The avian reovirus double-stranded RNA-binding and core protein σA has been crystallized in space group P1, with unit-cell parameters a = 103.2, b = 129.9, c = 144.0 Å, α = 93.8, β = 105.1, γ = 98.2°. A complete data set has been collected to 2.3 Å resolution and analyzed.
Keywords: σA, avian reovirus
Abstract
The avian reovirus protein σA plays a dual role: it is a structural protein forming part of the transcriptionally active core, but it has also been implicated in the resistance of the virus to interferon by strongly binding double-stranded RNA and thus inhibiting the double-stranded RNA-dependent protein kinase. The σA protein has been crystallized from solutions containing ammonium sulfate at pH values around 6. Crystals belonging to space group P1, with unit-cell parameters a = 103.2, b = 129.9, c = 144.0 Å, α = 93.8, β = 105.1, γ = 98.2° were grown and a complete data set has been collected to 2.3 Å resolution. The self-rotation function suggests that σA may form symmetric arrangements in the crystals.
1. Introduction
Avian reovirus is a non-enveloped virus and is a member of the Orthoreovirus genus of the Reoviridae family (Benavente & Martinez-Costas, 2007 ▶). The reovirus has been linked to various diseases in birds that account for considerable economic losses in the poultry industry (Jones, 2000 ▶). The virus has a double-stranded RNA genome consisting of ten segments which are encased by a double concentric icosahedral capsid (Spandidos & Graham, 1976 ▶). The inner capsid is made up of the λA, λC and σA proteins, which encapsulate up to 12 copies of the RNA-dependent RNA polymerase complex (consisting of λB and μA) together forming the viral ‘core’. Proteins μB, σB and σC are components of the outer shell (Martinez-Costas et al., 1997 ▶), with σC being responsible for primary host-cell attachment (Grande et al., 2000 ▶; Guardado-Calvo et al., 2005 ▶). The virus also encodes four nonstructural proteins which are involved in cell fusion (p10), viral assembly (μNS and σNS) or in as yet undetermined functions (p17) (Varela et al., 1996 ▶; Bodelon et al., 2001 ▶; Touris-Otero et al., 2004 ▶; Costas et al., 2005 ▶).
The avian reovirus protein σA plays a dual role. It functions primarily as a structural protein that forms part of the transcriptionally active core. In addition, it has also been implicated in the resistance of the virus to interferon by binding double-stranded RNA and thus inhibiting the double-stranded RNA-dependent protein kinase (Martinez-Costas et al., 2000 ▶; Gonzalez-Lopez et al., 2003 ▶). The structure of the mammalian reovirus core has been solved by X-ray crystallography (Reinisch et al., 2000 ▶). In this structure, the structural equivalent of avian reovirus σA, σ2, is present in 150 copies per core in three different locations (σ2-i, σ2-ii and σ2-iii) and acts as a core-stabilizing clamp. The protein interacts extensively with λ1 (the mammalian reovirus equivalent of λA) at two different binding sites (the binding sites for σ2-ii and σ2-iii are similar). There are only sparse contacts between σ2 monomers and between σ2 and the other core protein λ2 (the equivalent of avian reovirus λC). In agreement with the mammalian reovirus ‘core’ structure, cryo-electron microscopy of avian reovirions confirmed that they contain 150 σA ‘nodules’ contacting mainly λA, with minor contacts formed with the λC turrets and other σA molecules (Zhang et al., 2005 ▶). They also contact the μB proteins of the outer capsid (equivalent to μ1 in mammalian reovirus). However, with respect to double-stranded RNA binding, σA appears to play an analogous role to the mammalian reovirus protein σ3, as this protein (the structural equivalent of avian reovirus σB) exhibits double-stranded RNA-binding activity (Yue & Shatkin, 1997 ▶), albeit not as strongly as avian reovirus σA (Martinez-Costas et al., 2000 ▶).
In our ongoing efforts to obtain structural and functional information about the protein, we have expressed and purified σA in milligram amounts, crystallized it and collected a complete data set to 2.3 Å resolution.
2. Methods
The avian reovirus S1133 protein σA was expressed as a maltose-binding-protein fusion using the expression vector pMALCS2 described in Gonzalez-Lopez et al. (2003 ▶). Prior to proceeding with milligram-scale expression and purification for crystallization experiments, the sequence of the insert encoding σA was confirmed by DNA-sequence analysis (Sistemas Genomicos, Valencia, Spain).
For large-scale expression, the Escherichia coli strain XL1-Blue was freshly transformed with the pMALCS2 vector and four 750 ml cultures were grown aerobically at 310 K to an optical density of 0.6 measured at 600 nm in LB medium. Expression was induced by adding 1 mM isopropyl β-d-thiogalactopyranoside and allowed to proceed for 3 h. Harvested cells were resuspended in 40 ml lysis buffer (20 mM Tris–HCl pH 7.3, 200 mM sodium chloride, 1 mM dithiothreitol) containing a protease-inhibitor cocktail specific for bacterial cell extracts (Sigma–Aldrich, Madrid, Spain) and the suspension was frozen at 253 K. The cellular pellet was lysed by a double pass through an emulsifier (Avestin Emulsifier C5, Avestin Europe GmbH, Mannheim, Germany). After removal of the insoluble material by centrifugation, sodium chloride and poly(ethyleneimine) were added to final concentrations of 1 M and 0.1%(w/v), respectively, and the suspension was incubated at 277 K for 1 h to precipitate nucleic acids. Following a second centrifugation step, bovine ribonuclease A (>70 Kunitz units per milligram; Sigma–Aldrich, Madrid, Spain) was added to the supernatant to a final concentration of around 30 µg ml−1 and incubated for 30 min at room temperature. 3 ml suspended amylose resin (New England Biolabs, Ipswich MA, USA) was then added and gently mixed and the suspension was left for 10 min at room temperature before being poured into an empty column. The resin was washed three times with three column volumes of MBP buffer (20 mM Tris–HCl pH 7.5, 200 mM sodium chloride, 1 mM EDTA) and elution was performed by washing twice with 10 ml 10 mM maltose in MPB buffer. Calcium chloride and protease factor Xa (New England Biolabs, Ipswich MA, USA) were added to final concentrations of 1 µg ml−1 and 1 mM, respectively. Incubation was carried out at room temperature for 20 h. The digested protein was dialysed overnight against TE buffer (10 mM Tris–HCl pH 8.5, 1 mM EDTA) and applied onto a 1 ml Uno-Q column (Biorad, Barcelona, Spain). Pure σA eluted in the flowthrough, while maltose-binding protein and other impurities were retained on the column. The protein was concentrated with Centricon concentrators (Millipore, Madrid, Spain), using three washes with TE buffer to eliminate small-molecule impurities.
Crystallization was carried out by sitting-drop vapour diffusion at 278 K in CompactClover plates (Jena Biosciences, Jena, Germany), with 0.1–0.15 ml reservoirs and drops consisting of 2–5 µl protein solution (14–30 mg ml−1 protein in TE buffer) mixed with 2–5 µl reservoir solution, using equal volumes of both. Diffraction images were processed using MOSFLM (Leslie, 2006 ▶) and the crystallographic data were scaled using SCALA (Collaborative Computational Project, Number 4, 1994 ▶). Self-rotation functions were calculated using MOLREP (Vagin & Teplyakov, 2000 ▶).
3. Results and discussion
Recombinant σA was expressed as a fusion protein with maltose-binding protein. After lysis, excess nucleic acids were removed by precipitation and the preparation was extensively treated with RNase A to remove all traces of bound RNA (σA binds double-stranded RNA very tightly). The fusion protein was subsequently partially purified using amylose resin. Upon cleavage with a specific protease, σA was separated from maltose-binding protein and other impurities by anion-exchange chromatography; pure σA eluted in the flowthrough fraction. From the protease-cleavage step onwards, the procedures were performed at room temperature, since σA was found to precipitate in the cold in the absence of salt (albeit reversibly). This tendency to precipitate reversibly is an indication that the protein may form higher order aggregates in solution and the temperature-dependence of this aggregation suggests that polar interactions may predominate in the oligomerization process.
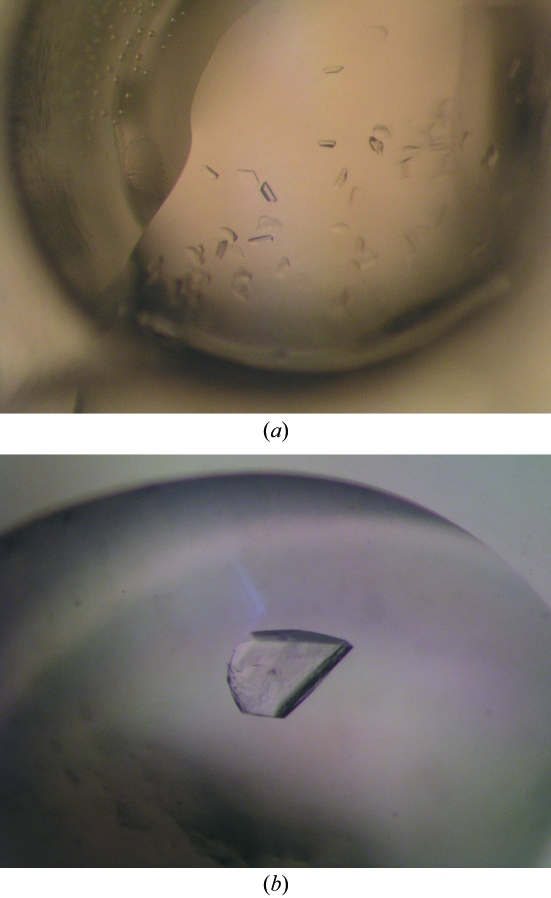
In vapour-diffusion experiments, sizable crystals were obtained after 6–12 months at pH 5.5–6.5 and 278 K using 100 mM sodium citrate, sodium phosphate or 2-(N-morpholino)ethanesulfonic acid–NaOH as a buffer and 0.1–0.6 M ammonium sulfate as precipitant (Fig. 1 ▶). For data collection, crystals were briefly incubated in mother liquor containing 25%(v/v) glycerol and then flash-frozen in liquid nitrogen.
Figure 1.
Crystals of the avian reovirus σA protein. (a) A typical crystallization drop containing multiple small crystals. (b) A single large crystal; this crystal is about 0.3 mm in width and 0.05 mm thick.
A high prevalence of multiple lattices was observed in the diffraction patterns obtained from these crystals. However, fairly complete data sets with reasonable statistics were collected from several crystals using synchrotron radiation, in the best case yielding a data set to 2.3 Å resolution (Table 1 ▶). For each crystal, 180 1° oscillation-angle images were collected, with exposure times of 1–2 s for each image, using the insertion-device beamline ID23-1 at the European Synchroton Radiation Facility. Data sets from different crystals were not merged, as the crystals were found to be insufficiently isomorphous to each other. The crystals were assigned to space group P1 and analysis of the Matthews coefficient suggested the presence of between six and 18 protein molecules in the asymmetric unit (with a solvent fraction of between 81 and 43%, respectively). We have recently obtained a convincing molecular-replacement solution containing 12 copies of a model based on the mammalian reovirus core protein σ2, which would give 62% solvent content.
Table 1. Statistics of crystallographic data collected from a single σA crystal.
Values in parentheses are for the highest resolution bin (where applicable).
| Beamline | ESRF ID23-1 |
| Wavelength (Å) | 0.954 |
| Detector | MAR Mosaic |
| Crystal-to-detector distance (mm) | 251 |
| Space group | P1 |
| Unit-cell parameters (Å, °) | a = 103.2, b = 129.9, c = 144.0, α = 93.8, β = 105.1, γ = 98.2 |
| Mosaicity (°) | 0.6 |
| No. of observed reflections† | 282973 (32868) |
| Resolution range (Å) | 30.0–2.34 (2.47–2.34) |
| Multiplicity | 2.0 (1.9) |
| Completeness (%) | 94.3 (74.8) |
| 〈I/σ(I)〉 | 7.7 (2.2) |
| Rsym‡ (%) | 10.5 (30.0) |
No σ-cutoff or other restrictions were used for the inclusion of observed reflections.
R
sym = 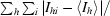
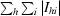 , where I
hi is the intensity of the ith measurement of the same reflection and 〈I
h〉 is the mean observed intensity for that reflection.
, where I
hi is the intensity of the ith measurement of the same reflection and 〈I
h〉 is the mean observed intensity for that reflection.
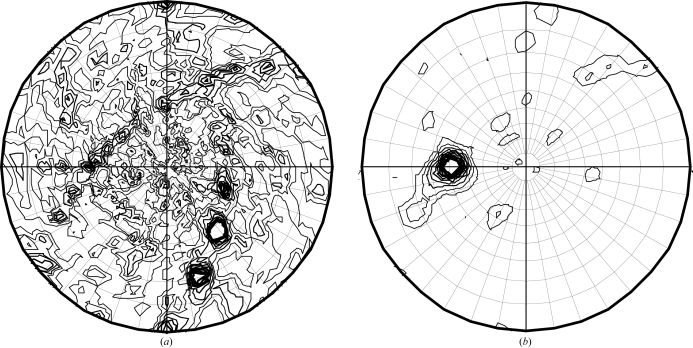
Self-rotation functions calculated from the different data sets consistently show peaks in the χ = 180 and 50° sections (Fig. 2 ▶). The χ = 50° section shows a single peak at θ = 49° and ϕ = −90°, while the more noisy χ = 180° section shows two rows of peaks of different intensities, indicating multiple twofold rotation axes perpendicular to a single higher symmetry axis. The θ = 49, ϕ = −90° peak is also present at χ = 100°, but not at χ = 150°, suggesting that this higher symmetry axis is not a complete sevenfold symmetry axis. The peak maximum is at a χ value of 49.7–50.2°, deviating by more than a degree from exact sevenfold symmetry. An exact complete sevenfold symmetry axis with perpendicular twofold axes would also not be consistent with the molecular-replacement solution, which contains only 12 molecules, not 14. Apparently, multiple σA molecules assemble into a regularly ordered assembly, which is likely to have importance for its function in sequestering double-stranded RNA. Structure solution, refinement and analysis are in progress and will be reported elsewhere.
Figure 2.
Self-rotation function calculated using the program MOLREP (Vagin & Teplyakov, 2000 ▶) from a complete σA data set. (a) χ = 180° section; (b) χ = 50° section, indicating the presence of multiple noncrystallographic twofolds and a single noncrystallographic higher symmetry axis.
Acknowledgments
We thank José Antonio Trillo, Salvador Blanco-Turnes and Rebeca Menaya-Vargas for technical assistance in the laboratory. Dave Hall (beamline ID23-1) helped with data collection at the European Synchrotron Radiation Facility. This research was sponsored by research grants BMC2002-02436, BFU2005-02974 and BFU2005-24982-E from the Spanish Ministry of Science and Technology and PGIDIT03PXIC20307PN from the Xunta de Galicia. As part of the European Science Foundation EUROCORES Programme EuroSCOPE, the work was also supported by funds from the European Commission’s Sixth Framework Programme under contract ERAS-CT-2003-980409.
References
- Benavente, J. & Martinez-Costas, J. (2007). Virus Res.123, 105–119. [DOI] [PubMed] [Google Scholar]
- Bodelon, G., Labrada, L., Martinez-Costas, J. & Benavente, J. (2001). Virology, 290, 181–191. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Costas, C., Martínez-Costas, J., Bodelon, G. & Benavente, J. (2005). J. Virol.79, 2141–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Lopez, C., Martinez-Costas, J., Esteban, M. & Benavente, J. (2003). J. Gen. Virol.84, 1629–1639. [DOI] [PubMed] [Google Scholar]
- Grande, A., Rodriguez, E., Costas, C., Everitt, E. & Benavente, J. (2000). Virology, 274, 367–377. [DOI] [PubMed] [Google Scholar]
- Guardado-Calvo, P., Fox, G. C., Hermo-Parrado, X. L., Llamas-Saiz, A. L., Costas, C., Martinez-Costas, J., Benavente, J. & van Raaij, M. J. (2005). J. Mol. Biol.354, 137–149. [DOI] [PubMed] [Google Scholar]
- Jones, R. C. (2000). Rev. Sci. Tech.19, 614–625. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed] [Google Scholar]
- Martinez-Costas, J., Gonzalez-Lopez, C., Vakharia, V. N. & Benavente, J. (2000). J. Virol.74, 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Costas, J., Grande, A., Varela, R., Garcia-Martinez, C. & Benavente, J. (1997). J. Virol.71, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinisch, K. M., Nibert, M. L. & Harrison, S. C. (2000). Nature (London), 960–967. [DOI] [PubMed] [Google Scholar]
- Spandidos, D. A. & Graham, A. F. (1976). J. Virol.19, 968–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touris-Otero, F., Martinez-Costas, J., Vakharia, V. N. & Benavente, J. (2004). Virology, 319, 94–106. [DOI] [PubMed] [Google Scholar]
- Vagin, A. & Teplyakov, A. (2000). Acta Cryst. D56, 1622–1624. [DOI] [PubMed] [Google Scholar]
- Varela, R., Martinez-Costas, J., Mallo, M. & Benavente, J. (1996). J. Virol.70, 2974–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, Z. & Shatkin, A. J. (1997). Virology, 234, 364–371. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Tang, J., Walker, S. B., O’Hara, D., Nibert, M. L., Duncan, R. & Baker, T. S. (2005). Virology, 343, 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]