Abstract
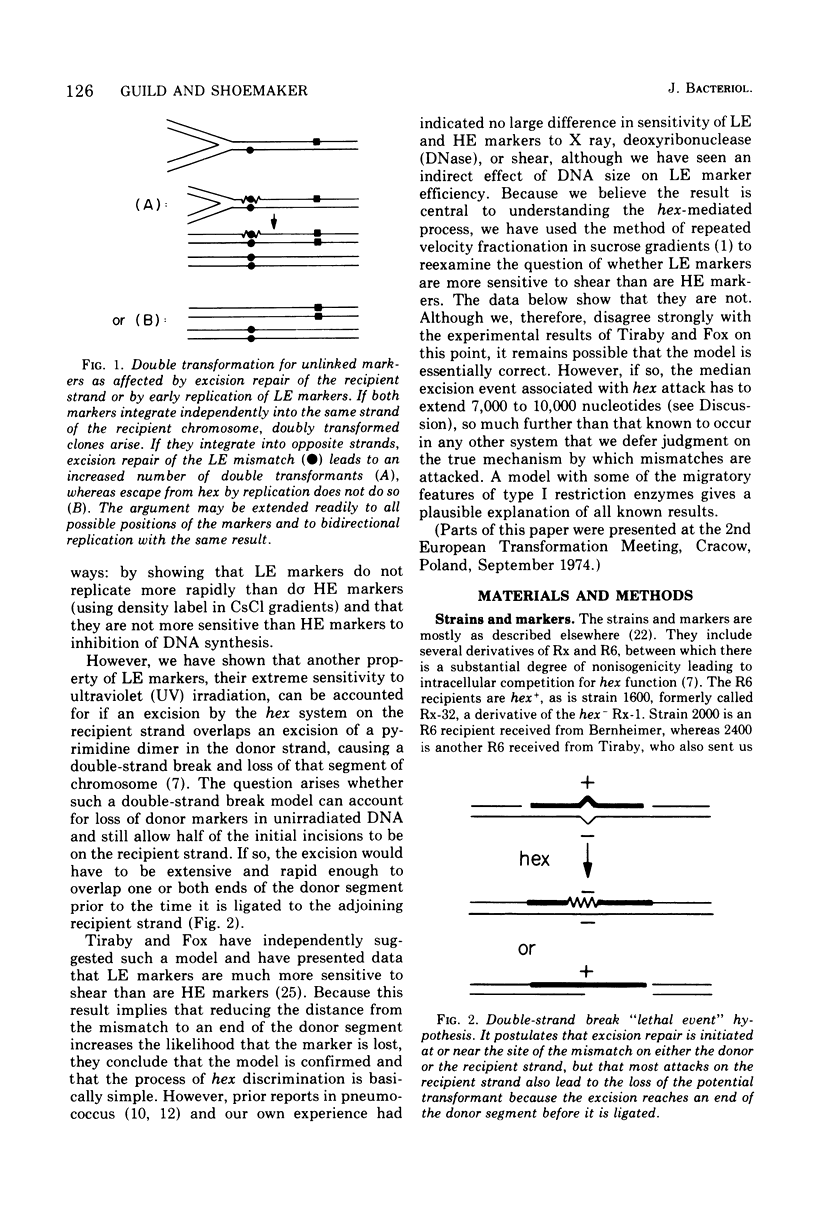
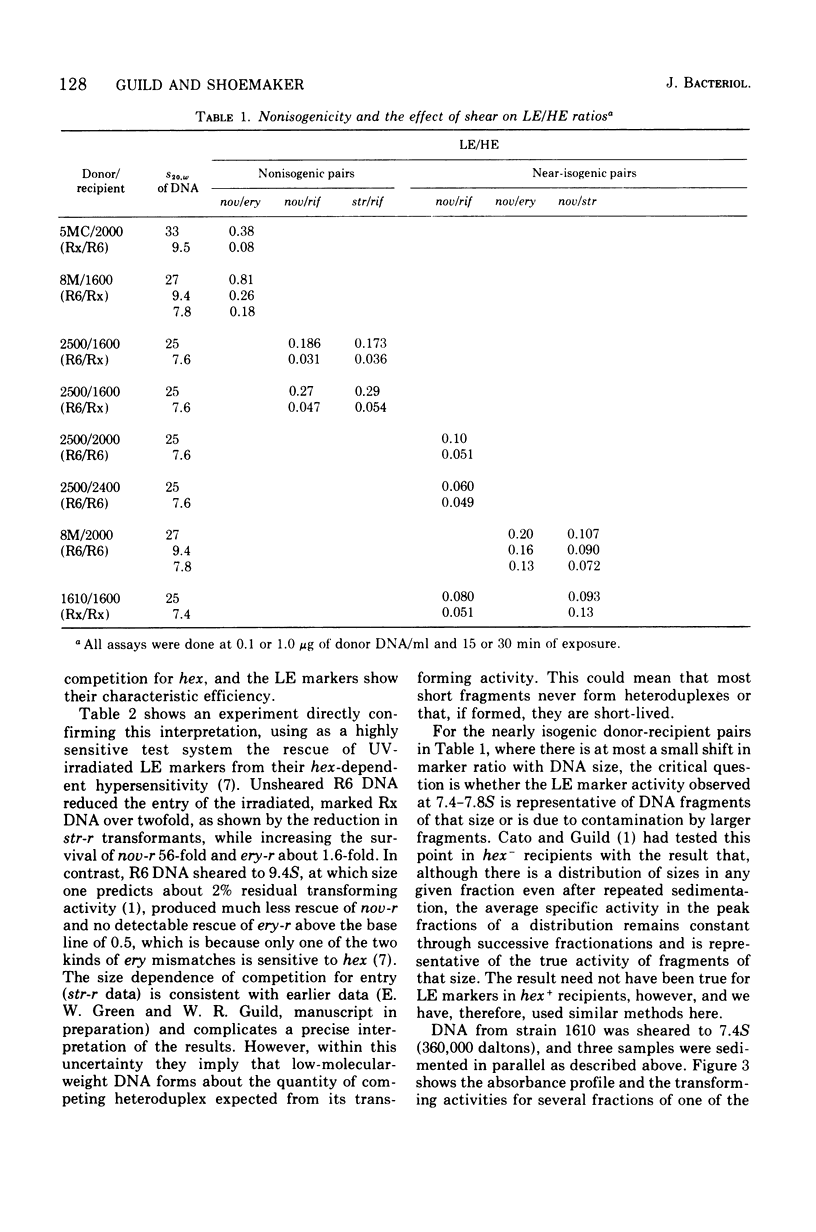
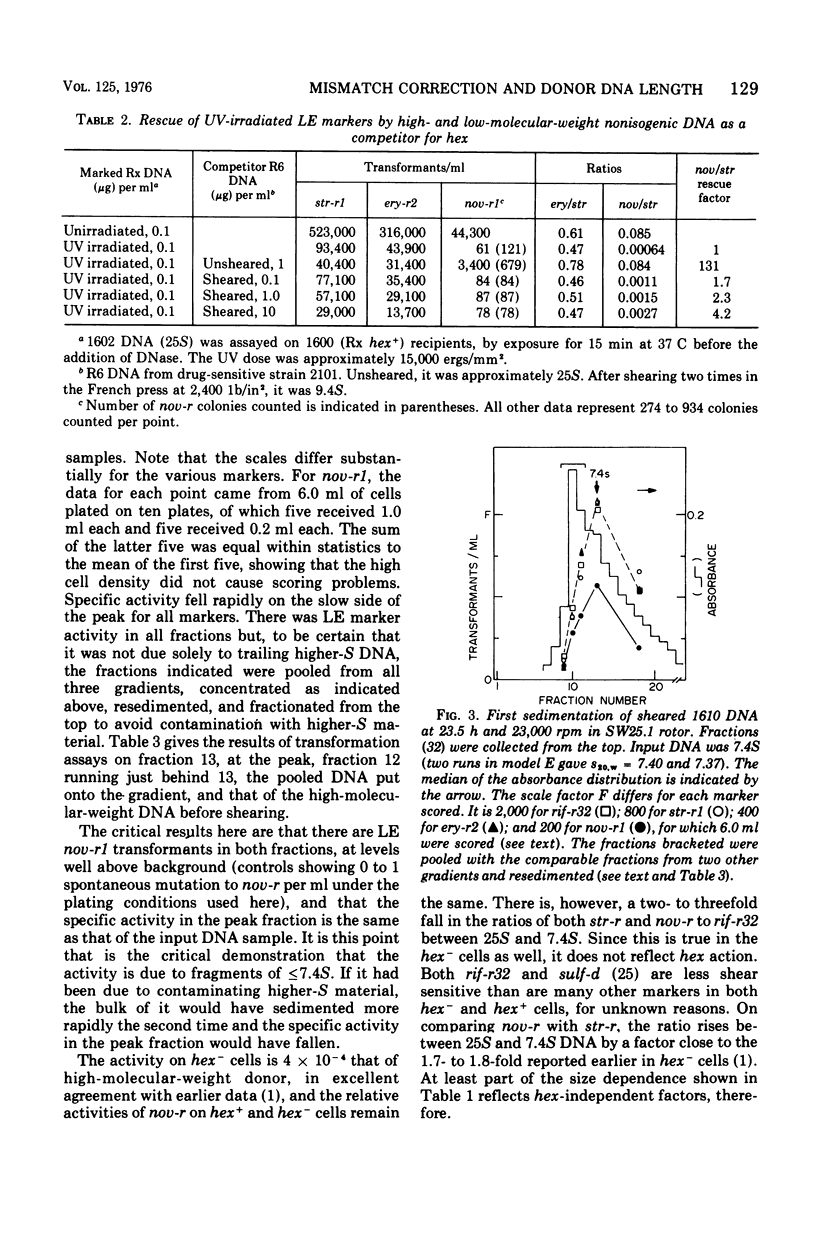
A hypothesis that preferential rejection of donor markers by the hex system of pneumococcus is due to lethal double-strand breaks has been examined in terms of its implications for the extent of the excision required. Experiments reported here were directed at asking whether hex-dependent marker efficiency depends on the length of the donor deoxyribonucleic acid (DNA). In the absence of intracellular competition for hex function, there was no detectable effect of DNA size on hex-dependent marker efficiency as donor DNA was sheared from greater than 1 x 107 daltons to 3.6 x 105 daltons. The latter DNA was purified by two successive velocity fractionations to ensure that the activity seen was representative of DNA of that size. Quantitative examination of the system shows that, for the lethal event hypothesis to be true, the excision step has to remove an average of 7,000 to 10,000 nucleotides. This figure is so much greater than that seen in other excision processes that alternate hypotheses should be considered. The presently known properties of the hex system can be accounted for by a model invoking the migratory features of type I restriction enzymes.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cato A., Jr, Guild W. R. Transformation and DNA size. I. Activity of fragments of defined size and a fit to a random double cross-over model. J Mol Biol. 1968 Oct 14;37(1):157–178. doi: 10.1016/0022-2836(68)90080-6. [DOI] [PubMed] [Google Scholar]
- Cooper P. K., Hanawalt P. C. Role of DNA polymerase I and the rec system in excision-repair in Escherichia coli. Proc Natl Acad Sci U S A. 1972 May;69(5):1156–1160. doi: 10.1073/pnas.69.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi-Taylor H. Genetic recombination in DNA-induced transformation of Pneumococcus. IV. The pattern of transmission and phenotypic expression of high and low-efficiency donor sites in the amiA locus. Genetics. 1966 Jul;54(1):211–222. doi: 10.1093/genetics/54.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi-Taylor H., Gray T. C. Genetic studies of recombining DNA in pneumococcal transformation. J Gen Physiol. 1966 Jul;49(6):211–231. doi: 10.1085/jgp.49.6.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREEN D. M. A host-specific variation affecting relative frequency of transformation of two markers in pneumococcus. Exp Cell Res. 1959 Nov;18:466–480. doi: 10.1016/0014-4827(59)90312-x. [DOI] [PubMed] [Google Scholar]
- GUILD W. R., ROBINSON M. Evidence for message reading from a unique strand of pneumococcal DNA. Proc Natl Acad Sci U S A. 1963 Jul;50:106–112. doi: 10.1073/pnas.50.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guild W. R., Shoemaker N. B. Intracellular competition for a mismatch recogition system and marker-specific rescue of transforming DNA from inactivation by ultraviolet irradiation. Mol Gen Genet. 1974;128(4):291–300. doi: 10.1007/BF00268517. [DOI] [PubMed] [Google Scholar]
- Gurney T., Jr, Fox M. S. Physical and genetic hybrids formed in bacterial transformation. J Mol Biol. 1968 Feb 28;32(1):83–100. doi: 10.1016/0022-2836(68)90147-2. [DOI] [PubMed] [Google Scholar]
- Howard L. V., Gooder H. Specificity of the autolysin of Streptococcus (Diplococcus) pneumoniae. J Bacteriol. 1974 Feb;117(2):796–804. doi: 10.1128/jb.117.2.796-804.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LERMAN L. S., TOLMACH L. J. Genetic transformation. II. The significance of damage to the DNA molecule. Biochim Biophys Acta. 1959 Jun;33(2):371–387. doi: 10.1016/0006-3002(59)90127-1. [DOI] [PubMed] [Google Scholar]
- Lacks S. Integration efficiency and genetic recombination in pneumococcal transformation. Genetics. 1966 Jan;53(1):207–235. doi: 10.1093/genetics/53.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S. Mutants of Diplococcus pneumoniae that lack deoxyribonucleases and other activities possibly pertinent to genetic transformation. J Bacteriol. 1970 Feb;101(2):373–383. doi: 10.1128/jb.101.2.373-383.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linn S., Lautenberger J. A., Eskin B., Lackey D. Host-controlled restriction and modification enzymes of Escherichia coli B. Fed Proc. 1974 May;33(5):1128–1134. [PubMed] [Google Scholar]
- Louarn J. M., Sicard A. M. Identical initial steps during transformation for high and low efficiency markers in Diplococcus pneumoniae. Biochem Biophys Res Commun. 1968 Aug 13;32(3):461–466. doi: 10.1016/0006-291x(68)90684-0. [DOI] [PubMed] [Google Scholar]
- Louarn J. M., Sicard A. M. Identical transformability of both strands of recipient DNA in Diplococcus pneumoniae. Biochem Biophys Res Commun. 1969 Jul 7;36(1):101–109. doi: 10.1016/0006-291x(69)90655-x. [DOI] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D. A., Guild W. R. Breakage prior to entry of donor DNA in Pneumococcus transformation. Biochim Biophys Acta. 1973 Apr 11;299(4):545–556. doi: 10.1016/0005-2787(73)90226-8. [DOI] [PubMed] [Google Scholar]
- Morrison D. A., Guild W. R. Transformation and deoxyribonucleic acid size: extent of degradation on entry varies with size of donor. J Bacteriol. 1972 Dec;112(3):1157–1168. doi: 10.1128/jb.112.3.1157-1168.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J. M., Guild W. R. Fractionated strands of bacterial deoxyribonucleic acid. 3. Transformation efficiencies and rates of phenotypic expression. J Bacteriol. 1968 Dec;96(6):1991–1996. doi: 10.1128/jb.96.6.1991-1996.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter R. D., Guild W. R. Number of transformable units per cell in Diplococcus pneumoniae. J Bacteriol. 1969 Mar;97(3):1033–1035. doi: 10.1128/jb.97.3.1033-1035.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Shoemaker N. B., Guild W. R. Destruction of low efficiency markers is a slow process occurring at a heteroduplex stage of transformation. Mol Gen Genet. 1974;128(4):283–290. doi: 10.1007/BF00268516. [DOI] [PubMed] [Google Scholar]
- Shoemaker N. B., Guild W. R. Kinetics of integration of transforming DNA in pneumococcus. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3331–3335. doi: 10.1073/pnas.69.11.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirotnak F. M., Hachtel S. L. Increased dihydrofolate reductase synthess in Diplococcus pneumoniae following translatable alteration of the structural gene. I. Genotype derivation and recombinational analyses. Genetics. 1969 Feb;61(2):293–312. doi: 10.1093/genetics/61.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Schueren E., Youngs D. A., Smith K. C. Sensitization of ultraviolet-irradiated Escherichia coli K-12 by different agars: inhibition of A rec and exr gene-dependent branch of the uvr gene-dependent excision-repair process. Photochem Photobiol. 1974 Jul;20(1):9–13. doi: 10.1111/j.1751-1097.1974.tb06541.x. [DOI] [PubMed] [Google Scholar]


