Abstract
LINEs are transposable elements, widely distributed among eukaryotes, that move via reverse transcription of an RNA intermediate. Mammalian LINEs have two ORFs (ORF1 and ORF2). The proteins encoded by these ORFs play important roles in the retrotransposition process. Although the predicted amino acid sequence of ORF1 is not closely related to any known proteins, it is highly basic; thus, it has long been hypothesized that ORF1 protein functions to bind LINE-1 (L1) RNA during retrotransposition. Cofractionation of ORF1 protein and L1 RNA in extracts from both mouse and human embryonal carcinoma cells indicated that ORF1 protein binds L1 RNA, forming a ribonucleoprotein particle. Based on UV crosslinking and electrophoretic mobility-shift assays using purified components, we demonstrate here that the ORF1 protein encoded by mouse L1 binds nucleic acids with a strong preference for RNA and other single-stranded nucleic acids. Furthermore, multiple copies of ORF1 protein appear to bind single-stranded nucleic acid in a manner suggesting positive cooperativity; such binding characteristics are likely to be facilitated by the protein–protein interactions detected among molecules of ORF1 polypeptide by coimmunoprecipitation. These observations are consistent with the formation of ribonucleoprotein particles containing L1 RNA and ORF1 protein and provide additional evidence for the role of ORF1 protein during retrotransposition of L1.
LINEs are long interspersed repeated DNA sequences that have achieved their high copy number in mammalian genomes by retrotransposition. As with other retrotransposons, the process of LINE-1 (L1) transposition must begin with an RNA transcript. Although the mechanism for L1 transposition is not known in detail, the proteins encoded by the two ORFs in L1 are essential for transposition (1). Unlike the familiar retroviruses and closely related retrotransposons, LINEs do not have long terminal repeat (LTR) sequences. This assures that there are fundamental differences between the life cycle of L1 and the relatively well-studied, LTR-containing retroelements (2–4). In marked contrast to the LTR-containing retroelements, cDNA synthesis and integration in LINEs is likely to occur simultaneously, using a 3′ hydroxyl from nicked genomic DNA as the primer for first-strand synthesis (5, 6).
L1 contains two ORFs, ORF1 and ORF2. A combination of evidence, based on deduction from sequence analysis and studies of purified components, has revealed that ORF2 encodes a reverse transcriptase (7), an endonuclease (6), and possibly RNase H (8). In contrast, sequence analysis of the ORF1 protein fails to expose homology with proteins of known enzymatic function; thus its functional role in L1 retrotransposition has not been explored previously using purified components. The deduced amino acid sequences of ORF1 from various mammalian species are characterized by a highly variable N-terminal domain, a relatively conserved C-terminal domain, and a high, overall net positive charge at neutral pH (9, 10). This high positive charge is characteristic of proteins that bind nucleic acids and suggests a role for ORF1 in binding L1 RNA, cDNA, and/or chromosomal DNA during the retrotransposition process.
In cytoplasmic extracts prepared from the mouse embryonal carcinoma cell line, F9, ORF1 protein cofractionates with full-length, sense-strand L1 RNA through sucrose density gradients. Examination of fractions that are enriched in full-length L1 RNA reveals that the RNA is present as part of a large ribonucleoprotein complex that is distinct from polyribosomes (11). Further evidence for interaction of the human L1 RNA and ORF1 protein was provided recently by using high-speed pellets obtained from cytoplasmic extracts of teratocarcinoma cells (12). These data are again consistent with a role for ORF1 in formation of a ribonucleoprotein complex during L1 retrotransposition. Because of the complexity of the preparations used in both of these studies, it was not possible to study the interaction of ORF1 protein with RNA specifically, and it remained formally possible that additional proteins are required.
In this report, we describe the results of experiments that directly examine the interaction of ORF1 protein with RNA by using ORF1 protein purified from bacteria and L1 RNA transcribed in vitro. The results obtained from these experiments demonstrate that ORF1 protein binds RNA and single-stranded DNA with no evidence for sequence-specificity. Furthermore, the binding profile of ORF1 protein to nucleic acid shows characteristics of positive-cooperativity. Such apparent cooperativity may be facilitated by protein–protein interactions among molecules of ORF1, which are demonstrated by coimmunoprecipitation. Taken together, these data contribute significantly toward understanding the functional role of ORF1 protein during the retrotransposition cycle of L1.
MATERIALS AND METHODS
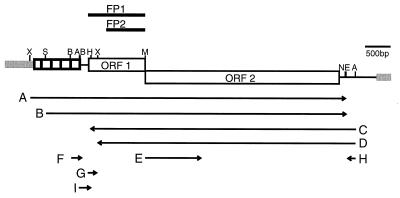
Constructs, Transcripts, and Fusion Proteins.
Plasmids (Table 1) were constructed for this study by cloning fragments from L1MdA2 (9) into pBluescript SK− (Stratagene) or pSP64 (Promega). Transcripts for ORF1 binding assays or for in vitro translation were synthesized in vitro using linearized plasmids (Fig. 1) and T7, T3, or SP6 polymerase, following the manufacturer’s recommendations (Promega). Following transcription, the DNA template was removed by digestion with RNase-free DNase (10–20 units, Boehringer Mannheim; 30 min at 37°C). The yield of RNA was measured by absorbance at 260 nm after purification of the product through a Sephadex G-50 (Pharmacia) spin column. Two transcripts with no L1 sequence were used: a 2.4-kb tnpA transcript (13) and the 149-nucleotide transcript from pGGJ122-1A (14). Some RNAs were radiolabeled during in vitro transcription with [32P]UTP (Amersham); these transcripts were separated from unincorporated isotope by gel filtration (Sephadex G-50; Pharmacia).
Table 1.
Cloned fragments used to generate transcripts A–I (see Fig. 1)
| Clone* | Fragment (coordinates†) | Transcript (coordinates†) |
|---|---|---|
| cD39 | PCR‡ (1,686–2,840) | G (1,686–1,854) |
| DB1 | ApaI–ApaI (1,490–7,001) | H (7,001–6,823) |
| I (1,490–1,720) | ||
| VK3 | XbaI–ApaI‡ (1,855–7,001) | D (7,001–1,859) |
| VK5 | XbaI–ApaI‡ (506–7,001) | A (506–6,829) |
| VK15 | SacII–ApaI‡ (893–7,001) | B (893–6,829) |
| C (7,001–1,725) | ||
| SH8 | BsmI–NcoI (2,860–6,818) | E (2,860–4,190) |
| 200.2 | BamHI–BamHI§ (1,311–1,508) | F (1,311–1,508) |
Figure 1.
Structure of mouse L1 and location of transcripts and ORF1 polypeptides. The structure of L1MdA2 (9) is depicted. Gray bars represent sequence outside of the boundaries of the L1 element as defined by target site duplications. The A-rich tail lies immediately preceding the 3′ end of the element where the black and gray lines meet; the tandem array of small boxes at the 5′ end of L1 represent the repeated, A-monomer motif. Locations of restriction sites are shown: A, ApaI; B, BamHI; E, Esp3I; M, BsmI; N, NcoI; H, NheI; S, SacII; X, XbaI. In vitro transcripts A–I (Table 1) are shown below the structure of L1, with arrows indicating the direction of transcription; the T7 gene10 fusion proteins, FP1 and FP2, are depicted above. The scale is indicated by a bar representing 500 bp.
The ORF1 fusion proteins, FP1 and FP2 (Fig. 1), were overexpressed in bacteria and purified from inclusion bodies; all solutions were at 4°C and incubations were on ice unless otherwise stated. The Escherichia coli cell pellet was washed in 1.0 culture volume of 0.1 M NaCl, 10 mM Tris⋅HCl (pH 8.0), and 1.0 mM EDTA, then resuspended in 0.8 (pellet) volume of 50 mM Tris⋅HCl (pH 8.0), 25% (wt/vol) sucrose, and 1.0 mM EDTA. An equal volume of the same sucrose buffer containing 10 mg/ml lysozyme was added. This solution was incubated for 30 min with occasional gentle vortex mixing. The solution was adjusted to 10 mM MgCl2, 1.0 mM MnCl2, and 10 μg/ml DNase I, mixed gently, and incubation was continued for about 30 min at room temperature until viscosity was reduced. A total of 2.0 ml/g of cell pellet of 0.2 M NaCl, 1.0% deoxycholic acid, 1.0% Nonidet P-40, 20 mM Tris⋅HCl (pH 7.5), and 2.0 mM EDTA was then added with thorough mixing. The resulting extract was spun at 5,000 × g for 10 min. The supernatantnt was removed, and the pellet containing insoluable fusion protein was washed three times in 0.5% Triton X-100 and 1.0 mM EDTA by pelleting as above. This pellet was resuspended in 25 mM Tris⋅HCl (pH 8.0), 10 mM EDTA, 10 mM DTT, and 8 M urea at 20°C, then fractionated by carboxymethyl Sepharose (Pharmacia) chromatography at room temperature using a 60–270 mM gradient of NaCl in the same buffer. Peak fractions containing FP1 or FP2 were identified by Western blot analysis and pooled. The urea was removed by dialysis into PBS, leaving the purified fusion protein in solution. The protein concentration was determined (BCA assay; Pierce), adjusted to 3.3 mg/ml, and stored in small aliquots at −80°C until use. Repeated freeze–thaw cycles of purified FP1 or FP2 were avoided because this was observed to lead to aggregation of the proteins. The N termini of FP1 and FP2 contain 10 and 12 amino acids from the N terminus of the gene10 leader peptide from T7, fused to either the entire ORF1 coding sequence or to amino acid 121, respectively.
UV Crosslinking.
FP1 (2–6 μg) was incubated with 10–20 ng RNA (1–2 × 106 cpm) in 20 μl UV crosslinking buffer containing 20 mM Hepes (pH 7.5), 100 mM NaCl, 2 mM MgCl2, 2 mM DTT, and 0.6 mM vanadyl ribonucleoside complex (Life Technologies, Gaithersburg, MD) and 5% glycerol. The RNA–protein complex was allowed to form for 10 min at 30°C, then the mixture was transferred to ice and irradiated with UV light (≈4,000 μW/cm2, Stratalinker UV crosslinker 2400, Stratagene) for 30 min. Following irradiation, all mixtures were treated with both RNase A (0.5 μg/μl; Amresco, Solon, OH) and RNase T1 (2 units/μl; Boehringer Mannheim) for 1 hr at 37°C in the presence of 1 mM phenylmethylsulfonyl fluoride. The proteins were fractionated by SDS/PAGE (9% acrylamide, 1:30 bisacrylamide/acrylamide). Gels were dried and proteins visualized following exposure of Hyperfilm-MP (Amersham).
Electrophoretic Mobility-Shift Assays (EMSAs).
FP1 and 32P-labeled in vitro RNA (≈2 ng or 50,000 cpm) were incubated in 15 μl of UV-crosslinking buffer containing 100 μg/ml bovine serum albumin for 20–25 min at 30°C. Salt conditions were optimized by titration of NaCl which revealed that complexes would form between 70 and 150 mM NaCl, with the maximum between 90 and 110 mM. No complexes were observed in the absence of NaCl or in the presence of 50 mM NaCl. When heparin was added, the 30°C incubation was extended for 10 min following the addition of heparin. The reactions were transferred to ice and 1.3 μl of dye solution (50% glycerol/0.1% bromophenol blue) were added. RNA–protein complexes were fractionated by electrophoresis (5 V/cm at 4°C, 16–20 hr) through 4.5% native polyacrylamide gels (1:50 bisacrylamide/acrylamide) with 1% glycerol in 0.5× TBE (1× TBE is 89 mM Tris/89 mM boric acid/2 mM EDTA). Some variability among experiments was observed regarding the exact amounts and distribution of the bound and free forms of the radiolabeled RNA. Because of this variability, conclusions are based on data generated within, and not between, the different experiments. Following electrophoresis, the gels were dried for autoradiography. PhosphorImager analysis (Molecular Dynamics with imagequant software) was used to quantitate the amount of complex formed and the disappearance of free (unbound) RNA. For competition experiments, various nucleic acid competitors were mixed with the radiolabeled transcript before FP1 was added.
For assays to examine binding of ORF1 protein to DNA, FP1 was incubated with 150 ng of single-stranded M13 DNA in UV-crosslinking buffer containing 100 μg/ml bovine serum albumin for 40 min at 30°C. The DNA protein complexes were fractionated by electrophoresis (2.5 V/cm at 4°C, 16–18 hr) through 0.75% agarose gels in 1× TBE. Following electrophoresis, DNA and DNA–protein complexes were visualized by autoradiography after Southern blot analysis and hybridization to a 32P-labeled M13 probe (15).
ORF1 Protein Coimmmunoprecipitation.
A total of 12 μl of in vitro translation reaction (Promega) was incubated with FP1 (1 μg), FP2 (0.7 μg), or control protein (1 μg of bacterial extract containing the T7⋅Tag epitope; Novagen) in 0.5 ml of RIPB (20 mM Tris⋅HCl, pH 8.0/130 mM NaCl/1 mM EDTA/0.15% Nonidet P-40) with 0.5 mM phenylmethylsulfonyl fluoride for 3–4 hr at 4°C. Both the N termini of FP1 and FP2 contain the gene10 leader peptide from T7. This peptide epitope is recognized by the monoclonal antibody, T7⋅Tag (Novagen). Complexes between in vitro translated ORF1 protein and either FP1 or FP2 were immunoprecipitated by adding 1–2 μg of the T7⋅Tag or the control [TGN38 (16)] antibody and continuing the incubation overnight at 4°C. Immunoprecipitates were recovered using protein A-Sepharose, washed three times in RIPB, and then analyzed by SDS/PAGE and autoradiography, as described (17).
RESULTS
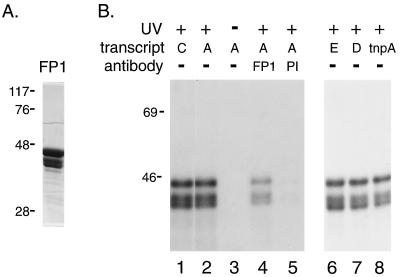
To examine whether ORF1 protein is capable of associating with RNA, FP1 was incubated with in vitro transcribed, 32P-labeled RNA prior to UV-induced photocrosslinking. Exposure to UV light causes bases to covalently link to amino acids, transferring 32P from RNA to ORF1 protein (Fig. 2B), indicating close contact between the two molecules (18). No labeling of the protein is detected in the absence of UV light (Fig. 2B, lane 3). The photolabeled proteins are immunoprecipitated by ORF1 antibody but not by the corresponding preimmune IgG (Fig. 2B, lanes 4 and 5, respectively). The three forms of protein detected after photocrosslinking are present in the original material (Fig. 2A). The two smaller polypeptides are believed to be proteolytic breakdown products of FP1 because they react with the FP1 antibody before (data not shown) and after (Fig. 2B, lane 4) photocrosslinking, and their presence in different preparations of FP1 is variable. The interaction of FP1 with L1 RNA appears to be identical on the sense and antisense strands of the RNA, and full-length RNA is not required for binding. Thus, in this UV-crosslinking assay, several, distinct 32P-labeled RNAs transfer 32P to L1 ORF1 protein (FP1, Fig. 2; FP2, data not shown) with no significant differences in efficiency, including the unrelated, trpA transcript (Fig. 2B, lane 8). These data suggest that L1 ORF1 protein binds RNA in a sequence-independent manner. Alternatively, is is possible that these results reflect recognition of a small, specific sequence that is fortuitously shared by these transcripts.
Figure 2.
UV-induced crosslinking of ORF1 protein to RNA. (A) Coomassie-stained SDS/PAGE of FP1 used for photocrosslinking. Numbers on the left indicate the position of marker proteins and their apparent molecular size (in kDa). (B) Autoradiogram of SDS/PAGE of FP1 following UV-induced crosslinking to the indicated 32P-labeled transcripts. Lanes 4 and 5 contain immunoprecipitates obtained using affinity-purified, anti-FP1 antibody (FP1) or its corresponding preimmune IgG (PI) after UV treatment.
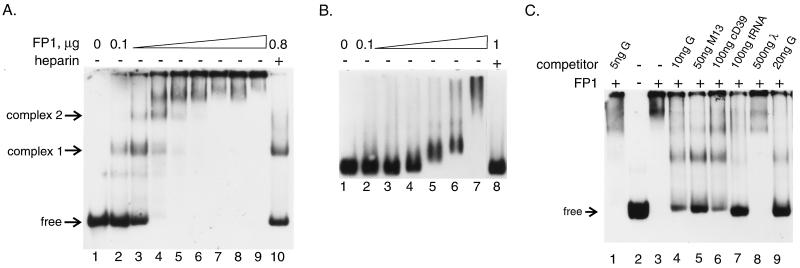
An EMSA was employed to study the interaction of ORF1 protein with RNA in more detail. At relatively low FP1 concentration, a discrete product is formed (complex 1, Fig. 3A), but increasing the amount of protein leads to the formation of additional complexes with mobility that is further reduced. When 300 ng or more was used in the assay, most of the RNA was shifted into complexes that either did not resolve or remained in the sample well under these electrophoresis conditions (Fig. 3A, lanes 4–9). It is likely that the binding of multiple ORF1 polypeptides to a single RNA molecule leads to a decrease in the electrophoretic mobility of the complex, not only because of the increase in molecular size, but also because there is a significant neutralization of the net negative charge of the RNA molecule due to the high positive charge of the ORF1 polypeptide. The addition of heparin (10 mg/ml; Fig. 3A, lane 10) disrupts these slowly migrating complexes and returns essentially all of the labeled RNA to complex 1 or the unbound form. This finding demonstrates that the observed loss of the free RNA band with high concentrations of FP1 is not due to degradation of the radiolabeled transcript, but instead reflects FP1 binding to the RNA. The amount of heparin used in this experiment was not sufficient to disrupt all of the ORF1 protein/RNA interactions. However, in other experiments where less FP1 was used, the same amount of heparin did fully disrupt the RNA–protein interactions, returning all of the labeled RNA to the free form. Transcripts G, H, and I (Fig. 1) were also used in the EMSA under similar conditions; all three gave equivalent results, as did assays using FP2 (data not shown).
Figure 3.
EMSA of FP1 binding to various nucleic acids. Autoradiograms show the following. (A) RNA binding. Lanes contain ≈2 ng of 32P-labeled in vitro transcript F in addition to increasing amounts (by 100 ng/lane) of FP1 and heparin. (B) Single-stranded DNA binding. Lanes contain 150 ng of single-stranded M13 DNA and increasing amounts of FP1 (lane 3 has 200 ng, each lane thereafter is with an additional 200 ng) and heparin. (C) Relative affinity of ORF1 protein for various nucleic acids. Lanes contain 32P-labeled transcript F (≈2 ng) and 200 ng FP1 as well as various, nonradioactive competitors as indicated. Single-stranded DNA is from M13 as in B, double-stranded DNA is purified plasmid cD39 (17) containing both relaxed circular and supercoiled forms (Qiagen), tRNA is total yeast tRNA, and bacteriophage λ DNA is uncut.
Because many RNA binding proteins also bind single-stranded DNA, the ability of ORF1 protein to interact with DNA was examined by agarose gel-retardation assays. The mobility of M13 DNA is reduced in the presence of increasing amounts of FP1 (Fig. 3B, lanes 4–7), and the complex is disrupted by the addition of heparin (Fig. 3B, lane 8). In contrast to the results obtained with RNA, however, we do not resolve discrete species of shifted M13 DNA as increasing amounts of FP1 are added. This is probably because the binding of a single molecule of ORF1 protein does not alter the mobility of the large M13 DNA significantly, leading to poor resolution between unbound, or multiple forms of bound, M13 DNA.
The relative affinity of ORF1 protein for a variety of nucleic acids was examined by competitions using the EMSA and radiolabeled RNA. For these experiments, labeled RNA was diluted with the unlabeled competitor (RNA, single- or double-stranded DNA), the mixture was incubated with FP1 and the complexes were resolved by electrophoresis. The amount of radioactivity recovered in the free RNA form and the RNA–protein complexes were determined by PhosphorImager analysis. Independent, nearly identical, results were obtained for radiolabeled transcripts G, H, and I. Titrations were used to determine the amount of competitor required to reduce or eliminate the radiolabeled transcripts from RNA–protein complexes. Shown here are examples chosen from an extensive series of titrations with these various competitor nucleic acids. For example, approximately 3-, 5-, and 10-fold excesses (by mass) of single-stranded M13 DNA, tRNA, and plasmid cD39 DNA (Fig. 3C, lanes 5, 7, and 6, respectively) were required to compete as effectively as cold transcript G for FP1 binding of radiolabeled transcript F. Approximately a 100-fold mass excess of intact λ DNA was required to show evidence of competition in this assay (Fig. 3C, lane 8). Similar results were obtained using the same competitors to disrupt binding to single-stranded M13 DNA (data not shown). Taken together, these data demonstrate that FP1 binds to RNA > single-stranded DNA > tRNA> double-stranded DNA.
It is apparent from the data presented in Fig. 3A that multiple copies of FP1 bind the radiolabeled RNA; at least four forms are resolved within the gel. It is also apparent that there is a dramatic increase in the fraction of RNA molecules that are bound over an extremely narrow range of increasing protein concentration. This narrow range of FP1 concentration over which the bound fraction of RNA increases from 10 to 90 percent (100–300 ng or 1.5–4.5 × 10−7 M; Fig. 3A, lanes 2–4) suggests that ORF1 protein binds to the RNA with positive cooperativity (19). Because of this apparent cooperativity, it is premature to attempt to use these data to calculate a precise binding constant for the ORF1 protein–RNA interaction. However, a rough estimate, based on PhosphorImager analysis of complex 1 formation at low protein concentrations (slower-migrating complexes are not detected, or are less than 10% of complex 1), suggests that the effective binding affinity for the interaction of a monomer of ORF1 protein with RNA lies within the micromolar range.
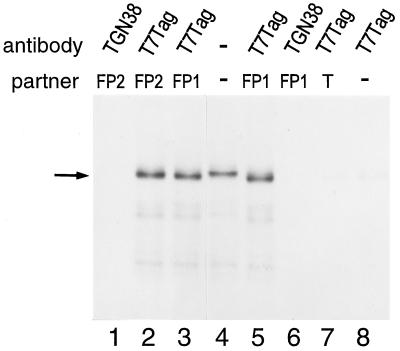
Cooperative binding of ORF1 protein to RNA would imply protein–protein interactions among molecules of the polypeptide. To determine whether molecules of ORF1 protein are capable of interacting with one another, we examined the ability of 35S-labeled ORF1 protein translated in vitro to interact with purified, unlabeled ORF1 purified from E. coli. This experiment was facilitated by the T7 epitope tag present on the N terminus of FP1 and FP2. Thus, after an incubation period where these two types of ORF1 proteins were allowed to interact, a monoclonal antibody, specific for the T7 epitope present on FP1 and FP2, was used to coimmunoprecipitate the radiolabeled ORF1 protein. Radiolabeled ORF1 protein could be coimmunoprecipitated from the in vitro translation reaction by both FP1 and FP2 using the monoclonal antibody to the T7 epitope (Fig. 4, lanes 2, 3, and 5). Radiolabeled ORF1 protein was not recovered in the immunoprecipitate when an unrelated, control antibody (TGN38) was used for the immunoprecipitation (Fig. 4, lanes 1 and 6), when a bacterial extract containing only the expressed T7 leader sequence was substituted for the ORF1 fusion proteins (Fig. 4, lane 7), or when proteins bearing the T7 epitope were omitted from the incubation altogether (Fig. 4, lane 8). Thus, L1 ORF1 protein is capable of interacting with at least one additional molecule(s) of ORF1 protein to form homodimers, or more complex multimers, in solution. Furthermore, because both FP1 and FP2 coimmunoprecipitated the ORF1 polypeptides translated in vitro, the N-terminal 120 amino acids of ORF1 protein are not required on both partners for this protein–protein interaction.
Figure 4.
ORF1 protein translated in vitro coimmunoprecipitates with FP1 and FP2. Autoradiogram of immunoprecipitated proteins fractionated by SDS/PAGE. Lane 4 contains 3 μl of the 35S-labeled in vitro translation reaction of transcript B without immunoprecipitation. Lanes 1–3 and 5–8 contain the proteins recovered following coimmunoprecipitation of 12 μl of that in vitro translation reaction with T7 epitope-tagged proteins, FP1, FP2, or bacterial extract (T). Proteins were immunoprecipitated using either the T7⋅Tag antibody or a control antibody, TGN38. The ORF1 in vitro translation product is indicated by an arrow.
DISCUSSION
These studies demonstrate that L1 ORF1 protein is a single-stranded, nucleic acid binding protein. It binds with no apparent sequence specificity to both RNA and single-stranded DNA. Competition experiments indicate that ORF1 protein binds double-stranded DNA with significantly reduced efficiency. The relatively weak competition observed with plasmid DNA (Fig. 3C, lane 6) may be due to single-stranded character assumed by the preponderance of supercoiled plasmid (20) in that population. This seems likely because in gel-shift experiments similar to those in Fig. 3B but with pBR322 DNA, the mobility of the supercoiled band is measurably retarded, but not the nicked-circular form (data not shown). Furthermore, bacteriophage λ DNA competes poorly for ORF1 binding to RNA, even when in vast excess (Fig. 3C, lane 8). ORF1 protein also appears to discriminate among different single-stranded nucleic acids because it reproducibly binds to mRNA with greater affinity than M13 single-stranded DNA or tRNA. The relatively weak binding to tRNA compared with the other RNAs tested may be a reflection of the extensive double-stranded character of tRNA.
In all experiments using EMSAs to examine ORF1 protein–RNA interactions, complex 1 appears to be relatively more stable than the other, higher order complexes. For example, in Fig. 3A (lane 10), a significant proportion of the labeled RNA is detected in complex 1, in addition to the free form, when the higher order complexes are eliminated by heparin. This suggests that complex 1 results from a higher affinity interaction between ORF1 protein and RNA compared with the interactions that lead to the other complexes. Although such an observation could reflect a sequence-specific binding site on the RNA, this seems unlikely because several different RNAs exhibit identical behavior in the assay. These RNAs differ dramatically in sequence composition, ranging between 46 and 61% GC (transcripts G and F, respectively). One property that all of the tested RNAs share, however, is that they have ends. Perhaps the “high” affinity site is found on one of the ends of the RNA molecule. This interpretation could also explain observations that M13 single-stranded DNA appears to be particularly ineffective at eliminating complex 1 (Fig. 3C, lane 5) and be involved in interactions with FP1 that are more sensitive to heparin (Fig. 3B).
There are no apparent, known RNA-binding motifs in the sequence of the mouse L1 ORF1, including the ribonucleoprotein particle (RNP) motif, arginine-rich motif, RGG box, or the KH motif (21). However, the ORF1 protein used for these studies, like the ORF1 encoding sequence from all mammalian LINEs, carries a high positive charge at neutral pH (Fig. 5). Its large positive charge may enable ORF1 protein to bind nucleic acids via nonspecific, electrostatic interactions with the negatively charged phosphosphodiester backbone. This general type of interaction is supported by the ability of ORF1 protein to bind a wide variety of transcripts in the UV-crosslinking and gel mobility-shift experiments and may facilitate the functional role of ORF1 protein during L1 retrotransposition. It is likely that many molecules of ORF1 protein “coat” the RNA to form the large, cytoplasmic RNPs detected in mammalian cells (11, 12, 22, 23); hence, the protein must be able to bind numerous sites along the RNA. However, this may be an incomplete representation of the mechanism of RNP formation in vivo. Our studies may not have detected an important, high-affinity binding site, with strong sequence or structural specificity, because the correct sequences were not present among the various transcripts tested. For example, the extreme 3′ end of L1Md was not present in any of the RNAs examined. Alternatively, the RNA transcribed in vitro may not have adopted the proper conformation for recognition by ORF1 protein, or the higher affinity of ORF1 protein for a hypothetical, specific site present in L1 RNA may have been masked by an excess of nonspecific binding sites.
Figure 5.
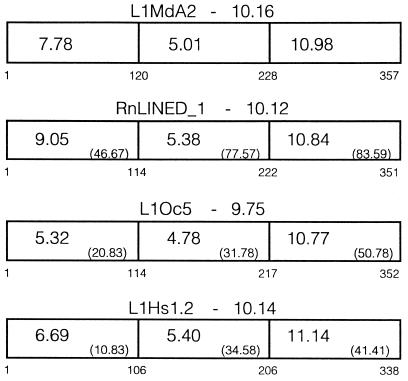
Comparisons of L1 ORF1 amino acid sequences from different mammals. ORF1 is broken into three domains; the first domain was chosen because it is missing in FP2, the second domain contains the region with an acidic pI, and the third domain contains the region with the most basic pI. The overall calculated pI from each ORF1 sequence is indicated above the schematic, and the regional pI is shown within it. All sequences were compared with L1MdA2, and the sequence identity is shown in parenthesis for each region. The numbers below each schematic indicate the amino acids aligned for each sequence. The sequences used were mouse L1MdA2 [sp|P11260 (9)], rat RnLINED_1 (pir|S21345), rabbit L1Oc5 (10), and human L1Hs1.2 [gp|M80343 (25)].
In addition to its ability to bind nucleic acids, ORF1 protein is capable of forming multimeric complexes through protein–protein interactions. Results obtained from the coimmunoprecipitation studies clearly indicate that at least two molecules of ORF1 protein can interact. In all experiments involving immunoprecipitation with ORF1 antibodies, it was necessary to use unusually low speeds for centrifugation (2,000 × g, this study and ref. 17) because we found that protein A Sepharose was not required for sedimentation following standard protocols for immunoprecipitation. This finding suggests that large aggregates of FP1 are also formed in the in vitro translation reactions. Furthermore, based on extensive efforts to express FP1 in E. coli in soluable form, all of the FP1 that can be detected is engaged in large complexes (these complexes are not inclusion bodies, but are excluded from an S-300 column; B. Lieberman, personal communication), again suggesting that protein–protein interactions among molecules of FP1 involve the formation of multimers and are not confined to dimers. Analogous observations have been made for protein–protein interaction among multiple copies of the ORF1 protein from human L1 (12).
To facilitate predictions about the regions of ORF1 protein that are involved in nucleic acid binding and protein–protein interaction, ORF1 amino acid sequences from four mammalian species were aligned, then subdivided into three regions. The N-terminal region, containing the first 120 amino acids of ORF1 from L1MdA2, is neither essential for RNA binding nor is it required on both partners for dimer formation, based on the observations that FP2 behaves like FP1 in both types of assay. In addition, this region is not conserved among the different ORF1 sequences compared, and its pI varies over a wide range (Fig. 5). Thus, it is unlikely that this region is crucial for the function of the ORF1 polypeptide during L1 retrotransposition, unless it functions in a highly species-specific manner. In contrast, the most C-terminal region is conserved among the different ORF1s, both at the sequence level and with respect to its pI. This region contains a large number of positively charged residues that are most likely to be essential for the RNA binding activity of the ORF1 protein. The central region is also relatively well conserved and maintains a constant (but acidic) pI among the sequences compared (Fig. 5), again indicative of functional importance.
One obvious model for the protein–protein interactions of ORF1 polypeptides is that they are based on electrostatic attraction between the positively (central region) and negatively (C-terminal region) charged domains. Alternatively, protein–protein interactions between molecules of ORF1 protein from human L1 appear to be mediated by a leucine-zipper motif (12). However, the absence of the putative leucine zipper in the mouse and rat ORF1 sequences indicate that there is not an absolute requirement for leucine-zipper-mediated, protein–protein interaction during retrotransposition of L1. It may be that the more general, universal requirement for protein–protein interaction among molecules of ORF1 protein for the purpose of RNP formation has been solved by independent mechanisms and/or sequences in different lineages of mammalian LINEs. Whatever the precise molecular mechanism involved, protein–protein interactions would facilitate the binding of multiple copies of ORF1 protein to an RNA molecule to form a coat around the RNA. The association of ORF1 polypeptides through protein–protein interactions could also explain the apparent cooperativity of ORF1 protein binding to RNA that was detected in the gel-shift experiments. Other RNA binding proteins, particularly from bacteriophage and viruses, are known to utilize protein–protein interactions to achieve cooperativity in binding of the protein to the RNA genome during encapsidation [e.g., the coat protein from R17 (24)].
These data represent a first step toward defining the functional properties of the ORF1-encoded polypeptide on a biochemical level, thereby enhancing our understanding of the biochemical basis for formation of L1 RNP in mouse and human cells. Future studies will dissect the protein–protein interaction domain from the RNA binding domain of the ORF1 protein. In addition to leading to a complete understanding of the basis of protein–protein interaction, a variant of the ORF1 polypeptide that fails to aggregate will greatly facilitate detailed analysis of its nucleotide binding properties, including accurate determination of binding and dissociation constants, as well as definition of the binding site. The properties of ORF1 protein revealed by this study, however, are consistent with the hypothesis that the RNP found in mouse and human cells are intermediates in the retrotransposition of L1. Recently, it was reported that short stretches of alanine substitutions in the conserved C-terminal, basic region of the human ORF1 protein reduced autonomous retrotransposition to less than 1% of wild type in cultured cells (1). It will be important to determine whether such mutations disrupt RNA binding and RNP formation to evaluate the significance of these biochemical properties of the ORF1 protein during L1 retrotransposition.
Acknowledgments
We thank D. Branciforte for purified FP1 and FP2, M. Zacharias for the 149-bp transcript from pGJ122-A, J. Saxton for the alignment and analysis of ORF1 amino acid sequences and help with preparing Fig. 5, and J. Li, B. Lieberman, S. Nordeen, and M. Zacharias for valuable discussion and helpful suggestions. This work was supported by Public Health Service Grant GM40367 to S.L.M. from the National Institutes of Health.
ABBREVIATIONS
- L1
LINE-1
- LTR
long terminal repeat
- RNP
ribonucleoprotein particle
- EMSA
electrophoretic mobility-shift assay
References
- 1.Moran J V, Holmes S E, Naas T P, DeBerardinis R J, Boeke J D, Kazazian H H., Jr Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 2.Hutchison C A, III, Hardies S C, Loeb D D, Shehee W R, Edgell M H. In: Mobile DNA. Berg D E, Howe M M, editors. Washington, DC: Am. Soc. for Microbol.; 1989. pp. 593–617. [Google Scholar]
- 3.Martin S L. Curr Opin Genet Dev. 1991;1:505–508. doi: 10.1016/s0959-437x(05)80199-6. [DOI] [PubMed] [Google Scholar]
- 4.Finnegan D J. Curr Opin Genet Dev. 1992;2:861–867. doi: 10.1016/s0959-437x(05)80108-x. [DOI] [PubMed] [Google Scholar]
- 5.Luan D D, Korman M H, Jakubczak J L, Eickbush T H. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 6.Feng Q, Moran J V, Kazazian H H, Jr, Boeke J D. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 7.Mathias S L, Scott A F, Kazazian H H, Jr, Boeke J D, Gabriel A. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 8.McClure M A. Mol Biol Evol. 1991;8:835–856. doi: 10.1093/oxfordjournals.molbev.a040686. [DOI] [PubMed] [Google Scholar]
- 9.Loeb D D, Padgett R W, Hardies S C, Shehee W R, Comer M B, Edgell M H, Hutchison C A., III Mol Cell Biol. 1986;6:168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demers G W, Matunis M J, Hardison R C. J Mol Evol. 1989;29:3–19. doi: 10.1007/BF02106177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin S L. Mol Cell Biol. 1991;11:4804–4807. doi: 10.1128/mcb.11.9.4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohjoh H, Singer M F. EMBO J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira A, Cuypers H, Gierl A, Schwarz-Sommer Z, Saedler H. EMBO J. 1986;5:835–841. doi: 10.1002/j.1460-2075.1986.tb04292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zacharias M, Hagerman P J. J Mol Biol. 1995;247:486–500. doi: 10.1006/jmbi.1995.0155. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Luzio J P, Brake B, Banting G, Howell K E, Braghetta P, Stanley K K. Biochem J. 1990;270:97–102. doi: 10.1042/bj2700097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolosha V O, Martin S L. J Biol Chem. 1995;270:2868–2873. doi: 10.1074/jbc.270.6.2868. [DOI] [PubMed] [Google Scholar]
- 18.Smith K C. In: Photochemistry and Photobiology of Nucleic Acids. Wang S Y, editor. Vol. 2. New York: Academic; 1976. pp. 187–218. [Google Scholar]
- 19.Carey J. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 20.Esposito F, Sinden R R. In: Oxford Surveys on Eukaryotic Genes. Maclean N, editor. Vol. 5. New York: Oxford Univ. Press; 1988. pp. 1–50. [PubMed] [Google Scholar]
- 21.Burd C G, Dreyfuss G. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 22.Martin S L, Branciforte D. Mol Cell Biol. 1993;13:5383–5392. doi: 10.1128/mcb.13.9.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Branciforte D, Martin S L. Mol Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Witherell G W, Wu H N, Uhlenbeck O C. Biochemistry. 1990;29:11051–11057. doi: 10.1021/bi00502a006. [DOI] [PubMed] [Google Scholar]
- 25.Dombrowski B A, Mathias S L, Nanthakumar E, Scott A F, Kazazian H H., Jr Science. 1991;254:1805–1808. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 26.Shehee W R, Chao S-F, Loeb D D, Comer M B, Hutchison C A, III, Edgell M H. J Mol Biol. 1987;196:757–767. doi: 10.1016/0022-2836(87)90402-5. [DOI] [PubMed] [Google Scholar]