Abstract
We have cloned Calx, a gene that encodes a Na-Ca exchanger of Drosophila melanogaster. Calx encodes two repeated motifs, Calx-α and Calx-β, that overlap domains required for exchanger activity and regulation. Calx has multiple transcripts in adults, including at least one expressed in the retina. The Calx genomic locus comprises ≥35 kb between the Atpα and rudimentary-like genes in chromosomal region 93B. In Xenopus oocytes, microinjected Calx cRNA induces calcium uptake like that of its homolog, the 3Na+-1Ca2+ exchanger of mammalian heart. Implications of Calx-α motifs for the mechanism of Na-Ca exchange are discussed.
Both animals and protists use cytoplasmic Ca2+ (Cai2+) as an excitatory signal, suggesting that cells co-opted this ion very early in life’s history (1). But excitation cannot recur without Cai2+ effluxes, and prolonged excess Cai2+ is lethal (2–5). Cells thus expel Cai2+ with ATP-driven pumps and Na-Ca exchangers (2). These are complementary: exchangers have a low affinity, but a high capacity, for intracellular calcium; pumps the reverse. Na-Ca exchangers are found in diverse metazoan tissues, including neurons, muscles, kidney, and blood cells (6). Both Limulus and Drosophila photoreceptors display sodium-dependent calcium export, and may use Na-Ca exchange to return to their resting state after activation by light (7–10).
Despite their ubiquity, genes experimentally proven to encode Na-Ca exchangers have been isolated only from mammals (11, 12); they fall into two classes, based on amino acid identity and stoichiometry of action. The cardiac exchanger (NCX1) trades three extracellular Na+ ions for one Cai2+ ion (13). Two other exchangers (NCX2 and NCX3), each expressed in brain and skeletal muscle, have 68–73% identity to NCX1 (14, 15). NCX2 and NCX3 behave similarly to cardiac NCX1 in patch clamps, and thus probably also perform 3Na+-1Ca2+ exchange (14–16). In contrast, the mammalian retinal exchanger NCKX1, expressed in rod photoreceptors, has only 21–25% identity to NCX1–NCX3, and has 4Na+-1Ca2+,1K+ stoichiometry (12, 13).
We and others have worked to identify components of Drosophila phototransduction by isolating visual cDNAs (17, 18). One such cDNA proved to arise from a Drosophila homolog of NCX1, and was the subject of a doctoral thesis of one of us (E.M.S.) in which it was designated Calx (19). We describe here, in abbreviated form, the cloning and analysis of the gene. Hryshko et al. (20) have used Calx in physiological experiments and shown that it has intriguing differences from the mammalian NCX1. Ruknudin et al. (21) have subsequently isolated this gene independently.
METHODS
Methods were essentially standard (22). An exhaustive description of all methods is provided by Schwarz (19).
RESULTS
Calx cDNAs.
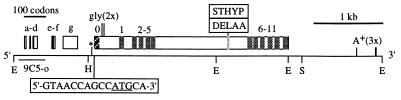
The original Calx cDNA, 9C5-o, is from a subtractive cDNA library and is 377 bp long (17). We isolated five more cDNAs from a λgt11 library (23) and sequenced the longest one (9C5-E). A schematic diagram of 9C5-E (5,408 bp) is shown in Fig. 1. Its major ORF of 950 residues (Fig. 2) has a complex pattern of similarities to known or suspected Na-Ca exchangers (Fig. 3). We thus rename 9C5 “Calx,” and its major ORF’s product “CALX.” The 5′ untranslated region of 9C5-E has seven minor nonoverlapping ORFs. The major ORF’s start site (5′-GTAACCAGCCATGCA-3′) differs from the Drosophila consensus (5′-CACAACCAAAATGGC-3′; ref. 25). While 9C5-E lacks a poly(A)+ tail, it does have three polyadenylation signals (26) near its oligo(A)+ 3′ terminus. Partial sequencing of 14 other cDNAs revealed five CALX residues subject to alternative splicing by a precise replacement of 5′-CGATGAATTGGCAG-3′ (in 9C5-E) with 5′-TTCCACTCACTACC-3′ (Fig. 1, ref. 19). They fall in a region poorly conserved between CALX and NCX1-NCX3; indeed, alternate splicing occurs in the same regions of NCX1 and NCX3 as in CALX (27, 28).
Figure 1.
Features of a Calx cDNA, 9C5-E. Along the cDNA’s lower side are shown: its overlap with the original 9C5-o clone (17); restriction sites important for subcloning (E, EcoRI; H, HindIII; and S, SspI); and the sequence context of its major initiation codon (offset box). Above the cDNA, nonoverlapping miniORFs in the 5′ untranslated region (a–g) and the major Calx ORF are shown as boxes. Two stop codons immediately 5′ from, and in frame with, the major ORF are marked (∗). Within the major ORF, a signal sequence (0) is striped; other transmembrane sequences (1–11) are shaded. Possible N-glycosylation sites are marked [gly (2×)]. Residues that vary between cDNAs (19) are shown with light shading and offset boxes above the cDNA; the “DELAA” mini-exon is found in 9C5-E. Three potential polyadenylation signals near the 3′ end of the cDNA are marked [A+ (3×)].
Figure 2.
The CALX major ORF. Amino acids are in one-letter code (22). The predicted precursor sequence is shown in lowercase type. Potential N-glycosylation substrates in the N-terminal half are underscored with carats. Twelve predicted transmembrane sequences are underlined. Residues identical to a consensus of Calx-α or Calx-β motifs are in boldface. A conserved protein kinase C substrate and acid cluster are denoted by underscoring with an asterisk and with italics.
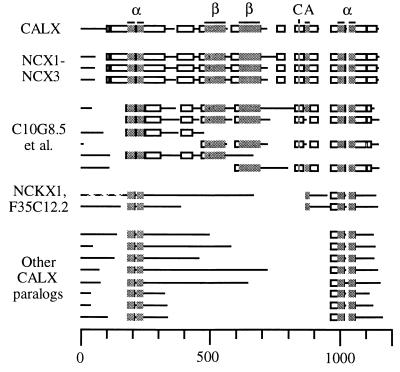
Figure 3.
Schematic alignment of CALX with its homologs. Five groups of sequences are shown, from top to bottom. First is CALX, with conserved motifs indicated above it (α, Calx-α; β, Calx-β; C, protein kinase C substrate; A, acid cluster). Second are CALX’s full-length mammalian orthologs NCX1-NCX3 (all from Rattus norvegicus; National Center for Biotechnology Information ID nos. gi|1346652, 1346653, and 1552526). Third are six partial ORFs also orthologous to CALX: C10G8.5, CENACAEX, ZC168.1 [5′], and ZC168.1 [3′] (C. elegans: gi|1572830, gi|1009384, gi|1246477, and gi|1246503); XLSOCALX2 and XLSOCALX1 (X. laevis: gi|1019101 and gi|1019099). Fourth are NCXK1 (Bos taurus: gi|108825) and F35C12.2 (C. elegans: gi|1813931). Fifth are paralogs with no clear function: C07A9.4 and C07A9.11 (C. elegans: gi|465770 and gi|465776); F7G19.17 (A. thaliana: gi|1922938); ORF_D1053 and YJ76_YEAST (S. cerevisiae: gi|1429350, gi|1352908); MJ0091 (M. jannaschii: gi|1590872); YRBG_ECOLI (E. coli: gi|1176841); and slr0681 (Synechocystis PCC6803: gi|1652037). Block-aligned regions shown as boxes, and unaligned ones as lines; gaps in the alignment are blank. Bar = residue length. The N terminus of NCKX1 has been truncated. The aligned regions have P values of 2.0⋅10−4 to <10−100 (24). Full sequences and references for each homolog are available under its National Center for Biotechnology Information ID no. via the Entrez server (http://www3.ncbi.nlm.nih.gov/Entrez).
Calx Tissue Expression.
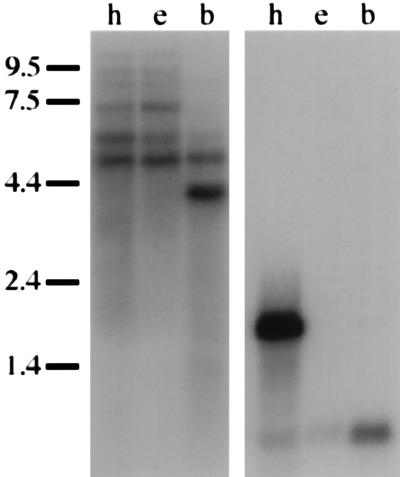
We tested the transcriptional polarity and tissue specificity of Calx by probing RNA blots with strand-specific 9C5-E probes (Fig. 4). Probes antisense to the major 9C5-E ORF revealed multiple transcripts in adult Drosophila; probes comprising the same strand as the major ORF yielded no signals, even against 10 μg of poly(A)+ RNA from adult heads (19). Calx poly(A)+ RNAs (Fig. 4) include a transcript in both heads and bodies (4.9 kb); a body-specific transcript (4.0 kb); and three head-specific transcripts that appear unaffected by the presence or absence of the visual system (8.2, 9.0, and ≥≈15 kb). A fifth head-specific transcript is enriched in eyeless heads, and thus appears to be primarily expressed outside the visual system (7.0 kb). A sixth transcript (5.6 kb, close in size to 9C5-E) appears to be expressed both within the visual system and elsewhere in the head. Spatially, Calx transcripts are ubiquitously present at all embryonic stages from syncytial to organogenesis (19), and in all tissues of the adult head (e.g., neurons: Fig. 5; ref. 19).
Figure 4.
Multiple Calx transcripts in adult tissues. In the left autoradiograph, antisense Calx probe is seen to hybridize to adult poly(A)+ RNAs. These consist of 5 μg from wild-type heads (h), 5 μg from eyeless (eyes absent) heads (e), and 10 μg from mixed-sex wild-type bodies (b). The positions of molecular weight markers are shown alongside the blot (in kb). In adult heads, transcripts with the following sizes are observed: ≥≈15, 9.0, 8.2, 7.0, 5.6, and 4.9 kb. The right autoradiograph is from the same blot as on the left, rehybridized with mixed ninaE (rhodopsin) and RpA1 probes. The retina-specific ninaE transcript (1.7 kb) and the ubiquitous RpA1 transcript (0.68 kb) are both seen.
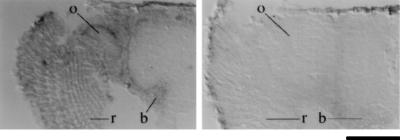
Figure 5.
Spatial pattern of Calx transcripts in the adult head. In the left in situ hybridization, digoxigenin-labeled antisense Calx probe is seen to hybridize to adult retina (r), optic ganglia (o), and brain (b). In the right in situ hybridization, sense (control) Calx probe gives only background signals in these tissues. B = 100 μm.
Sequence Analysis of the Calx ORF.
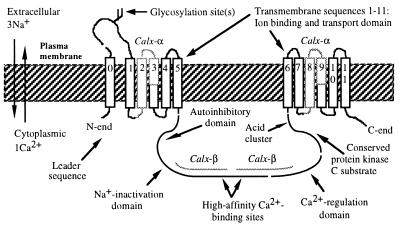
All mammalian Na-Ca exchangers have common features (13). They have two regions of six potential transmembrane sequences, split by a cytoplasmic domain. The first membrane sequence is transient, being proximal to a signal sequence cleavage site. The mature N terminus is followed by one or more extracellular glycosylation sites. The cytoplasmic domain bears many acidic residues; a prominent acid cluster sits at its C terminus. CALX is predicted to possess all of these traits (Fig. 2). Below, we report novel features of CALX and its homologs.
Multiple Alignment.
We used blastp (29) to iteratively screen the nonredundant protein database of the National Center for Biotechnology Information for CALX homologs. We used macaw (24) to align the homologs in ungapped blocks (Fig. 3). CALX is globally similar to NCX1, NCX2, and NCX3, having 55% identity out of 739 aligned amino acids. Six partial Caenorhabditis elegans ORFs and Xenopus laevis ORFs have subsets of the NCX1-NCX3 similarities along their lengths; these proteins, with NCX1-NCX3, appear to be CALX orthologs. Ten other proteins appear to be CALX paralogs. They significantly resemble CALX in four ungapped blocks totalling 123 residues: while their identity to CALX in this region is variable (15–37%), in pairwise alignments to CALX they have P values of 2⋅10−3 to 1⋅10−14 (30). CALX paralogs are found in metazoa, thale cress (Arabidopsis thaliana), yeast (Saccharomyces cerevisiae), an archaeon (Methanococcus jannaschii), and bacteria (Escherichia coli and Synechocystis PCC6803). While F35C12.2 is 45% identical to NCKX1 (and thus may be its C. elegans ortholog), the functions of most CALX paralogs are unknown.
The cytoplasmic domain of NCX1 has three regions defined by deletion analysis (16). The first two enable negative regulation (by an autoinhibitory peptide or elevated intracellular Na), and mediate high-affinity Ca2+ binding (31, 32). The third is required for activation by elevated Cai2+. The first two regions are well conserved between CALX and NCX1 (61% and 49% identity); the last is less well conserved (33% identity). Accordingly, CALX displays inhibition by autoinhibitory peptide and elevated intracellular sodium, but is partially inhibited by elevated Cai2+ (20). Protein kinase C stimulates calcium extrusion in Drosophila photoreceptors, which significantly depends on Na-Ca exchange (8); similar stimulation has also been seen for NCX1 (33). We checked for protein kinase C substrates conserved between CALX and all of its orthologs. One exists, next to the acid cluster (Figs. 2 and 3) in an NCX1 region not yet tested for function (16).
Intragenic Repeats.
We discovered two new motifs in CALX (Calx-α and Calx-β: Figs. 6, 7, 8). While first detected in CALX, they are also reiterated within CALX homologs (Figs. 7 and 8). Their statistical significance is demonstrable by P values for aligned pairs of motifs (30). For example, the Calx-α motifs of CALX itself have P values of <10−3 for 57/69 nonidentical pairings with other Calx-α motifs and 0.001–0.23 for 12/69 such pairings; those of NCKX1 have P values of <10−3 for 66/70 such pairings and 0.004–0.03 for 4/70. The Calx-β motifs of CALX have P values of <6⋅10−6 in all nonidentical Calx-β pairings.
Figure 6.
Schematic diagram of CALX. Ion flows during an outward exchange are shown to the left. Calx-α and Calx-β motifs map onto regions already known (in NCX1) to be necessary and sufficient for ion transport, high-affinity binding of intracellular calcium, inactivation by elevated intracellular sodium, and autoinhibition (16, 32). The motifs are precisely repeated: the Calx-α motifs occupy identical positions within their respective blocks of membrane sequences, while the Calx-β motifs are tandem.
Figure 7.
Sequence alignment of Calx-α. CALX sequences predicted to be membrane-spanning are underlined. Each instance of a motif is shown with: its protein’s name; the motif’s sequence, with aligned regions separated by indicated numbers of unaligned residues; and the location of the motif’s C terminus in the protein. Proteins here are described in Fig. 3. All aligned regions have P values of <10−100 (24). Residues that match a consensus residue or residue type (shown in the Consensus line) are in boldface type. Residue types are defined as: aromatic (F, W, Y, or H, denoted @); aliphatic (I, L, or V, denoted u); acidic (D or E, −); basic (H, K, or R, +); charged (acidic or basic, ∗); hydroxylic (S or T, $), methyl (A, S, or C, μ), or small (G, A, S, or C, denoted o). A “⋅” denotes sites with no particular consensus. Note that the Calx-α motifs of CALX orthologs vs. paralogs have slightly different consensuses. Calx-α residues in the “Notable” line are noted in the Discussion.
Figure 8.
Sequence alignment of Calx-β. Format, references, and P values are as in Fig. 7 unless noted. The N terminus of XLSOCALX1’s first Calx-β motif is assumed to be residues 400–416 of XLSOCALX2 (in brackets). “Pred. str.” denotes secondary structures predicted for Calx-β motifs by the PHD neural network (34). “E” and “L” denote residues predicted to be in an extended strand (e.g., a β-strand) or in a loop; those shown here were predicted with 82% accuracy for both N- and C-terminal Calx-β motifs. Most proteins here are described in Fig. 3, except for integrin-β4 (R. norvegicus: gi|1401303) and slr0408, slr1028, and slr1403 (Synechocystis PCC6803: gi|1653629, gi|1652242, and gi|1652714).
The Calx-α motif is duplicated in all known exchanger homologs, in equivalent locations within both transmembrane regions (Figs. 6 and 7). Calx-α motifs are predicted to bridge the membrane, comprising most of transmembrane sequences 2–3 and 8–9. Unlike the Calx-α motif, Calx-β is only found in exchangers showing global similarity to NCX1-NCX3 (Figs. 3 and 8). Yet an isolated Calx-β motif exists in mammalian integrin-β4 and in three Synechocystis proteins of unknown function. In both exchangers and mammalian integrins, Calx-β motifs are predicted to be cytoplasmic. Calx-β motifs are predicted by the PHD neural network (34) to have a series of β-strands and turns (Fig. 8); this is consistent with the Calx-β motif being a self-contained β-sheet.
Calx Genomic Region.
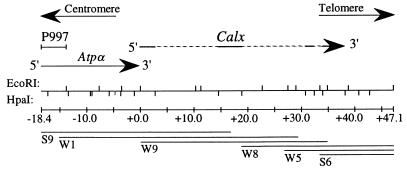
The Calx genomic locus is located in the 93B1,2 doublet of chromosome 3 (N. Bonini, personal communication); it is at least 32 kb long (Fig. 9). It is immediately downstream of the sodium pump α-subunit gene (Atpα), also located in 93B1–2 (35). The lethal P insertion P997 was generated in 93B by Wilson et al. (37); we mapped its insertion site within Atpα (Fig. 9), and found that it failed to complement l(3)01453 (mapped to 93B1-2 by the Berkeley Drosophila genome project; refs. 19 and 38). We probed rudimentary-like genomic phage DNA with a 5.0-kb EcoRI fragment from cosmid W5 (just off the 3′ end of the Calx region in Fig. 4); it hybridized to a genomic clone proximal to rudimentary-like, a UMP synthase gene in 93B6–7 (36). This physically links Atpα and rudimentary-like via Calx, and chromosomally orients both Calx and Atpα (Fig. 9).
Figure 9.
The Calx genomic region. The topmost line shows genomic DNA; upper ticks mark EcoRI sites and lower ticks mark HpaI sites. One HpaI site (at about −5.0 kb) separates two unordered HpaI-HpaI fragments 1.18 and 1.05 kb in size. Above this map, DNA fragments are shown that cross-hybridize to the P997 insertion site, the Na+/K+ pump α-subunit (Atpα) gene (35), and Calx cDNAs. The proximal end of the rudimentary-like genomic walk (36) begins with a 5.0-kb EcoRI fragment immediately to the right of the genomic DNA shown here. Chromosomal orientation is shown with arrows. DNA fragments cross-hybridizing to Calx cDNA are shown as solid lines; other fragments are stippled. Calx transcriptional polarity was deduced from cDNA probes and RNA blots (Fig. 4). Underneath the genomic map are shown distances in kb and six cosmids covering the region.
Direct Assays of Na-Ca Exchanger Activity.
To directly verify its functional homology to NCX1-NCX3, we assayed CALX by exploiting the reversibility of Na-Ca exchange in Xenopus oocytes (11). Capped Calx cRNAs were synthesized from pXexCalx, a HindIII/SspI subclone of 9C5-E containing the major ORF (Fig. 1). Plasmids encoding NCX1 and the Drosophila potassium channel Shaker (1) were used as templates for capped NCX1 and Shaker cRNAs (16, 39). Finally, we generated a translationally optimized Calx cRNA template (PCR-Calx) with PCR. All four templates were used to make standardized aliquots of cRNA.
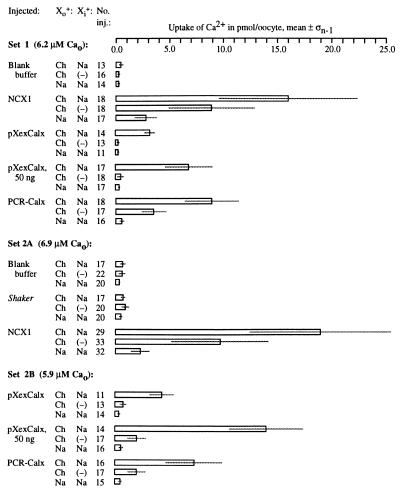
These cRNAs, in parallel with blank buffer, were microinjected into Xenopus oocytes (5 ng per oocyte). After 4–8 days of incubation, these oocytes were loaded with internal Na via nystatin (40), challenged with 45Ca2+, washed, and assayed for 45Ca2+ uptake. Calcium uptake was induced by Calx cRNAs (Fig. 10). This uptake occurred with external potassium (19) or choline; however, it was absent from oocytes challenged with external sodium, not loaded with internal sodium, or expressing an unrelated ion transporter. While pXexCalx-induced uptake (7 times background) was substantially weaker than that induced by NCX1 (30–35 times background), uptakes comparable to NCX1’s were induced by both PCR-Calx (12–19 times background) and 50 ng of pXexCalx cRNA (15–22 times background). Oocytes vigorously expressing exchangers showed some uptake even without sodium loading (Fig. 10). To test whether this might be due to residual sodium within the oocyte, we loaded Calx-expressing oocytes with lithium or potassium cations: these reduced calcium uptake to background levels (19).
Figure 10.
Expression of CALX in Xenopus oocytes. Sets of oocytes were microinjected, incubated for several days, loaded with intracellular sodium, challenged with 45Ca2+ along with a variable monovalent cation, and allowed to take up 45Ca2+ via reverse Na-Ca exchange for 10′. The concentration of 45Ca2+ used (5.9–6.9 μM) is indicated for each oocyte set. After uptake, oocytes were rinsed and individually assayed by scintillation counting. “Injected” refers to the substance injected; Xo+, the primary monovalent cation placed outside the oocytes being assayed for 45Ca2+ uptake; Xi+, the primary monovalent cation loaded into the oocytes; “No. inj.,” the number of oocytes injected with a particular solution; “Uptake of Ca2+ in pmol/oocyte, mean ± σn−1,” the mean quantity and SD of 45Ca2+ taken up per oocyte. “Ch” denotes cationic choline; (−) denotes oocytes left unloaded by outside cations.
DISCUSSION
Calx-α and Calx-β Motifs: Possible Functions.
Repeated motifs are thought to arise via intragenic duplications, and survive if they improve protein function (41). Calx-α motifs are duplicated in all full-length ORFs in which they are found at all, and exist in all three domains of life. This implies both that Calx-α motifs arose early in life’s history and that their existing as a tandem pair is essential for function. This is consistent with Calx-α motifs opposing one another symmetrically across an internal pore spanning the cell membrane (42). Both 3Na+-1Ca2+ and 4Na+-1Ca2+, 1K+ exchangers (i.e., NCX1 and NCKX1) appear to have two anions that neutralize either Ca2+ or 2Na+ during the Na-Ca exchange cycle (43, 44). However, no location for such an anion pair has so far been proposed for either NCX1 or NCKX1. Our analysis shows that only one anionic (D or E) residue is completely conserved in all Calx-α motifs (Fig. 7). This residue could be one member of a symmetrical pair of two invariant anions flanking the exchanger channel. This D/E residue is invariably adjacent to a helix-breaking residue (generally proline, occasionally glycine; Fig. 7). Such a P/G site could form a constricting bend in the channel, like that of acetylcholine receptors (45). We thus predict these D/E residues to be the two anions used in both 3Na+-1Ca2+ and 4Na+-1Ca2+,1K+ exchange. To fully account for its electrogenicity, NCKX1 requires an anion not found in NCX1 (44). One candidate is D1113 (Fig. 7): it is predicted to have roughly the same elevation within the membrane as the invariant D/E residues; it is found in the NCKX1 ortholog F35C12.2; and it exists in place of a strongly conserved asparagine residue. In NCX1, homologous residues in N- and C-terminal Calx-α motifs have quantitatively similar mutant phenotypes, and mutations of both invariant D/E sites abolish exchange (46). These data are consistent with our model for Calx-α function, though more data are needed to test the predictions here.
Extant data support several possibilities for Calx-β’s function. NCX1’s two Calx-β motifs contain a high-affinity Cai2+-binding site (Fig. 6; ref. 31). But, because this site only partly encompasses Calx-β motifs conserved for ≥≈540 My (Figs. 3 and 8), the motifs probably have further roles. They overlap a region required for inhibition of NCX1 by elevated intracellular sodium (16); the same region may also bind ankyrin (47). Any of these functions would be appropriate in integrin-β4, which connects hemidesmosomes with the cytoskeleton (48) and transduces intracellular signals (49).
Acknowledgments
We thank E. Eichenberger, L. Dowling, H. Davis, A. Gomez, and J. Gollub for technical assistance; the Indiana Drosophila Stock Center for Drosophila stocks; J. Tamkun, C. Labarca, and J. Rawls for genomic libraries and clones; N. Bonini for determining the chromosomal location of Calx; H. Lester and M. Quick for aiding our Xenopus expression work; D. Nicoll for NCX1 clones and reagents; members of the Benzer, Dreyer, and Lipshitz laboratories for cDNA libraries, protocols, and criticism; and K. Philipson for advice and encouragement. This work was supported by grants to S.B. from the National Science Foundation (MCB-9408718), the National Eye Institute (EY09278), the National Institute of Aging (AG12289), and the James G. Boswell Foundation, and by a grant to E.M.S. and S.B. from the Beckman Institute.
ABBREVIATIONS
- Cai2+
cytoplasmic calcium
- NCX1
-2, and -3, paralogous mammalian 3Na+-1Ca2+ exchangers
- NCKX1
4Na+-1Ca2+,1K+ mammalian retina exchanger
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF009897).
References
- 1.Hille B. Ionic Channels of Excitable Membranes. 2nd Ed. Sunderland, MA: Sinauer; 1992. [Google Scholar]
- 2.Carafoli E. Annu Rev Biochem. 1987;56:395–433. doi: 10.1146/annurev.bi.56.070187.002143. [DOI] [PubMed] [Google Scholar]
- 3.Choi D W. J Neurobiol. 1992;23:1261–1276. doi: 10.1002/neu.480230915. [DOI] [PubMed] [Google Scholar]
- 4.Nicotera P, Bellomo G, Orrenius S. Annu Rev Pharmacol Toxicol. 1992;32:449–470. doi: 10.1146/annurev.pa.32.040192.002313. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham K W, Fink G R. J Cell Biol. 1994;124:351–363. doi: 10.1083/jcb.124.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaustein M P, DiPolo R, Reeves J P, editors. Sodium-Calcium Exchange: Proceedings of the Second International Conference. New York: N. Y. Acad. Sci.; 1991. [PubMed] [Google Scholar]
- 7.O’Day P M, Gray-Keller M P, Lonergan M. J Gen Physiol. 1991;97:369–391. doi: 10.1085/jgp.97.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ranganathan R, Bacskai B J, Tsien R Y, Zuker C S. Neuron. 1994;13:837–848. doi: 10.1016/0896-6273(94)90250-x. [DOI] [PubMed] [Google Scholar]
- 9.Hardie R C. J Neurosci. 1995;15:889–902. doi: 10.1523/JNEUROSCI.15-01-00889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ranganathan R, Malicki D M, Zuker C S. Annu Rev Neurosci. 1995;18:283–317. doi: 10.1146/annurev.ne.18.030195.001435. [DOI] [PubMed] [Google Scholar]
- 11.Nicoll D A, Longoni S, Philipson K D. Science. 1990;250:562–565. doi: 10.1126/science.1700476. [DOI] [PubMed] [Google Scholar]
- 12.Reiländer H, Achilles A, Friedel U, Maul G, Lottspeich F, Cook N J. EMBO J. 1992;11:1689–1695. doi: 10.1002/j.1460-2075.1992.tb05219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philipson K D, Nicoll D A. Int Rev Cytol. 1993;137C:199–227. [PubMed] [Google Scholar]
- 14.Li Z, Matsuoka S, Hryshko L V, Nicoll D A, Bersohn M M, Burke E P, Lifton R P, Philipson K D. J Biol Chem. 1994;269:17434–17439. [PubMed] [Google Scholar]
- 15.Nicoll D A, Quednau B D, Qui Z, Xia Y R, Lusis A J, Philipson K D. J Biol Chem. 1996;271:24914–24921. doi: 10.1074/jbc.271.40.24914. [DOI] [PubMed] [Google Scholar]
- 16.Matsuoka S, Nicoll D A, Reilly R F, Hilgemann D W, Philipson K D. Proc Natl Acad Sci USA. 1993;90:3870–3874. doi: 10.1073/pnas.90.9.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyde D R, Mecklenburg K L, Pollock J A, Vihtelic T S, Benzer S. Proc Natl Acad Sci USA. 1990;87:1008–1012. doi: 10.1073/pnas.87.3.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith D P, Shieh D-H, Zuker C S. Proc Natl Acad Sci USA. 1990;87:1003–1007. doi: 10.1073/pnas.87.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwarz E M. Ph.D. thesis. Pasadena: California Institute of Technology; 1996. [Google Scholar]
- 20.Hryshko L V, Matsuoka S, Nicoll D A, Weiss J N, Schwarz E M, Benzer S, Philipson K D. J Gen Physiol. 1996;108:67–74. doi: 10.1085/jgp.108.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruknudin A, Valdivia C, Kofuji P, Lederer W J, Schulze D H. Am J Physiol. 1997;273:C257–C265. doi: 10.1152/ajpcell.1997.273.1.C257. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 23.Itoh N, Salvaterra P, Itakura K. Drosophila Inf Serv. 1985;61:89. [Google Scholar]
- 24.Schuler G D, Altschul S F, Lipman D J. Proteins. 1991;9:180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- 25.Cavener D R, Ray S C. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angelichio M L, Beck J A, Johansen H, Iveyhoyle M. Nucleic Acids Res. 1991;19:5037–5043. doi: 10.1093/nar/19.18.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S-L, Yu A S L, Lytton J. J Biol Chem. 1994;269:14849–14852. [PubMed] [Google Scholar]
- 28.Quednau B D, Nicoll D A, Philipson K D. Am J Physiol. 1997;272:C1250–C1261. doi: 10.1152/ajpcell.1997.272.4.C1250. [DOI] [PubMed] [Google Scholar]
- 29.Altschul S F, Boguski M S, Gish W, Wootton J C. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- 30.Pearson W R. Methods Enzymol. 1996;266:227–258. doi: 10.1016/s0076-6879(96)66017-0. [DOI] [PubMed] [Google Scholar]
- 31.Matsuoka S, Nicoll D A, Hryshko L V, Levitsky D O, Weiss J N, Philipson K D. J Gen Physiol. 1995;105:403–420. doi: 10.1085/jgp.105.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuoka S, Nicoll D A, He Z, Philipson K D. J Gen Physiol. 1997;109:273–286. doi: 10.1085/jgp.109.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iwamoto T, Pan Y, Wakabayashi S, Imagawa T, Yamanaka H I, Shigekawa M. J Biol Chem. 1996;271:13609–13615. doi: 10.1074/jbc.271.23.13609. [DOI] [PubMed] [Google Scholar]
- 34.Rost B, Sander C. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 35.Schubiger M, Feng Y, Fambrough D M, Palka J. Neuron. 1994;12:373–381. doi: 10.1016/0896-6273(94)90278-x. [DOI] [PubMed] [Google Scholar]
- 36.Eisenberg M, Gathy K, Vincent T, Rawls J. Mol Gen Genet. 1990;222:1–8. doi: 10.1007/BF00283015. [DOI] [PubMed] [Google Scholar]
- 37.Wilson C, Pearson R K, Bellen H J, O’Kane C J, Grossniklaus U, Gehring W J. Genes Dev. 1989;3:1301–1313. doi: 10.1101/gad.3.9.1301. [DOI] [PubMed] [Google Scholar]
- 38.Karpen G H, Spradling A C. Genetics. 1992;132:737–753. doi: 10.1093/genetics/132.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klaiber K, Williams N, Roberts T M, Papazian D M, Jan L Y, Miller C. Neuron. 1990;5:221–226. doi: 10.1016/0896-6273(90)90311-3. [DOI] [PubMed] [Google Scholar]
- 40.Longoni S, Coady M J, Ikeda T, Philipson K D. Am J Physiol. 1988;255:C870–C873. doi: 10.1152/ajpcell.1988.255.6.C870. [DOI] [PubMed] [Google Scholar]
- 41.Li W-H, Graur D. Fundamentals of Molecular Evolution. Sunderland, MA: Sinauer; 1991. [Google Scholar]
- 42.Kyte J. Nature (London) 1981;292:201–204. doi: 10.1038/292201a0. [DOI] [PubMed] [Google Scholar]
- 43.Hilgemann D W, Nicoll D A, Philipson K D. Nature (London) 1991;352:715–718. doi: 10.1038/352715a0. [DOI] [PubMed] [Google Scholar]
- 44.Perry R J, McNaughton P A. J Physiol (London) 1993;466:443–480. [PMC free article] [PubMed] [Google Scholar]
- 45.Unwin N. J Mol Biol. 1993;229:1101–1124. doi: 10.1006/jmbi.1993.1107. [DOI] [PubMed] [Google Scholar]
- 46.Nicoll D A, Hryshko L V, Matsuoka S, Frank J S, Philipson K D. J Biol Chem. 1996;271:13385–13391. doi: 10.1074/jbc.271.23.13385. [DOI] [PubMed] [Google Scholar]
- 47.Li Z, Burke E P, Frank J S, Bennett V, Philipson K D. J Biol Chem. 1993;268:11489–11491. [PubMed] [Google Scholar]
- 48.Spinardi L, Ren Y-L, Sanders R, Giancotti F G. Mol Biol Cell. 1993;4:871–884. doi: 10.1091/mbc.4.9.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mainiero F, Murgia C, Wary K K, Curatola A M, Pepe A, Blumemberg M, Westwick J K, Der C J, Giancotti F G. EMBO J. 1997;16:2365–2375. doi: 10.1093/emboj/16.9.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]