Abstract
CBP is a transcriptional coactivator required by many transcription factors for transactivation. Rubinstein–Taybi syndrome, which is an autosomal dominant syndrome characterized by abnormal pattern formation, has been shown to be associated with mutations in the Cbp gene. Furthermore, Drosophila CBP is required in hedgehog signaling for the expression of decapentapleigic, the Drosophila homologue of bone morphogenetic protein. However, no direct evidence exists to indicate that loss of one copy of the mammalian Cbp gene affects pattern formation. Here, we show that various abnormalities occur at high frequency in the skeletal system of heterozygous Cbp-deficient mice resulting from a C57BL/6-CBA × BALB/c cross. In support of a conserved signaling pathway for pattern formation in insects and mammals, the expression of Bmp7 was found to be reduced in the heterozygous mutants. The frequency of the different abnormalities was significantly lower in a C57BL/6-CBA background, suggesting that the genetic background is an important determinant of the variability and severity of the anomalies seen in Rubinstein–Taybi syndrome patients.
Keywords: coactivator, haploinsufficiency, pattern formation, bone morphogenetic protein
A protein that binds to the protein kinase A-phosphorylated form of cyclic AMP response element-binding protein (CREB) was originally identified and named CBP (1). CBP also binds to several components of the basal transcriptional machinery, including transcription factor IIB (2), RNA polymerase II holoenzyme complex (3), and the GCN5-like histone acetyltransferase, P/CAF (4), suggesting that CBP serves as a CREB coactivator. In addition, CBP binds to multiple kinases, such as S6 kinase pp90RSK (5) and cyclin-dependent kinases (6), and mediates the regulation of transcription by these kinases. The recent finding that CBP itself has histone acetyltransferase activity (7, 8) suggests that it contributes to transcriptional activation by disrupting the repressive chromatin structure. CBP interacts with not only CREB but also with many other transcription factors, including c-Jun (9), c-Fos (10), c-Myb (11, 12), nuclear hormone receptors (13, 14), Stat2 (15), and MyoD (16–18). CBP contributes to the transcriptional activation mediated by each of these factors. The Cbp gene family contains at least one other member, p300, that originally was identified through its ability to bind to the adenovirus E1A protein (19). No differences in the functional properties of CBP and p300 have been identified (20, 21).
Mutations in the human Cbp gene were reported to be associated with the Rubinstein–Taybi syndrome (RTS; ref. 22), a haploinsufficiency disorder characterized by mental retardation, craniofacial malformations, broad thumbs, and broad big toes (23, 24). However, it remains unclear how a 50% reduction in the amount of CBP causes the abnormal pattern formation observed in RTS. Transcription factors such as CREB, c-Myb, and AP-1, which use CBP, appear not to be involved in pattern formation during development, as judged from the results of gene knockout studies. CREB- (25), c-Jun- (26), c-Fos- (27, 28), or c-Myb-deficient mice (29) do not exhibit the abnormal pattern formation observed in RTS. Moreover, although craniofacial abnormalities are observed in RARα:RARγ double mutants (30), this phenotype is not found in homozygous mutants of either receptor alone or in trans-heterozygotes. Recently, we identified the Drosophila homologue of CBP (dCBP), and demonstrated by molecular and genetic studies that dCBP functions as a coactivator of the product of the segment polarity gene, cubitus interruptus (ci) (31). Ci functions in the hedgehog (hh) pathway and is a critical transcription factor for pattern formation (32). In response to Hh, Ci activates the decapentapleigic (dpp) and wingless (wg) genes, which encode the Drosophila homologue of transforming growth factor β/bone morphogenetic protein (BMP) and the Wnt family protein, respectively. These results raise the possibility that RTS is caused by a decrease in the activity of the mammalian homologue of Ci, the zinc finger-containing transcriptional activator, GLI (33, 34). Interestingly, mutations in one of the three members of the Gli gene family, Gli3, cause phenotypic changes through haploinsufficiency in mice (known as “extra-toes” (Xt) (35), and humans (Greig cephalopolysyndactyly syndrome) (36). These phenotypic changes resemble some of the abnormal traits of RTS. However, it has not yet been proved that the function of the mammalian GLI3 really requires the coactivator CBP.
Although mutations in the human Cbp gene were reported to be associated with RTS (22), no direct evidence exists to indicate that loss of one copy of the mammalian Cbp gene affects pattern formation. In addition, nothing excludes the possibility that other genes exist within the RTS critical region, as the map of this region is still far from complete. Moreover, the expected decrease in the level of normal CBP protein predicted to occur in hemizygous patients has yet to be demonstrated. To directly examine whether loss of one copy of the Cbp gene causes abnormal pattern formation, we have generated Cbp heterozygous mutant mice by gene targeting.
MATERIALS AND METHODS
Construction of the Targeting Vector.
The Cbp genomic clones were isolated from a library derived from TT2 cells by the standard plaque hybridization procedure. An 8.0-kb NotI–EcoNI genomic DNA subfragment was used to generate the targeting vector. The 1.0-kb AflII–EcoRI fragment containing the exon encoding amino acids 29–265 was deleted from the NotI–EcoNI fragment and replaced with a neomycin (neo) cassette driven by the phosphoglycerate kinase gene promoter. To increase the frequency of gene targeting, the diphtheria toxin-poly(A) signal cassette for negative selection (37) was used.
Generation of Cbp-Deficient Mutant Mice.
The NotI-linearized targeting vector (100 μg) was electroporated into 1.0 × 107 TT2 cells. Targeted clones were selected after 7–10 days growth in the presence of G418 (150 μg/ml) and then were expanded onto duplicate 96-well plates. The homologous nature of recombination was confirmed by Southern blot analysis. Chimeras were produced by injecting about 10 embryonic stem (ES) cells into 40 ICR eight-cell embryos and transplanting the embryos into the uterus of pseudopregnant females. Six- to eight-week-old male progeny with a high percent of chimerism were bred with BALB/c or C57BL/6 females to produce heterozygous mice capable of transmitting the targeted allele through the germ line. The mice were maintained by the Division of Experimental Animal Research, RIKEN.
Genotyping of ES Cells, Embryos, and Animals.
Genomic DNA was isolated from cultured cells, embryos, and tail clippings by digestion overnight at 55°C in lysis buffer (10 mM Tris⋅HCl, pH 7.5/100 mM NaCl/1 mM EDTA/0.5% SDS/0.5 mg/ml proteinase K/20 μg/ml RNase A) followed by phenolchloroform extraction and ethanol precipitation. For Southern blot analysis, genomic DNA (about 10 μg) was digested with SphI or BamHI and resolved on 0.8% or 0.5% agarose gels.
Detection of CBP Proteins.
The brains of 18.5 days postcoitus (dpc) fetuses were washed in PBS and resuspended in 300 μl of lysis buffer consisting of 20 mM Hepes (pH 7.9), 25% (vol/vol) glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, and 0.5 mM phenylmethylsulfonyl fluoride, and homogenized. After centrifugation, samples of lysates were resolved on a 7.5% SDS/PAGE gel. Proteins were transferred onto a nitrocellulose filter, and CBP or Hsp70 as a control were detected using the anti-CBP (Upstate Biotechnology: CBP-CT) or anti-Hsp70 (Santa Cruz Biotechnology: W27) antibodies and ECL detection reagents (Amersham).
Skeletal Analysis.
Bone and cartilage stained specimens were prepared essentially as described by Inouye (38). Fetuses on 18.5 dpc were recovered, and the tails were used for genotyping. Fetuses were skinned, eviscerated, and fixed in 95% ethanol. Nonskeletal tissues were digested with 2% potassium hydroxide. Cartilaginous tissues were stained by alcian blue 8GS, and ossified tissues were stained by alizarin red S.
In Situ Hybridization and Northern Blotting.
Whole-mount in situ hybridization using the 198-nucleotide digoxygenin-labeled BMP-7 RNA was performed essentially as described (39). RNA from the 11.5-dpc fetuses was isolated using Isogen (Nippon Gene, Toyama, Japan). RNA samples (20 μg) were run on 1% agarose-formaldehyde gels, blotted, and hybridized with BMP-7 or cytoplasmic β-actin probe.
RESULTS
Generation of Cbp-Deficient Heterozygous Mutants.
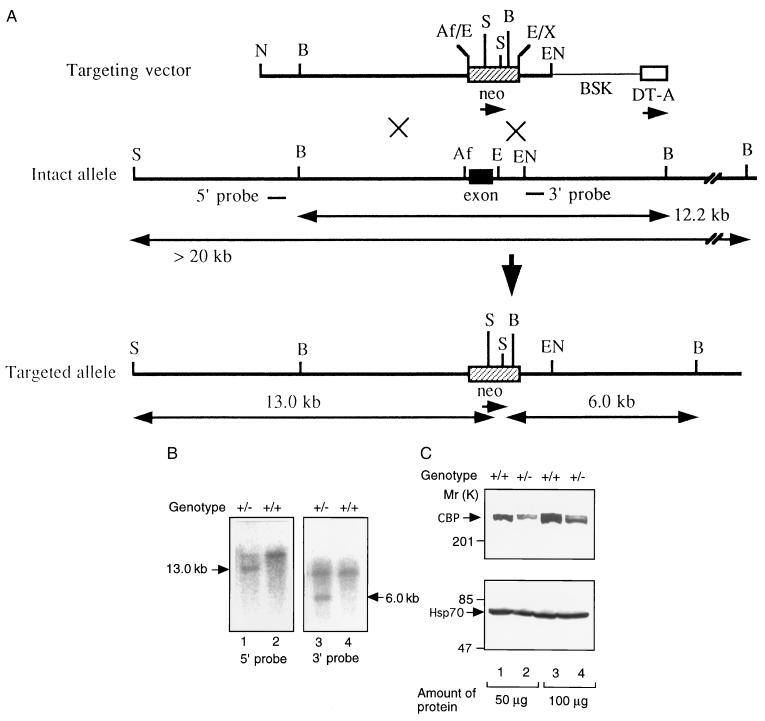
Cbp mutant mice were generated by homologous recombination in TT2 ES cells. Genomic Cbp DNA clones, which were isolated from the TT2 genomic library, were used to construct a gene targeting vector in which the exon encoding amino acids 29–265 was replaced by a neo cassette (Fig. 1A). We selected this exon to be replaced, because the replacement of the exon close to the amino terminus may severely affect the biosynthesis of full-length normal CBP protein. Homologous recombinants were characterized by the appearance of a 13.0-kb SphI fragment with the 5′ probe, and a 6.0-kb BamHI fragment with the 3′ probe (Fig. 1B). The chimeras normally were obtained from two independent mutant ES clones and were mated with C57BL/6 or BALB/c females to generate F1 heterozygous mutant mice. Heterozygous mice were identified by Southern blot analysis of genomic DNA isolated from tail or embryo samples of the offspring (Fig. 1B). Analysis of the genotype of embryos generated by intercross between the heterozygotes exhibited that homozygous mutants died at 8–10 dpc (the mechanism of embryonic lethality of homozygotes will be described elsewhere), but, in this paper, we will confine our description primarily to the abnormal pattern formation in the heterozygotes. To confirm that the level of CBP protein in the heterozygous mutants had decreased to half that of the wild-type level, Western blotting was performed using the anti-CBP antibody (Fig. 1C). The 265-kDa CBP protein was detected in the brains of wild-type fetuses of 18.5 dpc, whereas the expression level of CBP protein was about half that of the wild type in the heterozygous mutants. The levels of Hsp70 protein between wild type and the CBP heterozygote were not different.
Figure 1.
Generation of heterozygous Cbp-deficient mice. (A) Structure of the targeting construct, the Cbp locus, and targeted allele. The solid box represents the exon. Restriction enzyme sites are indicated (Af, AflII; B, BamHI; E, EcoRI; EN, EcoNI; N, NotI; S, SphI; X, XhoI). To construct the targeting vector, the AflII–EcoRI fragment containing the exon was replaced by the neomycin-gene cassette (neo). The location of two probes used for Southern blot analyses are given with the expected sizes of hybridizing fragments marked below. (B) Southern blot analysis. Genomic DNA extracted from the fetuses on 18.5 dpc was used for Southern blotting with the indicated probes. (C) Immunodetection of the CBP protein. Extracts (50 or 100 μg of protein) from the 18.5-dpc fetuses were used for Western blotting with the anti-CBP (Upper) or anti-Hsp70 antibodies (Lower).
Abnormal Skeletal Patterning in Cbp Heterozygous Mutants.
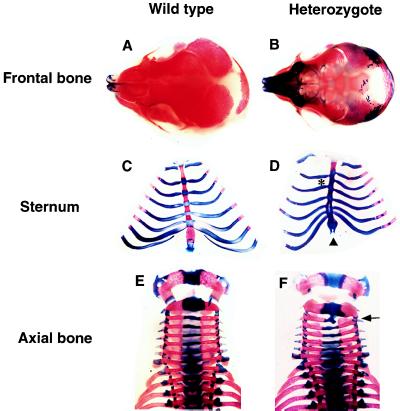
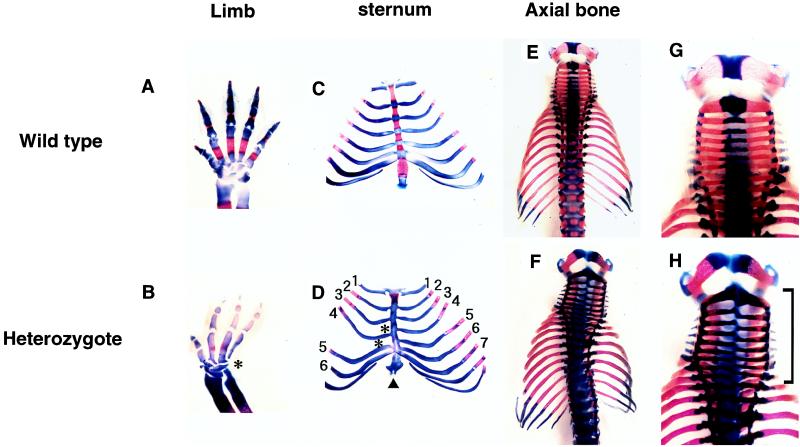
We first analyzed skeletal formation in the F1 heterozygous mutants obtained by mating with BALB/c mice. Because the TT2 ES cells that we used were derived from a F1 embryo between C57BL/6 and CBA mice (40), the F1 heterozygotes analyzed have a genetic background made up of 50% BALB/c, 25% C57BL/6, and 25% CBA. Bone and cartilage stained specimens of 21 heterozygous mutants and 30 wild-type fetuses of 18.5 dpc showed various skeletal abnormalities in discrete areas of the heterozygotes, namely the skull and the rib cage (Table 1). The most pronounced phenotype was delayed ossification of frontal bones (Fig. 2 A and B): an enlarged anterior frontanel was observed in two-thirds (14/21) of the heterozygous mutants, whereas this abnormality occurred in only one of 30 wild-type fetuses. A similar malformation of the anterior frontanel has been reported in 41% of RTS patients in the Netherlands (24). Abnormalities in the sternum and rib also were observed with lower frequency. Various anomalies of the sternum (extra, fusion, reduction, and asymmetry of the ossification with split in the sternum) were found in one-third (7/21) of the heterozygotes (Fig. 2 C and D). None of these abnormalities were observed in the 30 wild-type fetuses. Asymmetric sternocostal joints also were found in the heterozygotes with similar penetrance. An additional phenotype found in the heterozygotes was the malformation of the xiphoid process and vertebrae (Fig. 2 C–F). Distinct holes and fissures in the xiphoid process were found in 29% (6/21) of the heterozygotes, whereas only one of the 30 wild-type fetuses had this type of anomaly. One-third (7/21) of the heterozygotes exhibited severe growth retardation and delayed ossification. In particular, one of them displayed severe phenotypes of the axial bones (asymmetric cervical vertebrae, extra and split thoracic vertebrae, and scoliosis), the ribs (asymmetric ribs; reduction in the number on the right side and extra ribs on the left side), and oligodactyly (Fig. 3). Similar anomalies also have been reported for RTS patients (24). Abnormality of the limbs showed low penetrance in the heterozygotes. Thus, certain defects such as broad thumbs and broad halluces, which are the most frequent anomaly in RTS patients, were not observed in the heterozygotes.
Table 1.
Summary of the phenotypes of CBP heterozygous mutants
| Abnormalities | C57BL/6-CBA × BALB/c cross
|
C57BL/6-CBA × C57BL/6 cross
|
||
|---|---|---|---|---|
| No. of wild-type fetuses | No. of heterozygous fetuses | No. of wild-type fetuses | No. of heterozygous fetuses | |
| No. of litters | 10 | 6 | ||
| No. of fetuses analyzed | 30 | 21 | 23 | 24 |
| Severe growth retardation | ||||
| and delayed ossification | 0 | 7 | 0 | 0 |
| Large anterior frontanel | 1 | 14 | 1 | 6 |
| Asymmetric sternocostal | ||||
| joints | 1 | 7 | 1 | 1 |
| Abnormal ossification | ||||
| in sternum | 0 | 7a | 1 | 1 |
| Distinct holes in | ||||
| xiphoid process | 1 | 6 | 0 | 16 |
| Abnormalities in axial bones | 0 | 2b,c | 0 | 3g,h |
| Rib abnormalities | 5 | 4d,e | 0 | 2 |
| Limb abnormalities | 0 | 1f | 0 | 1i |
In some cases (marked by superior letters), the following severe phenotypes were observed:
Reduced and asymmetric ossification, and a split in the sternum.
Asymmetric cervical vertebrae, extra and split thoracic vertebrae, and scoliosis.
Extra thoracic vertebra.
Asymmetric ribs; reduction or increase in the number of ribs.
Extra 14 ribs of complete type and fusion of ribs.
Oligodactyly.
Deficiency of right third cervical vertebra.
Split of corpus in first lumber vertebra.
Triphalangia.
Figure 2.
Typical defects in skeletal formation observed in the heterozygous Cbp mutant. The skulls of wild type (A) and a Cbp heterozygote (B) of 18.5 dpc obtained by mating with BALB/c mice are shown. Note that delayed ossification of the frontal bones and large anterior frontanels were observed in Cbp heterozygotes. The sternum and ribs of the wild type (C) and a heterozygote (D) obtained by mating with BALB/c are indicated. Asymmetric ossification shown by ∗ and splits were observed in Cbp heterozygotes. Distinct holes and fissures in the xiphoid process observed in Cbp heterozygote are indicated by an arrowhead. The axial bones of wild type (E) and a heterozygote (F) obtained by mating with C57BL/6 are indicated. Deficiency of the third cervical vertebra observed in Cbp heterozygotes is indicated by an arrow.
Figure 3.
Severe abnormalities in the Cbp heterozygote of a BALB/c × C57BL/6-CBA background. Bone and cartilage stained specimens of the right forelimbs of the wild-type (A) and a Cbp heterozygote (B) exhibiting postaxial oligodactyly (∗). The sternums and ribs of a wild-type (C) and a Cbp heterozygote (D). Note six right ribs and misalignment of the rib pairs on the sternum (∗) in the Cbp heterozygote. Dorsal view of the vertebrae of a wild type (E) and a heterozygote (F). In the heterozygote, the presence of 10 ribs on the right side, 14 ribs on the left side, asymmetric cervical vertebrae, extra and split thoracic vertebrae, and scoliosis were observed. Higher magnification (×2) of E (G) and F (H). Asymmetric cervical vertebrae indicated by a bracket were observed in a Cbp heterozygote.
Effect of Genetic Background on the Phenotype of Cbp Heterozygous Mutants.
We also examined skeletal formation in the F1 heterozygotes obtained by mating with C57BL/6 (Table 1). This cross results in a genetic background made up of 75% C57BL/6 and 25% CBA. Although the abnormal skeletal patterning was quite similar, the frequencies were significantly different. Only 25% (6/24) of these heterozygotes exhibited a large anterior frontanel. Growth retardation was also milder than that seen in the BALB/c × C57BL/6-CBA background, but the heterozygotes were significantly smaller compared with wild-type fetuses. In addition, the frequency of abnormalities in the sternum was almost the same as that of the wild-type fetuses. Thus, the penetrance rate of abnormalities of the frontanel and the sternum was significantly lower compared with the rates observed in fetuses of a BALB/c × C57BL/6-CBA background. In contrast, the abnormality in the xiphoid process occurred more frequently, being observed in two-thirds (16/24) of the heterozygotes. The frequency of abnormalities in the axial bones, ribs, and limbs was low.
Decreased Bmp7 Expression in the Cbp Heterozygous Mutants.
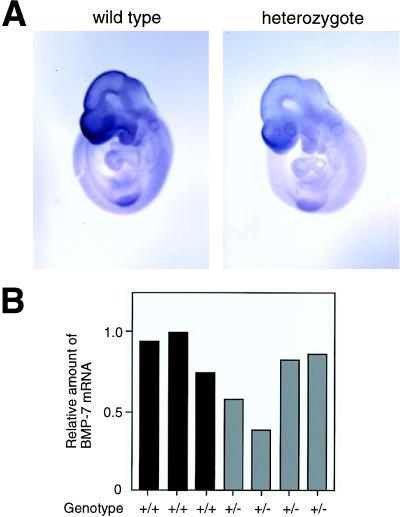
In Drosophila, dCBP is required for Hh-dependent expression of dpp, the Drosophila homologue of transforming growth factor β/BMP (31). Because the ectopic Sonic hedgehog (Shh) expression in the chicken limb locally induces Bmp2 (41, 42), this signaling cascade appears to be conserved during evolution, supporting the view that CBP also could be involved in the Shh-dependent induction of Bmp. However, compared with three dpp-related genes in Drosophila (dpp, screw, and 60A), more than 20 BMPs exist in vertebrates, and relatively little information is available on the comparative expression patterns on these BMPs. Although the information of mutant mice lacking Bmp is limited, some skeletal abnormalities in the sternum and rib found in the Cbp heterozygotes also were observed in the Bmp7- and Bmp5-deficient homozygotes (43, 44). In addition, similar abnormal skeletal patterning was seen in the Gli2- or Gli3-deficient homozygous mutant mice, and Gli3 heterozygotes (45, 46). Therefore, we examined the expression level of Bmp7 in the Cbp heterozygous mutants as one of the possible target genes whose expression could be regulated by CBP. The signal for Bmp7 mRNA detected by whole-mount in situ hybridization in the 9.5- and 10.5-dpc fetuses was significantly reduced in four of 10 heterozygotes of the BALB/c × C57BL/6-CBA background (Fig. 4A). In contrast, a similar decrease in expression was observed in only one of 27 wild-type fetuses. Northern blot analysis confirmed decreased Bmp7 expression in two of four heterozygotes (Fig. 4B). Thus, the level of Bmp7 expression was significantly reduced in the Cbp heterozygous mutants.
Figure 4.
Reduced expression of BMP-7 in the Cbp heterozygotes. (A) Detection of BMP-7 mRNA in the 10.5-dpc fetuses. Whole-mount in situ hybridization was done using the wild-type and Cbp heterozygous fetuses obtained by mating with BALB/c mice. Typical examples of the decreased and normal expression of Bmp7 in the heterozygotes and wild-type fetuses, respectively, are shown. (B) Northern blot analysis of BMP-7 mRNA. Northern blotting was done using the RNA samples from the wild-type and heterozygotes of 11.5 dpc. The amount of BMP-7 mRNA was normalized with respect to the β-actin mRNA level, and the relative amount is indicated as a bar graph.
DISCUSSION
Our study has demonstrated that a 50% decrease in the amount of CBP protein causes various skeletal abnormalities, and thereby provides direct evidence that loss of one copy of the Cbp gene affects pattern formation. We have carefully analyzed the expression of the truncated form of CBP in the heterozygous mutants, because the expression of a dominant negative form could lead to misinterpretation of the results. We failed to detect any truncated form of CBP specifically expressed in the heterozygous mutants, supporting the notion that loss of one copy of the Cbp gene really affects pattern formation. The 265-kDa CBP protein was easily detected in Western blotting using a small amount of lysates from the embryonic brain, indicating that the amount of CBP is much higher than that of many of the enhancer-binding transcription factors such as CREB. However, the abnormal pattern formation in the Cbp heterozygotes indicates that the amount of CBP protein in vivo is not excessive, but rather limiting. Because we observed that overexpression of dCBP in Drosophila embryos causes their death, the amount of CBP may be tightly regulated. Kamei et al. (13) suggested in a recent report that glucocorticoid inhibits AP-1 activity by recruiting limiting amounts of CBP to the glucocorticoid receptor. To examine whether this kind of regulation, involving competition between CBP-utilizing factors for binding to limiting amounts of CBP, really occurs in vivo, Cbp heterozygous mutants expressing half of the wild-type level of CBP should be extremely useful.
In the Cbp heterozygous mutants, the level of Bmp7 expression was obviously reduced. Although it is not clear whether CBP directly affects the transcription of Bmp7 gene, this raises the possibility that CBP contributes to the expression of dpp/Bmp in both Drosophila and mouse, and that a conserved signaling pathway is used for pattern formation in insects and mammals. However, it is unlikely that the 50% reduction in Bmp7 expression levels can account for the bone defects, because the heterozygous Bmp7 mice do not show any skeletal defects (43). In addition, the Bmp7-deficient homozygous mice show different skeletal defects than the one observed in the Cbp heterozygous mice, although some skeletal abnormalities, such as asymmetric pairing of ribs and malformation of the xiphoid processes, were observed in both Cbp heterozygotes and Bmp7-deficient homozygotes (43, 47). For instance, Bmp7 homozygous mutants show preaxial polydactyly, whereas the Cbp heterozygotes show a reduced number of digits. Furthermore, a defect in the patterning of the axial skeleton was observed in the Cbp heterozygotes, but not in the Bmp7 homozygotes. These results could be explained by the speculation that the activity of multiple BMPs is partially reduced in the Cbp heterozygotes, although we have examined the expression level of only Bmp7 among many members of the Bmp gene family. In addition to the decreased activity of various BMPs, some other target gene(s) could be affected by loss of one copy of Cbp. In fact, we recently found that not only the Ci activity but also the Dorsal activity, a Drosophila homologue of transcription factor NF-κB, is decreased in the dCBP mutant (48). Due to the decreased Dorsal activity, the twist (twi) expression, which is a target gene of Dorsal, is blocked in the dCBP mutant. If this signaling pathway also is conserved between Drosophila and vertebrates, the level of twi expression could be reduced in the Cbp heterozygotes. Loss of one copy of the human twi gene causes Saethre–Chotzen syndrome, which is an autosomal dominant craniosynostosis with brachdactyly (49, 50), raising the possibility that a partial decrease in the twi expression in the Cbp heterozygotes also may contribute to the skeletal abnormalities. Thus, the phenotype of Cbp heterozygotes may be resulted from a reduced expression of various target genes of CBP.
The frequencies of the various anomalies in the Cbp heterozygotes were different depending on the genetic background, indicating that the nature of the genetic background may potentiate or attenuate the Cbp phenotype. Thus, the genetic background of patients suffering from RTS also might be partly responsible for the high degree of diversity of RTS-associated symptoms. Although certain malformations, such as an anterior frontanel, were seen in both Cbp heterozygous mutants and RTS patients, the most obvious anomalies, such as broad thumbs and broad halluces, were not observed in the heterozygous mice. These latter malformations have been reported in 87% and 100% of RTS patients in the Netherlands, respectively (24). Although this also may be a reflection of the effect of the genetic background, it is possible that mutations in other unidentified gene(s) also contribute to the phenotype of RTS patients. Nevertheless, our data obtained by using the Cbp heterozygotes gives a clue to understanding the molecular mechanism of RTS.
Acknowledgments
We thank Shin-ich Aizawa for the TT2 genomic library, Richard Goodman for mouse CBP cDNA, Elizabeth Robertson and Brigid Hogan for mouse BMP-7 cDNA, and Tamio Hirabayashi for encouragement.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: BMP, bone morphogenetic protein; CBP, CREB-binding protein; CREB, cyclic AMP response element-binding protein; dCBP, Drosophila homologue of CBP; dpc, days postcoitus; ES, embryonic stem; RTS, Rubinstein–Taybi syndrome.
References
- 1.Chrivia J C, Kwok R P S, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 2.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G R, Green M R, Goodman R H. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 3.Kee B L, Arias J, Montminy M R. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 4.Yang X-J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M R. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 6.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 7.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 8.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 9.Arias J, Alberts A S, Brindle P, Claret F X, Smeal T, Karin M, Feramisco J, Montminy M. Nature (London) 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 10.Bannister A J, Kouzarides T. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai P, Akimaru H, Tanaka Y, Hou D-X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 12.Oelgeschläger M, Janknecht R, Krieg J, Schreek S, Lüscher B. EMBO J. 1996;15:2771–2780. [PMC free article] [PubMed] [Google Scholar]
- 13.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 14.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I G, Juguilon H, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 15.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D’Andrea A, Livingston D M. Nature (London) 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 16.Eckner R, Yao T-P, Oldread E, Livingston D M. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 17.Puri P L, Avantaggiati M L, Balsano C, Sang N, Graessmann A, Giordano A, Levrero M. EMBO J. 1997;16:369–383. doi: 10.1093/emboj/16.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sartorelli V, Huang J, Hamamorti Y, Kedes L. Mol Cell Biol. 1997;17:1010–1026. doi: 10.1128/mcb.17.2.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckner R, Ewen M E, Newsome D, Gerdes M, DeCapiro J A, Lawrence J B, Livingston D M. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 20.Lundblad J R, Kwok R P S, Laurance M E, Harter M L, Goodman R H. Nature (London) 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 21.Arany Z, Newsome D, Oldread E, Livingston D M, Eckner R. Nature (London) 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 22.Petrij F, Giles R H, Dauwerse H G, Saris J J, Hennekam R C M, Masuno M, Tommerup N, van Ommen G B, Goodman R H, Peters D J M, Breuning M H. Nature (London) 1995;376:348–351. doi: 10.1038/376348a0. [DOI] [PubMed] [Google Scholar]
- 23.Rubinstein J H, Taybi H. Am J Dis Child. 1963;105:588–608. doi: 10.1001/archpedi.1963.02080040590010. [DOI] [PubMed] [Google Scholar]
- 24.Hennekam R C M, van den Boogaard M J, Sibbles B J, van Spijker H G. Am J Med Genet Suppl. 1990;6:17–29. doi: 10.1002/ajmg.1320370604. [DOI] [PubMed] [Google Scholar]
- 25.Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva A J. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 26.Hilberg F, Aguzzi A, Howells N, Wagner E F. Nature (London) 1993;365:179–181. doi: 10.1038/365179a0. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z-Q, Ovitt C, Grigoriadis A E, Möhle-Steinlein U, Rüther U, Wagner E F. Nature (London) 1992;360:741–745. doi: 10.1038/360741a0. [DOI] [PubMed] [Google Scholar]
- 28.Johnson R S, Spiegelman B M, Papaioannou V. Cell. 1992;71:577–586. doi: 10.1016/0092-8674(92)90592-z. [DOI] [PubMed] [Google Scholar]
- 29.Mucenski M L, McLain K, Kier A B, Swedlow S H, Schreiner C M, Miller T A, Pietryga D W, Scott W J, Potter S S. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 30.Lohnes D, Mark M, Mendelsohn C, Dollé P, Dierich A, Gorry P, Gansmuller A, Chambon P. Development (Cambridge, UK) 1994;120:2723–2748. doi: 10.1242/dev.120.10.2723. [DOI] [PubMed] [Google Scholar]
- 31.Akimaru H, Chen Y, Dai P, Hou D-X, Nonaka M, Smolik S M, Armstrong S, Goodman R, Ishii S. Nature (London) 1997;386:735–738. doi: 10.1038/386735a0. [DOI] [PubMed] [Google Scholar]
- 32.Domínguez M, Brunner M, Hafen E, Basler K. Science. 1996;272:1621–1625. doi: 10.1126/science.272.5268.1621. [DOI] [PubMed] [Google Scholar]
- 33.Kinzler K W, Vogelstein B. Mol Cell Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orenic T V, Slusarski D C, Kroll K L, Holmgren R A. Genes Dev. 1990;4:1053–1067. doi: 10.1101/gad.4.6.1053. [DOI] [PubMed] [Google Scholar]
- 35.Hui C-C, Joyner A L. Nat Genet. 1993;3:241–245. doi: 10.1038/ng0393-241. [DOI] [PubMed] [Google Scholar]
- 36.Vortkamp A, Gessler M, Grzeschik K-H. Nature (London) 1991;352:539–540. doi: 10.1038/352539a0. [DOI] [PubMed] [Google Scholar]
- 37.Yagi T, Nada S, Watanabe N, Tamemoto H, Kohmura N, Ikawa Y, Aizawa S. Anal Biochem. 1993;214:77–86. doi: 10.1006/abio.1993.1459. [DOI] [PubMed] [Google Scholar]
- 38.Inouye M. Congenital Anomalies. 1976;16:171–173. [Google Scholar]
- 39.Masuya H, Sagai T, Wakana S, Moriwaki K, Shiroishi T. Genes Dev. 1995;9:1645–1653. doi: 10.1101/gad.9.13.1645. [DOI] [PubMed] [Google Scholar]
- 40.Yagi T, Tokunaga T, Furuta Y, Nada S, Yoshida M, Tsukada T, Saga Y, Takeda T, Ikawa Y, Aizawa S. Anal Biochem. 1993;214:70–76. doi: 10.1006/abio.1993.1458. [DOI] [PubMed] [Google Scholar]
- 41.Francis P H, Richardson M K, Brickell P M, Tickle C. Development (Cambridge, UK) 1994;120:209–218. doi: 10.1242/dev.120.1.209. [DOI] [PubMed] [Google Scholar]
- 42.Laufer E, Nelson C E, Johnson R L, Morgan B A, Tabin C. Cell. 1994;79:993–1003. doi: 10.1016/0092-8674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 43.Luo G, Hofmann C, Bronckers A L J J, Sohocki M, Bradley A, Karsenty G. Genes Dev. 1995;9:2808–2820. doi: 10.1101/gad.9.22.2808. [DOI] [PubMed] [Google Scholar]
- 44.Kingsley D M, Bland A E, Grubber J M, Marker P C, Russel L B, Copeland N G, Jenkins N A. Cell. 1992;30:399–410. doi: 10.1016/0092-8674(92)90510-j. [DOI] [PubMed] [Google Scholar]
- 45.Mo R, Freer A M, Zinyk D L, Crackkower M A, Michaud J, Heng H H-Q, Chik K W, Shi X-M, Tsui L-C, Cheng S H, Joyner A L, Hui C-C. Development (Cambridge, UK) 1997;124:113–123. doi: 10.1242/dev.124.1.113. [DOI] [PubMed] [Google Scholar]
- 46.Johnson D R. J Embryol Exp Morph. 1967;17:543–581. [PubMed] [Google Scholar]
- 47.Dudley A T, Lyons K M, Robertson E J. Genes Dev. 1995;9:2795–2807. doi: 10.1101/gad.9.22.2795. [DOI] [PubMed] [Google Scholar]
- 48.Akimaru, H., Hou, D.-X. & Ishii, S. (1997) Nat. Genet., in press. [DOI] [PubMed]
- 49.Howard T D, Paznekas W A, Green E D, Chiang L C, Ma N, Luna R I O D, Delgado C G, Gonzalez-Ramos M, Kline A D, Jabs E W. Nat Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- 50.Ghouzzi V E, Merrer M L, Perrin-Schmit F, Lajeunie E, Benit P, Renier D, Bourgeois P, Bolcato-Bellemin A-L, Munnich A, Bonaventure J. Nat Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]