Preliminary crystallographic data are reported for the third SRP GTPase FlhF from Bacillus subtilis.
Keywords: FlhF, GTPase domains
Abstract
The Gram-positive bacterium Bacillus subtilis contains three proteins belonging to the signal recognition particle (SRP) type GTPase family. The well characterized signal sequence-binding protein SRP54 and the SRP receptor protein FtsY are universally conserved components of the SRP system of protein transport. The third member, FlhF, has been implicated in the placement and assembly of polar flagella. This article describes the overexpression and preliminary X-ray crystallographic analysis of an FlhF fragment that corresponds to the well characterized GTPase domains in SRP54 and FtsY. Three crystal forms are reported with either GDP or GMPPNP and diffract to a resolution of about 3 Å.
1. Introduction
Motile bacteria move by the means of flagella. The biosynthesis of a flagellum is a complex process (reviewed in Macnab, 2003 ▶) and requires a minimal set of 30 genes in Bacillus subtilis (Bischoff & Ordal, 1992 ▶). Most of the genes responsible for chemotaxis and flagella assembly are located in the che/fla operon. Among these genes, flhf encodes a basic polypeptide of 41 kDa which contains a putative GTP-binding motif (Andersen et al., 2006 ▶; Carpenter et al., 1992 ▶). Studies investigating the function of FlhF implicate a role in flagellar placement (Correa et al., 2005 ▶; Pandza et al., 2000 ▶), assembly (Carpenter et al., 1992 ▶; Niehus et al., 2004 ▶), motility (Carpenter et al., 1992 ▶; Niehus et al., 2004 ▶; Shen et al., 2001 ▶) and virulence of pathogenic bacteria (Correa et al., 2005 ▶; Niehus et al., 2004 ▶). The protein associates with the plasma membrane (Nowalk et al., 2006 ▶) and localizes at the cell poles (Murray & Kazmierczak, 2006 ▶).
FlhF shows significant sequence homology to the signal recognition particle (SRP) GTPases SRP54 and FtsY (SRPDB; Andersen et al., 2006 ▶; Carpenter et al., 1992 ▶) and was thus termed the third SRP GTPase (Andersen et al., 2006 ▶; Zanen et al., 2004 ▶). SRP54 and FtsY are well characterized as essential components of SRP-mediated protein targeting (reviewed in Luirink & Sinning, 2004 ▶). However, FlhF seems to be dispensable for protein secretion (Zanen et al., 2004 ▶). SRP GTPases are multidomain proteins and share conserved NG domains (Freymann et al., 1997 ▶; Montoya et al., 1997 ▶). The N domain comprises a bundle of four α-helices which is structurally and functionally linked to the GTPase domain (G domain).
Here, we describe the overexpression, purification, crystallization and the preliminary crystallographic analysis of a fragment containing the NG domain of FlhF from B. subtilis. A number of crystal forms were obtained in the presence of different nucleotides.
2. Materials and methods
2.1. Cloning and protein expression
A gene fragment encoding amino acids 79–366 (NG-FlhF) of B. subtilis FlhF (gi|544314) was designed by multiple sequence alignments, amplified by polymerase chain reaction (PCR) using the Expand High Fidelity PCR system (Roche) and cloned into a pET24d vector (Novagen) via the NcoI/BamHI restriction sites. The construct was verified by sequencing and contains a 6×His tag at the N-terminus of the protein. Escherichia coli strain BL21 (DE3) (Novagen) was used for expression of NG-FlhF. Cells were grown in lysogeny broth medium (LB) complemented with 1.5%(w/v) d(+)-lactose monohydrate for 16 h at 303 K.
2.2. Protein purification
Cell pellets were resuspended in 10 ml lysis buffer per gram of cells and passed through an M1-10L Microfluidizer (Microfluidics). The lysis buffer contained 20 mM Na HEPES pH 8.0, 350 mM NaCl, 10 mM MgCl2, 10 mM KCl. The lysate was clarified by centrifugation (125 000g for 30 min at 277 K) using a Ti-45 rotor (Beckmann) and applied onto a 1 ml HisTrap HP column (GE Healthcare). The column was initially washed with five column volumes of lysis buffer containing 40 mM imidazole pH 8.0. The protein was eluted in lysis buffer containing 500 mM imidazole pH 8.0. Subsequently, the protein was concentrated to approximately 30 mg ml−1 using an Amicon Ultracel-10K (Millipore) and underwent size-exclusion chromatography using an S75/26–60 column (GE Healthcare) in the same buffer as above but without imidazole. Protein-containing fractions were pooled and concentrated to 10 mg ml−1. Prior to crystallization experiments, either 5 mM GDP (guanosine 5′-diphosphate) or 5 mM of the nonhydrolysable GTP analogue GMMPNP [guanosine 5′-(β,γ-imido)-triphosphate] were added to the protein solution.
2.3. Crystallization and X-ray analysis
Crystallization of NG-FlhF was performed using the hanging-drop method in MD3-11 XRL plates (Molecular Dimensions) using Index Screens I and II (Hampton Research). For initial trials, equal volumes (1–2 µl) of protein solution (10 mg ml−1) and crystallization screening buffer were mixed on a cover slip and suspended over a reservoir containing 500 µl crystallization screening solution. Experiments were performed at 291 K. Prior to X-ray analysis, crystals were flash-cooled in liquid nitrogen after cryoprotection by transfer into a cryosolution containing mother liquor and 20–25%(v/v) glycerol.
Diffraction data were measured on beamlines ID29, ID14-1 and ID14-2 at the European Synchrotron Radiation Facility (ESRF) under cryogenic conditions (100 K; Oxford Cryosystems Cryostream). Data were recorded with ADSC Q315r CCD (beamline ID29), ADSC Q210 CCD (beamline 14-1) and ADSC Q4R CCD (beamline 14-2) detectors. Data were processed and scaled with the CCP4-implemented programs MOSFLM and SCALA, respectively (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
FlhF is a member of the SRP GTPase subfamily of small G proteins. The NG domain of FlhF (NG-FlhF) was predicted to include amino acids 79–366 by multiple sequence alignments and secondary-structure prediction using ClustalX (Thompson et al., 1997 ▶) and ProteinPredict (Rost et al., 2004 ▶), respectively. NG-FlhF was expressed and typical yields were about 50 mg per litre of culture (corresponding to 8 g of cells). Protein purification took place according to standard protocols as described above. The protein purity prior to crystallization was >90% as judged from Coomassie-stained SDS–PAGE (not shown).
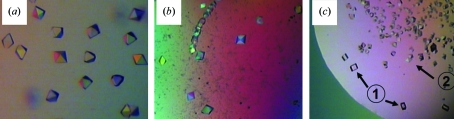
Initial crystals of NG-FlhF grew in the presence of 5 mM GDP from hanging drops with a reservoir solution containing 10%(w/v) PEG 6000, 4%(v/v) 2-methyl-2,4-pentanediol and 0.1 M Tris pH 7.5. Numerous bipyramidal crystals appeared within a few hours and grew to full size within 2 d (Fig. 1 ▶ a, crystal form A). The crystals were not very stable and decomposed after about one week. Only a fraction of them were suitable for X-ray analysis; they diffracted to 3.2 Å resolution at beamline ID29 at the ESRF in Grenoble. When NG-FlhF was crystallized in the presence of 5 mM GMPPNP with a similar reservoir solution consisting of 13%(w/v) PEG 6000, 4%(v/v) 2-methyl-2,4-pentanediol and 0.1 M MOPS pH 7.5, a large number of crystals appeared within 2 d. They exhibited a similar bipyramidal shape (Fig. 1 ▶ b, crystal form B) and were unstable; similar to the crystals obtained with GDP, only a few of them showed diffraction. A data set could be collected to 3.2 Å resolution. However, these crystals did not decompose and after three to five weeks a change in crystal morphology was observed (Fig. 1 ▶ c). These new crystals (crystal form C) were of homogenous quality and diffracted to a resolution of 3.0 Å. It is important to note that the addition of GDP or GMPPNP was a prerequisite for successful crystallization, as no crystals were obtained in the absence of nucleotide.
Figure 1.
Various crystals of B. subtilis FlhF. (a) Bipyramidal crystals grown in the presence of 5 mM GDP (crystal form A, Table 1 ▶). (b) Crystals grown in the presence of 5 mM GMPPNP show a similar bipyramidal shape (crystal form B, Table 1 ▶). (c) Bipyramidal crystals (labelled ‘2’) grown in the presence of 5 mM GMPPNP transform after 3–5 weeks into cuboids (labelled ‘1’; crystal form C, Table 1 ▶). The size of the crystals is typically about 30–70 µm.
For X-ray data collection, crystals were transferred to a glycerol-containing cryoprotection solution and subsequently flash-cooled as described above. The use of various cryoprotection agents such as ethylene glycol, medium light paraffin oil or sucrose did not improve diffraction. Data collection was performed at beamlines ID29, ID14-1 and ID14-2 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. Three data sets were collected; statistics and crystal data are summarized in Table 1 ▶. Notably, crystal forms A and B are basically identical in unit-cell parameters and diffraction quality, despite being obtained in the presence of GDP or GMPPNP, respectively. This suggests that the addition of these nucleotides does not influence crystal packing. Crystal form C belongs to a different space group and the Matthews coefficient (Matthews, 1968 ▶) suggests the presence of eight molecules in the asymmetric unit, while crystal forms A and B contain one molecule in the asymmetric unit (Table 1 ▶). As the transition from crystal form B to C only occurs with the nonhydrolysable GTP analogue GMPPNP, it might be a consequence of slow nucleotide binding or exchange. Analysis of the self-rotation function of the data from crystal form C shows that the crystal forms A (or B) and C are not related (not shown).
Table 1. Summary of preliminary crystallographic analysis.
Values in parentheses are for the highest resolution shell.
| Crystal form | A | B | C |
|---|---|---|---|
| Wavelength (Å) | 0.979 | 0.934 | 0.933 |
| Space group | P41212 or P43212 | P41212 or P43212 | P21 |
| Unit-cell parameters (Å, °) | a = b = 80.96, c = 126.4 | a = b = 79.5, c = 127.5 | a = 104.65, b = 111.37, c = 112.1, β = 89.78 |
| Resolution (Å) | 50.0–3.2 (3.31–3.2) | 79.6–3.2 (3.37–3.2) | 50–3.0 (3.16–3.0) |
| Completeness (%) | 99.9 (99.9) | 99.9 (100) | 99.1 (99.8) |
| Rsym† (%) | 8.8 (40.8) | 11.0 (51.6) | 6.7 (51.9) |
| I/σ(I) | 19.4 (4.2) | 10.5 (2.9) | 12.3 (2.2) |
| Redundancy | 7.3 | 4.1 | 3.0 |
| No. of molecules in ASU‡ | 1 | 1 | 8 |
Our study describes the expression, purification and preliminary crystallographic analysis of B. subtilis FlhF. We expect that the structure will be solvable by molecular replacement using SRP54 or FtsY as a search model. With the structure of FlhF, all three members of the SRP GTPases will be characterized. The structure should provide insights into the regulation of SRP GTPases in general and provide a molecular basis for the role of the third SRP GTPase in flagellar assembly.
Acknowledgments
We thank G. W. Ordal for kindly providing the DNA coding for B. subtilis FlhF. We thank A. Hendricks for excellent technical assistance, E. Söllner for her contribution at the beginning of the project and R. Parlitz for stimulating discussions. This work was supported by a grant from the Deutsche Forschungsgemeinschaft (SFB638, A3) to IS. We acknowledge access to the beamlines at the ESRF in Grenoble and the support of the beamline scientists.
References
- Andersen, E. S., Rosenblad, M. A., Larsen, N., Westergaard, J. C., Burks, J., Wower, I. K., Wower, J., Gorodkin, J., Samuelsson, T. & Zwieb, C. (2006). Nucleic Acids Res.34, D163–D168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff, D. S. & Ordal, G. W. (1992). Mol. Microbiol.6, 23–28. [DOI] [PubMed] [Google Scholar]
- Carpenter, P. B., Hanlon, D. W. & Ordal, G. W. (1992). Mol. Microbiol.6, 2705–2713. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Correa, N. E., Peng, F. & Klose, K. E. (2005). J. Bacteriol.187, 6324–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymann, D. M., Keenan, R. J., Stroud, R. M. & Walter, P. (1997). Nature(London), 385, 361–364. [DOI] [PubMed]
- Luirink, J. & Sinning, I. (2004). Biochim. Biophys. Acta, 1694, 17–35. [DOI] [PubMed] [Google Scholar]
- Macnab, R. M. (2003). Annu. Rev. Microbiol.57, 77–100. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Montoya, G., Svensson, C., Luirink, J. & Sinning, I. (1997). Nature (London), 385, 365–368. [DOI] [PubMed] [Google Scholar]
- Murray, T. S. & Kazmierczak, B. I. (2006). J. Bacteriol.188, 6995–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehus, E., Gressmann, H., Ye, F., Schlapbach, R., Dehio, M., Dehio, C., Stack, A., Meyer, T. F., Suerbaum, S. & Josenhans, C. (2004). Mol. Microbiol.52, 947–961. [DOI] [PubMed] [Google Scholar]
- Nowalk, A. J., Gilmore, R. D. Jr & Carroll, J. A. (2006). Infect. Immun.74, 3864–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandza, S., Baetens, M., Park, C. H., Au, T., Keyhan, M. & Matin, A. (2000). Mol. Microbiol.36, 414–423. [DOI] [PubMed] [Google Scholar]
- Rost, B., Yachdav, G. & Liu, J. (2004). Nucleic Acids Res.32, W321–W326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y., Chern, M., Silva, F. G. & Ronald, P. (2001). Mol. Plant Microbe Interact.14, 204–213. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997). Nucleic Acids Res.25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanen, G., Antelmann, H., Westers, H., Hecker, M., van Dijl, J. M. & Quax, W. J. (2004). J. Bacteriol.186, 5956–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]