992;255:1121–1125. doi: 10.1126/science.1546312. [DOI] [PubMed] [Google Scholar]
- 25.Park S J, Hou Y-M, Schimmel P. Biochemistry. 1989;28:2740–2746. doi: 10.1021/bi00432a056. [DOI] [PubMed] [Google Scholar]
- 26.Shi J-P, Francklyn C, Hill K, Schimmel P. Biochemistry. 1990;29:3621–3626. doi: 10.1021/bi00467a005. [DOI] [PubMed] [Google Scholar]
- 27.McClain W H, Foss K, Jenkins R A, Schneider J. Proc Natl Acad Sci USA. 1991;88:9272–9276. doi: 10.1073/pnas.88.20.9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Asahara H, Himeno H, Hasegawa T, Shimizu M. J Mol Evol. 1991;4:129–132. [Google Scholar]
- 29.Liu H, Yap L-P, Musier-Forsyth K. J Am Chem Soc. 1996;118:2523–2524. [Google Scholar]
- 30.Jasin M, Regan L, Schimmel P. Nature (London) 1983;306:441–447. doi: 10.1038/306441a0. [DOI] [PubMed] [Google Scholar]
- 31.Ho C, Jasin M, Schimmel P. Science. 1985;229:389–393. doi: 10.1126/science.3892692. [DOI] [PubMed] [Google Scholar]
- 32.Jasin M, Regan L, Schimmel P. J Biol Chem. 1985;260:2226–2230. [PubMed] [Google Scholar]
- 33.Hou Y-M, Schimmel P. Biochemistry. 1992;31:10310–10314. doi: 10.1021/bi00157a019. [DOI] [PubMed] [Google Scholar]
- 34.McClain W H. J Mol Biol. 1993;234:257–280. doi: 10.1006/jmbi.1993.1582. [DOI] [PubMed] [Google Scholar]