Abstract
The RNA phage Qβ requires for the replication of its genome an RNA binding protein called Qβ host factor or Hfq protein. Our previous results suggested that this protein mediates the access of replicase to the 3′-end of the Qβ plus strand RNA. Here we report the results of an evolutionary experiment in which phage Qβ was adapted to an Escherichia coli Q13 host strain with an inactivated host factor (hfq) gene. This strain initially produced phage at a titer ≈10,000-fold lower than the wild-type strain and with minute plaque morphology, but after 12 growth cycles, phage titer and plaque size had evolved to levels near those of the wild-type host. RNAs isolated from adapted Qβ mutants were efficient templates for replicase without host factor in vitro. Electron microscopy showed that mutant RNAs, in contrast to wild-type RNA, efficiently interacted with replicase at the 3′-end in the absence of host factor. The same set of four mutations in the 3′-terminal third of the genome was found in several independently evolved phage clones. One mutation disrupts the base pairing of the 3′-terminal CCCoh sequence, suggesting that the host factor stimulates activity of the wild-type RNA template by melting out its 3′-end.
Bacteriophage Qβ RNA (4217 nucleotides) is replicated by Qβ replicase in a two-step process involving, first, the synthesis of complementary minus strands, which in a second step direct the synthesis of more viral plus strands (reviewed in refs. 1 and 2). Plus strand synthesis from the minus strand template takes place efficiently by the core form of replicase (3–7) consisting of the three subunits β (phage-coded), γ, and δ (the host elongation factors EF-Tu and EF-Ts). In contrast, efficient synthesis of minus strands from the plus strand template requires the presence of replicase holoenzyme containing the additional host-derived α subunit (ribosomal protein S1) and of an RNA binding protein (an oligomer of 11-kDa subunits) called host factor (8–11) or Hfq protein. In our previous work, we found that protein S1 mediates the recognition of Qβ plus strand by replicase via binding interactions at two internal RNA sites (the M and the S site) but that the interaction at the 3′-end, where minus strand synthesis begins, is mediated by the host factor protein (12–14).
In the host cell, the Hfq protein functions as a pleiotropic regulator of many genes, including the rpoS σ factor (15–17) and the DNA repair gene mutS (G. Feng, H.-C.T.T., and M.E.W., unpublished work). Its function appears to be translational (15, 16) and may involve interactions with mRNA secondary structures near the ribosomal binding sites (17, 18). An Escherichia coli strain carrying an inactivating insertion in the hfq gene was described recently (19). It was found to support the replication of phage Qβ at a level 5000-fold lower than the parental wild-type strain (20). Because of their high mutation rates (≈10−4; ref. 21), RNA phages have an amazing facility of evolutionary recovery after a variety of site-directed mutational changes (see, for example, ref. 22). We therefore undertook this study to test if the efficiency of Qβ replication in the hfq mutant would improve spontaneously during repeated growth cycles, in the hope that the expected adaptive mutations might yield clues on the mechanism of action of Hfq in Qβ RNA replication.
MATERIALS AND METHODS
The host factor-deficient E. coli strain NU426 hfq1 has been described (19). The Hfr strain E. coli Q13 hfq1 was constructed by phage P1vir transduction (23) from strain TX2808 (19) into E. coli Q13 (24).
Phage RNA was prepared as described (25). Template activity of the RNAs with replicase in vitro was determined, in the presence or absence of host factor, by two-step replication assays measuring the efficiency of initiation of minus strand synthesis (14). Initiation was allowed to take place for 45 s at 37°C in the presence of GTP and ATP, followed by elongation of the product RNA to full size in the presence of CTP, [α-32P]UTP, and polyethylene sulfonate, an inhibitor of initiation (26).
Electron microscopy of phage RNA complexes with replicase or host factor was carried out as described (13, 14, 27).
Full-length, infectious cDNA clones were obtained from the mutant RNA preparations by reverse transcription and PCR using rTth DNA polymerase (Perkin-Elmer) according to the protocol provided by the supplier. The cDNAs were inserted into a vector derived from plasmid pBRT7Qβ (13). Sequencing was carried out on an Applied Biosystems 310 Genetic Analyzer.
Secondary structure models were generated using the program mfold of the GCG software. The 3′-terminal foldings of wild-type and mutant sequences were analyzed over several segment lengths up to sizes of 1317 nt, with consistent results for the structures in the terminal 160 nt.
RESULTS AND DISCUSSION
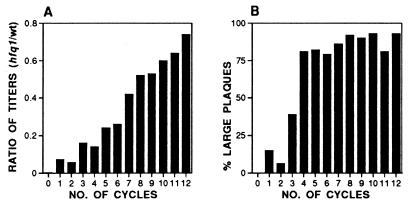
To start the evolutionary experiment with a homogeneous Qβ nucleotide sequence, we derived the initial phage preparation from a cloned, infectious genomic Qβ plasmid (28). Phage were recovered from the supernatants of three cultures derived from single colonies of the F− strain NU426 hfq1 harboring the infectious genomic plasmid pBRT7Qβ and were plated on the Hfr strain Q13 hfq1. Two of the resulting minute plaques from each culture (labeled 1.1, 1.2, 2.1, 2.2, 3.1, and 3.2) were amplified by confluent lysis on plates, followed by 11 lytic cycles in liquid culture at a multiplicity of infection of ≈5. For cultures 1.1 and 1.2, the total phage populations generated in cycles 2–10 rose from 7 × 1010 to 7 × 1011. Cultures 2.1, 2.2, 3.1, and 3.2 were grown at higher dilutions, and the respective population numbers ranged from 3 × 109 to ≈1011. In the last two cycles, all six cultures were scaled up to population sizes of approximately 5 × 1013. The ratio of phage titers determined in parallel on the hfq1 and the wild-type host was initially about 10−4 and rose in the successive lysates to values between 0.4 and 0.8 (Fig. 1A). Similarly, the plaque size increased on the hfq1 host to a range approaching that found on wild-type Q13 (Fig. 1B). As judged from these parameters, the course of the adaptation was similar in the six amplification experiments. The titers reached with the adapted phage after infection of wild-type host cells were two to three times lower than after infection with wild-type Qβ under identical conditions, indicating that the adaptation caused a moderate loss of fitness with regard to growth on the wild-type host.
Figure 1.
Course of the adaptation as followed by plaque-forming characteristics. The data shown are from adaptation experiment 3.2 and are representative for all others. (A) Relative plating efficiency. Ratios of the phage titers as determined on E. coli Q13 hfq1 and on wild-type E. coli Q13 for the successive lytic cycles. In the initial lysate (cycle 0) this ratio was 10−4. (B) Plaque morphology. Percentage of plaques with a diameter of 1 mm or more on E. coli Q13 hfq1 in the successive lytic cycles. For wild-type phage plated on wild-type E. coli Q13, this fraction is 95–100%.
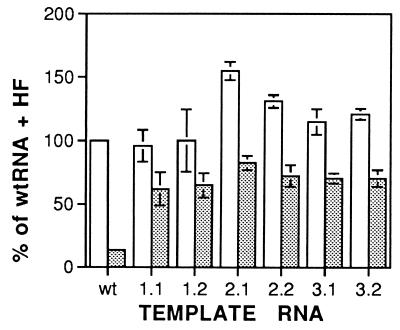
Genomic RNAs were prepared from the six evolved phage populations and were tested for activity as templates for Qβ replicase in vitro by assays measuring the amount of initiation of minus strand RNA synthesis in the presence and absence of host factor. As shown in Fig. 2, the template efficiency of the evolved Qβ RNAs in the presence of host factor is similar or slightly higher than that of wild-type Qβ RNA. However, whereas omission of the host factor reduces the template activity of wild-type RNA to 8–15%, the reduction on the evolved RNA templates is much slighter, to 50–65%, indicating that the evolved Qβ RNAs are much better replicase templates in the absence of host factor than wild-type RNA. The replicase preparation used originated from cells expressing the wild-type replicase gene (29), so the observed difference in template characteristics must be due to changes in the RNA sequence and structure.
Figure 2.
Template activity of adapted phage RNAs with Qβ replicase in the presence and absence of host factor in vitro. Empty columns, with host factor; stippled columns, without host factor. The data are averages of at least four determinations. SD are indicated by error bars. The 100% value of the wild-type Qβ RNA template corresponds to a fraction of 6.2% of enzyme molecules initiating synthesis in the 45-s initiation period.
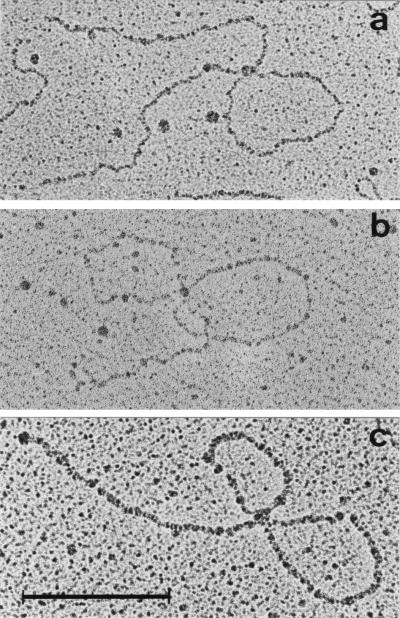
Wild-type Qβ RNA interacts with Qβ replicase mainly at two internal sites called the “S site” and the “M site,” located within the segments comprising nucleotides 1247–1344 and 2545–2867 from the 5′-end, respectively (12). These interactions can be visualized after chemical crosslinking by electron microscopy using a protein-free spreading method (27, 13, 14). The resulting Qβ RNA-replicase complexes appear at a frequency of ≈30% as filamentous structures consisting of an interior loop and two tails of approximate equal length (Fig. 3A). Complexes with two loops in which an RNA end (probably the 3′-end) is bound to replicase are found relatively rarely (0–8%). In contrast, host factor frequently (15–30%) forms double-looped complexes (Fig. 3B), in which the M and S sites (or adjacent sites) are linked to the protein together with a terminal site identified as the 3′-end (13, 14). These findings suggested that the role of the host factor consists of mediating the interaction of replicase with the 3′-end of the plus strand, where minus strand synthesis initiates. We now analyzed replicase complexes with Qβ RNAs adapted to host factor-less host cells (only samples 1.1 and 1.2) by electron microscopy. We found 20% double-looped and 20% single-looped structures with sample 1.1 and 30% double-looped and 18% single-looped structures with sample 1.2. Within the accuracy of the determination, the sizes of loops and tails of mutant RNAs were identical to those described for the wild type (13, 14). A representative picture of a double-looped replicase complex from RNA sample 1.2 is shown in Fig. 3C. Evidently replicase interacts with the 3′-end of the adapted Qβ RNA much more readily than with that of the wild type, in agreement with the suggested role of host factor.
Figure 3.
Electron micrographs of the frequently observed species of double-looped RNA–protein complexes. (A) Replicase with wild-type Qβ RNA. (B) Host factor with wild-type Qβ RNA. (C) Replicase with adapted Qβ RNA (sample 1.2). (Bar = 700 nt.)
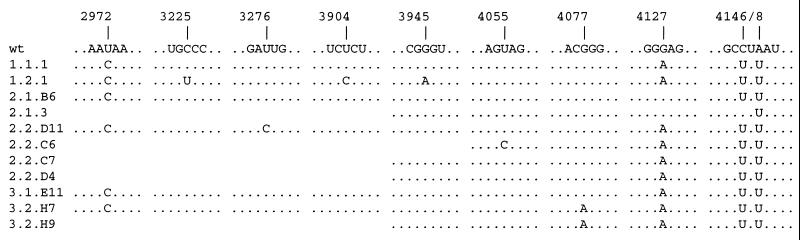
To identify the mutations responsible for the adapted phage phenotype, full length, infectious cDNA plasmids were constructed by reverse transcription and PCR amplification of each of the six separately adapted Qβ RNA preparations. The relevant sequence changes were mapped to the genome region downstream of position 2940 by exchanging cDNA segments of increasing length from the 5′-end against those of wild-type Qβ and by observing the plaque phenotypes. The complete sequence between position 2940 and the 3′-end was determined for one representative clone of each of the six cDNAs, and a number of additional clones was partially sequenced. Except for two isolates exhibiting only a partially adapted plaque phenotype (2.1.B6 and 2.1.3), all sequences (Fig. 4) showed the same set of four base substitutions, along with a few additional scattered point mutations in some of the clones. The experiment had been carried out in triplicate, but completely independently, from three identical plasmid clones, so the four common substitutions must have been selected independently in the three groups of isolates. This established the adaptation as a response of surprising specificity. The fact that both plasmid isolates derived from experiment 2.1 exhibited an incomplete adaptation may be due to a sampling coincidence because neither the plaque phenotype nor the template activity data of this experiment differed significantly from those of the other five. The finding, however, illustrates a heterogeneity in the adapted phage populations, as expected in view of their high mutation rate (21, 30).
Figure 4.
Mutations identified in adapted Qβ phage RNAs by sequencing of cDNA clones.
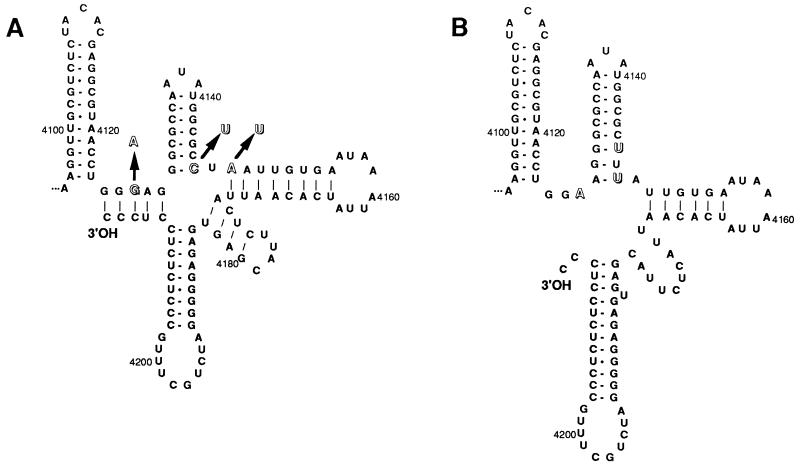
The possible consequences of the three mutations near the 3′-end of the RNA from the adapted phages on the 3′-terminal secondary structure were evaluated using the program mfold of the GCG software. These calculations indicated that, in the wild-type RNA, the essential 3′-terminal CCCoh sequence and two preceding nucleotides are engaged in long range base pairing with nucleotides 4125–4129 as shown in Fig. 5A, an interaction supported by sequence comparison with other phage RNAs (31, 32). The three mutations G4127A, C4146U, and A4148U cause substantial changes in the 3′-terminal secondary structure (Fig. 5B), most notably the destabilization of the 3′-terminal long range interaction and the release of the two terminal nucleotides in an unpaired form. In fact, the G4127A mutation alone would be sufficient to destabilize the 3′-terminal base pairing, and preliminary data show that it contributes highly to the adapted phenotype. We are presently investigating the effects of the other three mutations.
Figure 5.
Secondary structure models of the 3′-terminal region of wild-type and adapted Qβ RNA. (A) Optimal predicted secondary structure of the wild-type Qβ RNA sequence. (B) Optimal predicted secondary structure of the mutated sequence found in Qβ phage adapted to host factor-less host. The three mutations identified in this region are indicated by outline letters.
Our results strongly suggest that the release of the 3′-end from base pairing is an important factor for the facilitated access of replicase to the 3′-end of the template and for its independence from host factor. At the same time, they agree with the proposed function of host factor of making the 3′-end of Qβ RNA available to replicase (12–14). The cellular role of the Hfq protein recently has been suggested to be that of an “RNA chaperone” modifying the secondary structure of cellular mRNAs (17). On Qβ RNA, it appears to do this by melting out the base-paired 3′-terminus.
Acknowledgments
We thank Drs. Christof Biebricher for highly purified Qβ replicase, Charles Weissmann, Martin Billeter, and Imma Barrera for discussions, and Ms. Heidi Mayer-Rosa for photographic reproductions. This work was supported by the Schweizerische Nationalfonds, the Kanton Zürich (Switzerland), and Grant NP-926 from the American Cancer Society to M.E.W.
References
- 1.Blumenthal T, Carmichael G G. Annu Rev Biochem. 1979;48:525–548. doi: 10.1146/annurev.bi.48.070179.002521. [DOI] [PubMed] [Google Scholar]
- 2.van Duin J. In: The Bacteriophages. Calendar R, editor. New York: Plenum; 1988. pp. 117–167. [Google Scholar]
- 3.Kondo M, Gallerani R, Weissmann C. Nature (London) 1970;228:525–527. doi: 10.1038/228525a0. [DOI] [PubMed] [Google Scholar]
- 4.Kamen R. Nature (London) 1970;228:527–533. doi: 10.1038/228527a0. [DOI] [PubMed] [Google Scholar]
- 5.Kamen R, Kondo M, Römer W, Weissmann C. Eur J Biochem. 1972;31:44–51. doi: 10.1111/j.1432-1033.1972.tb02498.x. [DOI] [PubMed] [Google Scholar]
- 6.Blumenthal T, Landers T A, Weber K. Proc Natl Acad Sci USA. 1972;69:1313–1317. doi: 10.1073/pnas.69.5.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wahba A J, Miller M J, Niveleau A, Landers T A, Carmichael G, Weber K, Hawley D A, Slobin L I. J Biol Chem. 1974;249:3314–3316. [PubMed] [Google Scholar]
- 8.Franze de Fernandez M T, Eoyang L, August J T. Nature (London) 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- 9.Franze de Fernandez M T, Hayward T S, August J T. J Biol Chem. 1972;247:824–831. [PubMed] [Google Scholar]
- 10.Carmichael G G, Weber K, Niveleau A, Wahba A J. J Biol Chem. 1975;250:3607–3612. [PubMed] [Google Scholar]
- 11.Kajitani M, Ishihama A. Nucleic Acids Res. 1991;19:1063–1066. doi: 10.1093/nar/19.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer F, Weber H, Weissmann C. J Mol Biol. 1981;153:631–660. doi: 10.1016/0022-2836(81)90411-3. [DOI] [PubMed] [Google Scholar]
- 13.Barrera I, Schuppli D, Sogo J M, Weber H. J Mol Biol. 1993;232:512–521. doi: 10.1006/jmbi.1993.1407. [DOI] [PubMed] [Google Scholar]
- 14.Miranda G, Schuppli D, Barrera I, Hausherr C, Sogo J M, Weber H. J Mol Biol. 1997;267:1089–1103. doi: 10.1006/jmbi.1997.0939. [DOI] [PubMed] [Google Scholar]
- 15.Muffler A, Fischer D, Hengge-Aronis R. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 16.Brown L, Elliott T. J Bacteriol. 1996;178:3763–3770. doi: 10.1128/jb.178.13.3763-3770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muffler A, Traulsen D R, Fischer D, Lange R, Hengge-Aronis R. J Bacteriol. 1997;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown L, Elliott T. J Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsui H-C T, Leung H-C E, Winkler M E. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 20.Qiu S, Schuppli D, Tsui H-C T, Winkler M E, Weber H. Virology. 1997;227:211–214. doi: 10.1006/viro.1996.8302. [DOI] [PubMed] [Google Scholar]
- 21.Batschelet E, Domingo E, Weissmann C. Gene. 1976;1:27–32. doi: 10.1016/0378-1119(76)90004-4. [DOI] [PubMed] [Google Scholar]
- 22.Olsthoorn R C L, van Duin J. Proc Natl Acad Sci USA. 1996;93:12256–12261. doi: 10.1073/pnas.93.22.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. A Short Course in Bacterial Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. [Google Scholar]
- 24.Reiner A M. J Bacteriol. 1969;97:1431–1436. doi: 10.1128/jb.97.3.1431-1436.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weissmann C, Colthart L, Libonati M. Biochemistry. 1968;7:865–874. doi: 10.1021/bi00842a045. [DOI] [PubMed] [Google Scholar]
- 26.Kondo M, Weissmann C. Biochim Biophys Acta. 1972;259:41–49. doi: 10.1016/0005-2787(72)90472-8. [DOI] [PubMed] [Google Scholar]
- 27.Vollenweider H J, Koller Th, Weber H, Weissmann C. J Mol Biol. 1976;101:367–377. doi: 10.1016/0022-2836(76)90153-4. [DOI] [PubMed] [Google Scholar]
- 28.Taniguchi T, Palmieri M, Weissmann C. Nature (London) 1978;274:223–228. doi: 10.1038/274223a0. [DOI] [PubMed] [Google Scholar]
- 29.Biebricher C K, Eigen M, Luce R. Nature (London) 1986;321:98–91. doi: 10.1038/321089a0. [DOI] [PubMed] [Google Scholar]
- 30.Domingo E, Sabo D, Taniguchi T, Weissmann C. Cell. 1978;13:735–744. doi: 10.1016/0092-8674(78)90223-4. [DOI] [PubMed] [Google Scholar]
- 31.Adhin M R, Alblas J, van Duin J. Biochim Biophys Acta. 1990;1050:110–118. doi: 10.1016/0167-4781(90)90150-z. [DOI] [PubMed] [Google Scholar]
- 32.Beekwilder M J, Nieuwenhuizen R, van Duin J. J Mol Biol. 1995;247:903–917. doi: 10.1006/jmbi.1995.0189. [DOI] [PubMed] [Google Scholar]