Abstract
Eukaryotic genome similarity relationships are inferred using sequence information derived from large aggregates of genomic sequences. Comparisons within and between species sample sequences are based on the profile of dinucleotide relative abundance values (The profile is ρ*XY = f*XY/f*Xf*Y for all XY, where f*X denotes the frequency of the nucleotide X and f*XY denotes the frequency of the dinucleotide XY, both computed from the sequence concatenated with its inverted complement). Previous studies with respect to prokaryotes and this study document that profiles of different DNA sequence samples (sample size ≥50 kb) from the same organism are generally much more similar to each other than they are to profiles from other organisms, and that closely related organisms generally have more similar profiles than do distantly related organisms. On this basis we refer to the collection {ρ*XY} as the genome signature. This paper identifies ρ*XY extremes and compares genome signature differences for a diverse range of eukaryotic species. Interpretations on the mechanisms maintaining these profile differences center on genome-wide replication, repair, DNA structures, and context-dependent mutational biases. It is also observed that mitochondrial genome signature differences between species parallel the corresponding nuclear genome signature differences despite large differences between corresponding mitochondrial and nuclear signatures. The genome signature differences also have implications for contrasts between rodents and other mammals, and between monocot and dicot plants, as well as providing evidence for similarities among fungi and the diversity of protists.
Local and global compositional heterogeneity is recognized on many scales in eukaryotic genomes, including variation in G+C content (e.g., isochore compartments, coding vs. noncoding), mobile insertion elements, characteristic centromeric satellite and telomeric repeats, CpG (=CG) suppression in vertebrates, and methylation patterns. Our recent studies of genomic sequence data have demonstrated that (i) the dinucleotide relative abundance values (defined below) of different sequence samples of DNA from the same organism are generally much more similar to each other than they are to sequence samples from different organisms and (ii) related organisms generally have more similar dinucleotide relative abundance values than do distantly related organisms (1). Dinucleotide relative abundance values are equivalent to the “general designs” derived from biochemical nearest-neighbor frequency analysis (2, 3). These highly stable DNA doublet forms suggest that there may be genome-wide factors, such as functions of the replication and repair machinery, context-dependent mutation rates, DNA modifications, and base-step conformational tendencies that impose limits on the compositional and structural patterns of a genomic sequence. The set of dinucleotide relative abundance values constitutes a “genomic signature” (1, 4) that may reflect the influence of such factors.
Relative Abundance Values
A standard assessment of dinucleotide bias is through the odds ratio ρXY = fXY/fXfY, where fX denotes the frequency of the nucleotide X and fXY denotes the frequency of the dinucleotide XY. The formula for ρXY is modified for double-stranded DNA by calculating the odds ratio ρ*XY for the given DNA sequence concatenated with its inverted complementary sequence. The set of ρ*XY values is referred to as the dinucleotide relative abundance profile.
A measure of difference between two sequences f and g (from different organisms or from different regions of the same genome) is the average absolute dinucleotide relative abundance difference calculated as δ*(f, g) = 1/16 ∑XY|ρ*XY(f) − ρ*XY(g)|, where the sum extends over all dinucleotides.
DNA Sequence Samples
Large collections of genomic DNA sequences were extracted from GenBank (Release 101.0, June 1997) for 32 diverse eukaryotic organisms (see legend to Table 1). We restricted attention to species for which at least 100 kb of nonredundant sequence data was available. Most sequence aggregates include several contigs ≥10 kb and often ≥30 kb. Apart from complete chromosomes, the sequences in each species ensemble were culled of duplications. It appears that simple eukaryotes like yeast, nematodes, and Arabidopsis consist mostly of closely juxtaposed gene sequences interrupted with rare mobile DNA elements. The current human collection (all contigs ≥80 kb) tends to be biased toward genes of medical interest. The D. melanogaster data set is replete with developmental genes. The data collections are certainly unrepresentative of the scope of invertebrate, plant, and protist taxa. For example, the dicots, apart from A. thaliana, cover only a few species and the monocots are restricted to a few grasses.
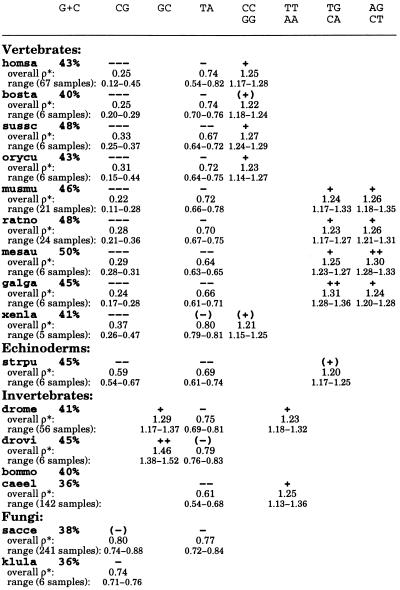
Table 1.
Genome signature extremes for diverse eukaryotes
Each sequence collection was cleaned by removing duplicate copies of the same gene as identified in the sequence annotations. Species names are abbreviated as follows: Vertebrates: homsa (Homo sapiens, human, aggregate sequence length 3,326 kb, minimum contig size 80 kb); bosta (Bos taurus, bovine, 136, 10); sussc (Sus scrofa, pig, 142, 10); orycu (Oryctolagus cuniculus, rabbit, 165, 5); musmu (Mus musculus, mouse, 1,065, 50); ratno (Rattus norvegicus, rat, 656, 10); mesau (Mesocricetus auratus, hamster, 101, 5); galga (Gallus gallus, chicken, 297, 10); xenla (Xenopus laevis, African clawed frog, 251, 5). Echinoderms: strpu (Strongylocentrotus purpuratus, sea urchin, 127, 3). Invertebrates: drome (Drosophila melanogaster, 2,779, 50); drovi (Drosophila virilis, 162, 5); bommo (Bombyx mori, silkworm, 140, 4); caeel (Caenorhabditis elegans, chromosomes II and III). Fungi: sacce (Saccharomyces cerevisiae, complete genome); klula (Kluyveromyces lactis, 117, 3); canal (Candida albicans, 165, 3); schpo (Schizosaccharomyces pombe, 1,582, 20); neucr (Neurospora crassa, 148, 5); emeni (Emericella nidulans, 222, 5); aspni (Aspergillus niger, 119, 3); ustma (Ustilago maydis, 100, 3). Plants: arath (Arabidopsis thaliana, thale cress, 1,989, 80); nicta (Nicotiana tabacum, tobacco, 98, 5); soltu (Solanum tuberosum, potato, 138, 3); solly (Solanum lycopersicum, tomato, 135, 5); zeama (Zea mays, maize, 287, 5); horvu (Hordeum vulgare, barley, 114, 5); orysa (Oryza sativa, rice, 197, 5). Protists: plafa (Plasmodium falciparum, malaria parasite, 260, 5); trybr (Trypanosoma brucei, 207, 5); dicdi (Dictyostelium discoideum, 100, 5). No extremes for the dinucleotides AT, AC/GT, and GA/TC occur in the species at hand. The ranges were determined from ≈50 kb disjoint sequence samples. For sequence collections of aggregate length ≥≈200 kb, the available contigs were concatenated in a random order and the resulting sequence partitioned into equal lengths of ≈50 kb.
Dinucleotide Compositional Extremes
From statistical theory and data experience, a dinucleotide relative abundance may be conservatively described as significantly low if ρ*XY ≤ 0.78 and significantly high if ρ*XY ≥ 1.23 (4). We distinguish extremes of dinucleotide relative abundances as follows: extremely high, symbolically +++, ρ*XY ≥ 1.50; very high, ++, 1.30 ≤ ρ*XY < 1.50; significantly high, +, 1.23 ≤ ρ*XY < 1.30; marginally high, (+), 1.20 ≤ ρ*XY < 1.23; extremely low, −−−, ρ*XY ≤ 0.50; very low, −−, 0.50 < ρ*XY ≤ 0.70; significantly low, −, 0.70 < ρ*XY ≤ 0.78; marginally low, (−), 0.78 < ρ*XY ≤ 0.81.
The following trends were observed (see Table 1).
(i) TA is broadly underrepresented in eukaryotes (and prokaryotes) generally in the range ρ*TA ≈ 0.61–0.81. The lowest average value of ρ*TA among the eukaryotes is found in the fungus U. maydis (0.50). Interestingly, TA occurrences are in the random (normal) range in animal mitochondrial (mt) and chloroplast genomes (5). Possible reasons for TA underrepresentation may be its low thermodynamic stacking energy, which is the lowest among all dinucleotides (6), the high degree of degradation of UA dinucleotides by ribonucleases in mRNA tracts (7), and the presence of TA as part of many regulatory signals (e.g., TATA box, transcription terminators, and polyadenylation signal AATAAA). In this last context, TA suppression may help to avoid inappropriate binding of regulatory factors.
(ii) CG is the most underrepresented dinucleotide with drastic suppression in vertebrates on the one hand and significant overrepresentation in some α-proteobacterial and halobacterial sequences on the other hand (8, 9). Overall, ρ*CG values in vertebrates range from 0.23 to 0.37. A pronounced CG underrepresentation at a similar level is found in the archaeon Methanococcus jannaschii (0.32) and also in Sulfolobus sequences (8, 9). CG is strongly suppressed in the sea urchin, S. purpuratus (0.59), in various yeasts (S. cerevisiae, K. lactis, and C. albicans), and in dicot plants, but is only marginally low to low normal in monocot plants (Table 1). CG is suppressed in animal mitochondria (these ρ* values are mostly in the range 0.50–0.65), whereas it is in the normal range in plant chloroplast genomes (5). CG has normal representations in insects, worms, and most fungi. CG suppression has usually been ascribed to the classical methylation/deamination/mutation scenario causing mutation of CG to TG/CA. However, this hypothesis cannot account for the pervasive CG suppression in animal mitochondria that lack the standard methylase activity. Moreover, some mammalian genomes and all animal mt genomes have CC/GG high but TG/CA in the normal range suggesting a possible CG → CC/GG mutation bias. We have proposed that CG deficiencies may in some circumstances be due to structural constraints related to high dinucleotide stacking energy, supercoiling, and chromatin packing (1).
(iii) The dinucleotides CC/GG, TG/CA, and AG/CT, all a single base mutation from CG, are (except for dicot plants) only overrepresented in genomes with strong CG suppression. Interestingly, scrutiny of Table 1 reveals that these dinucleotide relative abundances separate rodents possessing TG/CA and AG/CT of significantly high representations and CC/GG in the normal range from the nonrodents (primates, artiodactyls, and lagomorphs) that possess relative high abundances of CC/GG, but TG/CA and AG/CT in the normal range.
(iv) Other dinucleotide biases in eukaryotes include overrepresentation of GC in Drosophila species but apparently not in other higher eukaryotes. GC is significantly abundant in most γ-proteobacteria (8, 9).
(v) No dinucleotide extremes were found in the moth B. mori or in barley (H. vulgare). Protists form a diverse group with no consistent pattern of dinucleotide relative abundances.
Dinucleotide Relative Abundance δ*-Differences Among Eukaryotes
The sequence collections were organized into distinct ≈50 kb sequence samples. The average δ*-differences between samples from the same genome (within-species δ*) or from two different genomes (between-species δ*) are exhibited in Table 2. It is useful to distinguish distinct levels of δ*-differences like random (δ* < 0.018), very close (0.020 < δ* < 0.030), close (0.035 < δ* < 0.050), moderately similar (0.055 < δ* < 0.075), weakly similar (0.080 < δ* < 0.115), distantly similar (0.120 < δ* < 0.150), distant (0.160 < δ* < 0.200), and very distant (δ* > 0.200; cf. ref. 8).
Table 2.
Average δ*-differences (multiplied by 1,000) within and between eukaryotes based on ≈50 kb sequence samples
See legend to Table 1 for the list and sizes of the sequence samples.
The following relations are evident in Table 2.
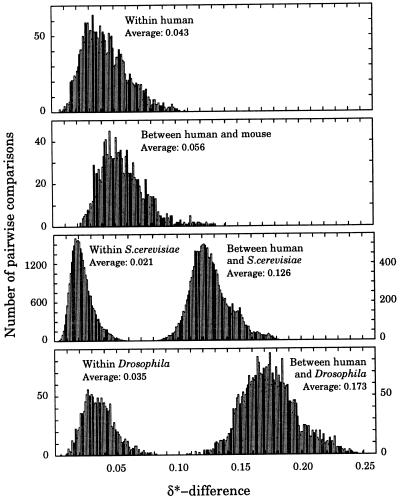
(i) Within-species δ*-differences are with very few exceptions lower than between-species δ*-differences, reflecting the validity of the genome signature (1, 8, 9). The most homogeneous genomes occur among fungi (see Fig. 1 for S. cerevisiae), whereas the most variable genomes are found among protists. The DNA sequence samples of P. falciparum are the most heterogeneous (average within-species δ* = 0.059). Fig. 1 shows distributions of all pairwise δ*-differences between 50 kb sequence samples from several species. The distribution of the δ*-differences between human and mouse sequence samples is only slightly shifted relative to δ*-differences within human sequence samples, reflecting the close relatedness of human and mouse (Fig. 1). On the other hand, the δ*-differences between human and S. cerevisiae, and between human and D. melanogaster are substantially higher than all within-species δ*-differences.
Figure 1.
Distribution of δ*-differences within and between selected species based on all pairwise comparisons of ≈50-kb disjoint sequence samples.
(ii) The δ*-differences were determined for all 50 kb samples between 15 large human contigs each of length at least 80 kb. This collection included Alzheimer disease (STM2) gene (87 kb, on chromosome 1), class II major histocompatibility complex region (198 kb, chromosome 6), T-cell receptor β-chain (685 kb, chromosome 7), tyrosine protein kinase gene (84 kb, chromosome 9), chromosome 12p13 gene cluster (117 kb, chromosome 12), retinoblastoma susceptibility gene (180 kb, chromosome 13), BRCA2 gene region (773 kb, chromosome 13), neurofibromatosis 1 region (101 kb, chromosome 17), breakpoint cluster region (152 kb, chromosome 22), region between the human RCP/GCP and G6PD loci (219 kb, chromosome X), iduronate 2-sulfatase gene region (206 kb, chromosome X), fragile X (FMR-1) gene (153 kb, chromosome X), chromosome X cosmid (106 kb, chromosome X), cosmid J136O17 (85 kb, chromosome X), and cosmid J30E17 (82 kb, chromosome X). The average within-contig δ*-differences range from 0.013 to 0.046 and between contigs range from 0.020 to 0.081, which indicates very close to moderate similarity among the human contigs. The highest δ*-differences are revealed between the breakpoint cluster region 152 kb contig of chromosome 22 and other contigs. The ρ*CG values of these human contigs are drastically low with only one (the 219 kb contig of the X chromosome) above 0.35. This contig is annotated with 17 CpG islands, which may in part explain the marginally higher value ρ*CG = 0.41 versus most values of the other human contigs about 0.25. As with all vertebrates, all contigs are significantly underrepresented in TA at the level ρ*TA from 0.58 to 0.79 and marginally to significantly overrepresented in CC/GG at the level ρ*CC/GG ≈ 1.20–1.26. The dinucleotide relative abundances of TG/CA and AG/CT are also high normal to significantly high. In ref. 10 we introduced the codon signature, defined as the dinucleotide relative abundances at the distinct codon positions {1,2}, {2,3}, and {3,4} (4 = 1 of the next codon). For large collections of genes (50 or more), we found that the codon signature, like the genome signature, is essentially invariant. Moreover, the codon signature in mammals largely parallels the genome signature but also accommodates amino acid constraints (10).
(iii) The available mammals split into two groups: rodents and nonrodents (human, artiodactyls, and rabbit). The mutual δ*-differences within each group are close but δ*-differences between rodents and nonrodent mammals show only moderate similarity, with hamster somewhat more distant from the nonrodents. The chicken and X. laevis sequences are weakly or moderately similar to mammals and the sea urchin is weakly similar. In fact, chicken is somewhat closer to rodents than to nonrodent mammals, whereas X. laevis is somewhat closer to nonrodent mammals than to rodents.
(iv) Insects form a diverse group with mutual δ*-differences in the range 0.071–0.118 (weakly similar). Interestingly, the insects tend to be weakly similar to monocots but generally distantly similar to dicots.
(v) Fungi constitute a coherent group with most δ*-differences in the range from 0.035 to 0.075, close or moderately similar. Exceptions are C. albicans and the smut U. maydis, both distant from other fungi. The δ*-differences between the 16 chromosomes of the yeast S. cerevisiae are remarkably close. The within-chromosome δ*-differences range is 0.018 to 0.024 except for chromosome 1 (average δ*, 0.035), and the between-chromosome range is 0.018 to 0.030, indicating that all chromosomes are very close to each other (see Fig. 1). δ*-differences within and between chromosomes II and III of C. elegans are remarkably similar with mean δ*-differences about 0.038, marginally higher than the within whole yeast genome.
(vi) In the group of plants, the three dicot species (tobacco, potato, and tomato, all in the Solanaceae family) are very close (0.027–0.032) and their δ*-differences to the dicot A. thaliana are at the level of moderate similarity (0.058–0.063). Our samples from monocots are mutually very close or close and only moderately similar to dicots. Plants (equally with respect to both dicots and monocots) are close to S. cerevisiae (δ*-differences 0.036–0.054) and also to the yeast-like K. lactis, but only moderately or weakly similar to other fungi. Thus, in terms of DNA normalized doublet comparisons of whole genomes, fungi appear to be closer to plants than to animals, a result that is at variance with some protein sequence comparisons (11). Dicot plants are weakly similar to mammals (δ*-differences 0.070–0.132). On the other hand, insects are distant or very distant to mammals.
(vii) Protists are a diverse group. All δ*-differences from P. falciparum to other eukaryotes exceed 0.090 and mostly exceed 0.120. T. brucei is moderately similar to some insects [the closest δ*(trybr,bommo) = 0.068], some fungi [δ*(trybr,schpo) = 0.059], and monocot plants (δ*-differences 0.058–0.062). D. discoideum is weakly similar to yeast and dicot plants.
Comparisons of mt Genomes Between Species
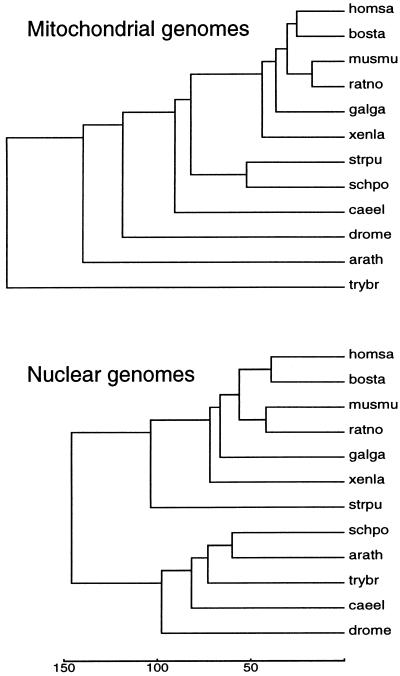
The δ*-differences between various mt-genomes and separately between the nuclear genomes of the corresponding host species are given in Table 3. The principal observation is that levels of similarity among mt genomes assessed by δ*-differences largely parallel levels of evolutionary relatedness assessed by the genome signature δ*-differences among the corresponding host genomes (Fig. 2). That is, species exhibiting small (large) δ*-differences between their nuclear genomes generally have relatively small (large) δ*-differences between corresponding mt genomes and vice versa. However, comparing the mt genomic signature with its host genome signature shows δ*-differences generally distant to very distant (δ* values mostly 0.130–0.230, see Table 3) with no discernable pattern among the various eukaryotic kingdoms.
Table 3.
δ*-differences between mitochondrial and the host nuclear genomes (multiplied by 1,000)
| homsa | bosta | musmu | ratno | galga | xenla | strpu | drome | caeel | sacce | schpo | arath | trybr | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 145 | 25 | 26 | 33 | 36 | 50 | 101 | 139 | 85 | 468 | 76 | 158 | 184 | homsa |
| 37 | 135 | 23 | 40 | 48 | 36 | 91 | 130 | 91 | 457 | 68 | 145 | 188 | bosta |
| 56 | 55 | 153 | 17 | 34 | 37 | 96 | 120 | 87 | 457 | 72 | 154 | 177 | musmu |
| 56 | 52 | 40 | 146 | 26 | 44 | 97 | 112 | 88 | 461 | 72 | 154 | 174 | ratno |
| 69 | 72 | 59 | 59 | 165 | 52 | 93 | 130 | 101 | 487 | 68 | 150 | 183 | galga |
| 61 | 55 | 77 | 75 | 88 | 85 | 81 | 106 | 81 | 465 | 66 | 124 | 172 | xenla |
| 110 | 93 | 108 | 98 | 116 | 89 | 172 | 116 | 98 | 486 | 52 | 94 | 177 | strpu |
| 173 | 169 | 196 | 197 | 182 | 144 | 147 | 175 | 96 | 385 | 116 | 163 | 167 | drome |
| 169 | 153 | 183 | 177 | 172 | 156 | 117 | 99 | 228 | 439 | 92 | 168 | 131 | caeel |
| 126 | 109 | 138 | 136 | 133 | 98 | 78 | 91 | 75 | 527 | 496 | 542 | 388 | sacce |
| 142 | 131 | 157 | 156 | 142 | 113 | 105 | 77 | 72 | 36 | 130 | 84 | 188 | schpo |
| 124 | 103 | 131 | 125 | 132 | 100 | 66 | 122 | 78 | 42 | 59 | 83 | 240 | arath |
| 154 | 147 | 165 | 165 | 152 | 124 | 107 | 90 | 92 | 67 | 59 | 84 | 219 | trybr |
Included are all eukaryotes for which both the complete mitochondrial genome and a sufficient sample of nuclear genomic DNA is available. δ*-differences between mitochondrial genomes are given in the upper right triangle, δ*-difference between the mitochondrial genome and the corresponding host (boldface) are shown in the diagonal, and average δ*-difference between corresponding nuclear genomes (italics) in the lower left triangle.
Figure 2.
Evolutionary tree derived from δ*-differences between mt (Upper) and nuclear (Lower) genomes. The trees were generated by the UPGMA (average linkage) method. The scale (δ*-differences multiplied by 1,000) is shown at the bottom.
Organellar genomes (mitochondria and chloroplasts) are widely accepted as bacterial endosymbionts in that these genomes constitute the remnants of once free-living cellular organisms (12). There is great diversity among mitochondria, including substantial size variation and contrasting patterns of mt genome organization and gene expression relative to animal, plant, fungal, and protist lineages (12). A central unresolved problem concerns whether mt evolution (primary and/or secondary endosymbiont events) is monophyletic or polyphyletic.
The reported D. melanogaster mt genome (19,517 bp) contains about 4 kb (15,566-end) consisting of 12 copies, each about 350 bp long, of the NADH–ubiquinone oxidoreductase chain 6 of unusual composition. This 4 kb section was removed and the remaining sequence used as the adjusted D. melanogaster mt genome. The S. cerevisiae mtDNA (≈78 kb) composition is an extreme anomaly. This is attested to by the exorbitant δ*-differences from all other mt or nuclear genomes (Table 3), in part due to the more than 100 G+C-rich clusters, each about 50 bp long, separated by A+T-rich spacer regions and to the numerous transposable elements. The δ*-differences (Table 3) reflected in the tree (Fig. 2) place Trypanosoma far out. It is possible that the mt–Trypanosoma endosymbiont reflects a distinct event from that of the other organisms or, like the S. cerevisiae endosymbiont, possesses highly anomalous nucleotide organization due to invasion by transposons or other drastic mutational events.
Animal (vertebrate and invertebrate) mt sequences show significant underrepresentations of CG dinucleotides, ρ*CG ≈ 0.40 to 0.60 (13), almost to the same extent as occurs in vertebrate genomic sequences. The adjusted D. melanogaster and (unadjusted) D. yakuba mt genomes entail ρ*CG = 0.73 and 0.68, respectively (13). The fungal S. pombe has ρ*CG = 0.54 typical of animal mitochondria. However, the Podospora anserina fungal mt CG representation is in the normal range. The mt genome of A. thaliana has ρ*CG = 0.73, significantly low. The single persistent significantly high ρ* value occurs for ρ*CC/GG ≥ 1.30 in animal and fungal mt sequences. Intriguingly, the chloroplast genomes are all significantly high in ρ*CC/GG, which is the only consistent extreme among currently available chloroplast sequences (5).
Molecular Evolutionary Implications
Based on comparisons of both the dinucleotide relative abundance extremes (genome signature) and δ*-differences we venture some interpretations of molecular evolutionary relationships among eukaryotic nuclear and mt genomes. In this context, we hypothesize that specificity in replication and repair machinery and context-dependent mutation biases largely maintain the homogeneity of the whole genome of an organism as reflected in the constancy of dinucleotide relative abundances and that differences in this machinery produce the differences in dinucleotide relative abundances among species (1, 9, 14).
(i) Rodents are somewhat exceptional among mammals in δ*-differences. In particular, the nonrodent mammals (human, artiodactyls, rabbit) and rodents (mouse, rat, hamster) constitute two coherent groups but show only moderate intergroup similarity. What can account for this separation? Rodents tend to have a higher mutation rate and shorter generation time than many nonrodent mammals (15). Moreover, it is established that rodents are inefficient in global repair of cyclobutane thymidine dimers compared with humans and probably also in repair of other forms of oxidative DNA damage (16). These differences relate principally to replication and repair mechanisms and context-dependent mutation tendencies, consistent with our hypothesis that such molecular differences could produce the observed differences in the genome signature (1, 9). One might inquire about the reasons for differences in repair proficiency of thymidine dimers between human and rodents. We speculate that the rodents analyzed live in more secluded (often underground or nocturnal) habitats with lesser exposure to sunlight and other sources of radiation damage. In these environments, we could expect that natural selection has attenuated relevant molecular mechanisms required for efficient repair of cyclobutane dimer adducts. However, embryonic rodent and human cells do not show differences in repair processes and transcription-coupled repair processes are largely similar in humans and rodents (16). The relative abundance value of TT/AA in mouse and rat sequences is about 1.06 (1.03 in hamster) compared with 1.12–1.14 in human, cow, pig, and rabbit. On this basis, there are fewer occurrences of TT (thymidine dimer) in rodents compared with nonrodent mammals.
The foregoing discussion suggests a testable hypothesis. There are rodents active in relatively exposed environments, including the grey squirrel and especially capybara. In these cases, one could expect an efficient cyclobutane dimer repair system. On the other hand, purely nocturnal mammals (e.g., the owl monkey) are conceivably inefficient in thymidine dimer adduct repair.
(ii) δ*-differences in the dicots (tobacco, potato, tomato) are mutually close and similarly for the monocots (maize, barley, rice), whereas the monocot and these dicot sequences are only moderately similar (between-group δ*-differences larger by a factor of about two than within-group δ*-differences). Thus, the fundamental monocot/dicot morphological distinction parallels a significant genome signature difference. Intriguingly, the fungi are equally distant to monocots and dicots, whereas the insects are closer to monocots over dicots by a factor of about 1.4. The separation of dicots and monocots (the grasses considered to have arisen from a subgroup of dicots) may have occurred about 200 million years ago (17).
(iii) The genome signature comparisons are in agreement with the classic division of most metazoan phyla into the deuterostomes (e.g., vertebrates, echinoderms) and protostomes (insects, worms), since the sea urchin is weakly similar to vertebrates but the protostomes are distant or very distant from the vertebrates.
(iv) Compositional biases across bacterial genomes were discussed in Karlin et al. (9). The dinucleotide relative abundance values (genomic signature) of bacterial genomes place the Sulfolobus-like sequences (eocyte phylogeny) closer to vertebrates than are all other bacterial genomes, and cyanobacteria closer to fungi and plants (4, 8). Enigmatically Haemophilus influenzae is moderately similar to D. melanogaster sequences.
(v) A challenging question concerns reasons and mechanisms to account for the qualitative concordance between the evolutionary development of host nuclear genomes and the development of mt organelle genomes despite the pronounced difference between the mt and host nuclear genome signatures. The mt and nuclear genomes for animal and fungal organisms use independent DNA polymerase machinery (e.g., γ vs. α, ɛ, δ subunits, respectively). Also, the methods of replication and the nature of the replication origins are fundamentally different. Specifically, the animal and fungal mt transcription-primed replication machinery is distinctive in that most of the heavy strand is synthesized first and the light strand subsequently, whereas the nuclear genomes are replicated analogously to eubacteria synchronized over multiple replication origins.
What about influences of repair processes? There appears to be no DNA excision repair mechanism to deal with cyclobutane dimers in the mitochondrion and apparently bulky lesions are not repaired (18). mtDNA in animals and fungi shows elevated levels of single- and double-strand breaks, mismatches, and generally corrupted base pairings (19). This may be due to a paucity of abasic site correction facilities and mismatch repair capacity in mt genomes (19). Moreover, repair may be less urgent for mt activity because each cell has many mitochondria (hundreds or thousands) and a modicum of impaired organelles may not significantly curtail energy production.
Notably, virtually all mitochondria maintain normal representations of TA dinucleotides, whereas nuclear DNA overwhelmingly tends to have TA in low relative abundance, suggesting that mtDNA may be less thermodynamically stable than nuclear DNA because the dinucleotide TA has the lowest stacking energies compared with all other base steps (6).
Acknowledgments
We thank Drs. B. E. Blaisdell, A. M. Campbell, and C. Burge for helpful comments on the manuscript. This work was supported in part by National Institutes of Health Grants 2R01GM10452–33 and 5R01HG00335–09, and National Science Foundation Grant 9403553–002.
ABBREVIATION
- mt
mitochondrial
References
- 1.Karlin S, Burge C. Trends Genet. 1995;11:283–290. doi: 10.1016/s0168-9525(00)89076-9. [DOI] [PubMed] [Google Scholar]
- 2.Josse J, Kaiser A D, Kornberg A. J Biol Chem. 1961;263:864–875. [PubMed] [Google Scholar]
- 3.Russell G J, Subak-Sharpe J H. Nature (London) 1977;266:533–535. doi: 10.1038/266533a0. [DOI] [PubMed] [Google Scholar]
- 4.Karlin S, Cardon L R. Annu Rev Microbiol. 1994;48:619–654. doi: 10.1146/annurev.mi.48.100194.003155. [DOI] [PubMed] [Google Scholar]
- 5.Karlin S, Campbell A M. Proc Natl Acad Sci USA. 1994;91:12842–12846. doi: 10.1073/pnas.91.26.12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breslauer K J, Frank R, Blöcker H, Marky L A. Proc Natl Acad Sci USA. 1986;83:3746–3750. doi: 10.1073/pnas.83.11.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beutler E, Gelbart T, Han J, Koziol J A, Beutler B. Proc Natl Acad Sci USA. 1989;86:192–196. doi: 10.1073/pnas.86.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlin S, Mrázek J. In: Bacterial Genomes: Physical Structure and Analysis. de Bruijn F J, Lupski J, Weinstock G, editors. New York: Chapman & Hall; 1997. pp. 196–212. [Google Scholar]
- 9.Karlin S, Mrázek J, Campbell A M. J Bacteriol. 1997;179:3899–3913. doi: 10.1128/jb.179.12.3899-3913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlin S, Mrázek J. J Mol Biol. 1996;262:459–472. doi: 10.1006/jmbi.1996.0528. [DOI] [PubMed] [Google Scholar]
- 11.Baldauf S L, Palmer J D. Proc Natl Acad Sci USA. 1993;90:11558–11562. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray M W. Int Rev Cytol. 1992;141:233–357. doi: 10.1016/s0074-7696(08)62068-9. [DOI] [PubMed] [Google Scholar]
- 13.Cardon L, Burge C, Clayton D A, Karlin S. Proc Natl Acad Sci USA. 1994;91:3799–3803. doi: 10.1073/pnas.91.9.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaisdell B E, Campbell A M, Karlin S. Proc Natl Acad Sci USA. 1996;93:5854–5859. doi: 10.1073/pnas.93.12.5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li W-H, Ellsworth D L, Krushkal J, Chang B H-J, Hewett-Emmett D. Mol Phylogenet Evol. 1996;5:182–187. doi: 10.1006/mpev.1996.0012. [DOI] [PubMed] [Google Scholar]
- 16.Bohr V A, Phillips D H, Hanawalt P C. Cancer Res. 1987;47:6426–6436. [PubMed] [Google Scholar]
- 17.Savard L, Li P, Strauss S H, Chase M W, Michaud M, Bousquet J. Proc Natl Acad Sci USA. 1994;91:5163–5167. doi: 10.1073/pnas.91.11.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shadel G S, Clayton D A. Annu Rev Biochem. 1997;66:409–434. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- 19.Yakes F M, Van Houten B. Proc Natl Acad Sci USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]