S. cerevisiae Atg10, an E2-like enzyme that mediates the conjugation reaction between Atg12 and Atg5, was crystallized and diffracted to 2.3 Å resolution.
Keywords: Atg10, autophagy
Abstract
Atg10 is an E2-like enzyme that catalyzes the conjugation reaction between Atg12 and Atg5. The Atg12–Atg5 conjugate is essential for autophagy, the bulk degradation process of cytoplasmic components by the vacuolar/lysosomal system. Microcrystals of Saccharomyces cerevisiae Atg10 were obtained by the free-interface diffusion method using polyethylene glycol and sodium acetate as precipitants. Using these precipitants, large crystals suitable for data collection were obtained using the sitting-drop vapour-diffusion method. The crystals belong to space group P41212 or P43212, with unit-cell parameters a = b = 51.61, c = 256.16 Å, and are estimated to contain two protein molecules per asymmetric unit. A native data set was collected to 2.3 Å resolution from a single crystal.
1. Introduction
Ubiquitin is conjugated to its target proteins by the following sequential reactions. Firstly, ubiquitin is processed by a specific protease to expose a glycine residue at its carboxy-terminus. The exposed glycine is activated by an E1 enzyme and is then transferred to an E2 enzyme. Finally, usually supported by an E3 enzyme, ubiquitin is conjugated to its target proteins (Varshavsky, 1997 ▶). Many ubiquitin-like modifiers have been reported and they all seem to be conjugated to their targets via a mechanism similar to ubiquitination (Welchman et al., 2005 ▶). Autophagy, the bulk degradation process of cytosolic components by the vacuolar/lysosomal system (Ohsumi, 2001 ▶), has also been shown to have similar modification systems, such as ubiquitination in the yeast Saccharomyces cerevisiae (Mizushima et al., 1998 ▶; Ichimura et al., 2000 ▶). A 21 kDa protein, Atg12, is one such modifier essential for autophagy (Mizushima et al., 1998 ▶). The calboxy-terminal glycine of Atg12 is activated by Atg7, an E1-like enzyme, through ATP hydrolysis (Tanida et al., 1999 ▶) and Atg12 is then transferred to Atg10, an E2-like enzyme (Shintani et al., 1999 ▶). Finally, an isopeptide linkage is formed between the carboxy-terminal glycine of Atg12 and the Lys149 side chain of Atg5. The Atg12 conjugation system is conserved in eukaryotes, including mammals, and has been shown to be essential for autophagy in both yeast and mammals.
In contrast to ubiquitin, which has many target proteins, the target for Atg12 is restricted to Atg5 alone. Furthermore, no E3-like enzyme has been reported for the Atg12 conjugation system. Therefore, Atg10 seems to recognize both Atg12 and Atg5 directly and to catalyze their conjugation reaction. Atg12 has a ubiquitin fold despite having low sequence homology with ubiquitin (Suzuki et al., 2005 ▶). Atg5 has also been shown to possess ubiquitin folds (Matsushita et al., 2007 ▶). Structural information on Atg10 would be helpful in order to understand how this enzyme recognizes two ubiquitin-fold proteins simultaneously and catalyzes their conjugation reaction. In this report, we describe the purification, crystallization and preliminary crystallographic analysis of S. cerevisiae Atg10.
2. Expression and purification
Using the NdeI–EcoRI restriction sites, the region encoding full-length Atg10 (residues 1–167) was inserted into the pGEX-6P vector (GE Healthcare), which has an NdeI site introduced upstream of the BamHI site. pGEX-6p-Atg10 has a PreScission protease-cleavage sequence between glutathione-S-transferase (GST) and Atg10. GST-fused Atg10 with a PreScission cleavage site was expressed in Escherichia coli BL21 DE3. After cell lysis, the GST-fused Atg10 was first purified by affinity chromatography using a glutathione Sepharose 4B column (GE Healthcare). The GST was then excised from Atg10 with PreScission protease (GE Healthcare) at 277 K, resulting in Atg10 with a Gly-Pro-His sequence at its N-terminus (molecular weight 20 500 Da). After changing the buffer to 20 mM Tris buffer pH 8.0, 2 mM DTT and 150 mM NaCl using a HiTrap desalting column (GE Healthcare), the protein was applied onto a glutathione Sepharose 4B column and was eluted with 20 mM Tris buffer pH 8.0, 2 mM DTT and 150 mM NaCl in order to remove GST. Finally, the eluted protein was applied onto a Superdex 75 gel-filtration column (GE Healthcare) and was eluted with 20 mM Tris buffer pH 8.0, 150 mM NaCl and 2 mM DTT. The purified protein was concentrated to 15 mg ml−1 for crystallization.
3. Crystallization
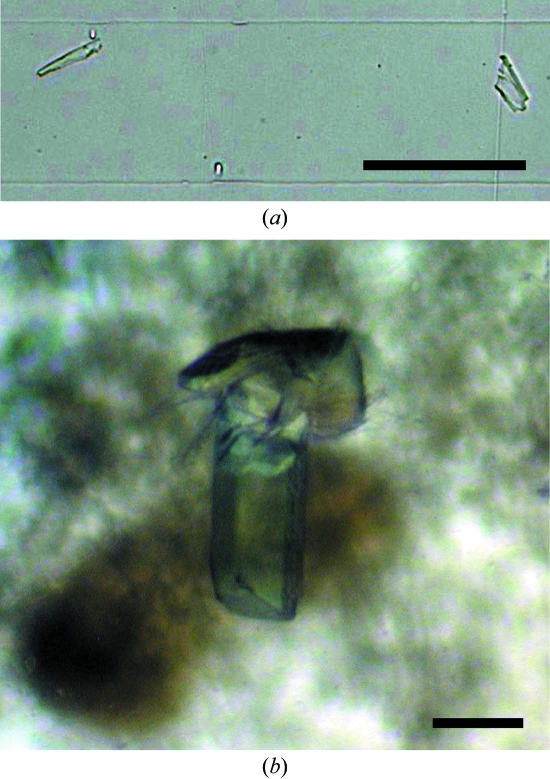
The free-interface diffusion method was initially applied for crystallization trials using TOPAZ screening chips (Fluidigm). OptiMix-1 and OptiMix-4 (Fluidigm) were used as crystallization reagents. A total of 3 µl of 15 mg ml−1 Atg10 in 150 mM NaCl, 20 mM Tris–HCl pH 8.0 and 2 mM DTT was loaded into 192 diffusion chambers within two chips and was mixed with 192 varieties of OptiMix reagents by diffusion at 293 K. Small but well shaped crystals of Atg10 were obtained using a solution consisting of 0.5 M sodium acetate and 27% polyethylene glycol 4000 (Fig. 1 ▶ a). Next, using these reagents as reservoir solutions, crystallization trials were performed using the sitting-drop vapour-diffusion method. 1.0 µl drops of 15 mg ml−1 Atg10 in 150 mM NaCl, 20 mM Tris–HCl pH 8.0 and 2 mM DTT were mixed with equal amounts of reservoir solution and equilibrated against 100 µl of the same reservoir solution by vapour diffusion at 293 K. After optimization of crystallization conditions, large crystals of Atg10 were obtained using a reservoir solution consisting of 0.4 M sodium acetate, 17% polyethylene glycol 3350 and 0.1 M CHES buffer pH 9.5 (Fig. 1 ▶ b). The crystals grew to dimensions of about 0.3 × 0.1 × 0.1 mm in a week.
Figure 1.
Crystals of Atg10. (a) Microcrystals obtained using the free-interface diffusion method. (b) Crystals obtained using the sitting-drop vapour-diffusion method. The black scale bar is 100 µm in length.
4. Preliminary crystallographic analysis
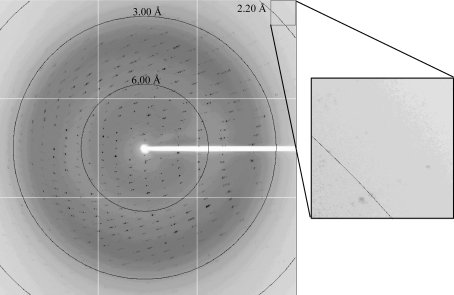
Crystals were immersed for several seconds into reservoir solution supplemented with 10% glycerol as a cryoprotectant and then flash-cooled and kept in a stream of nitrogen gas at 100 K during data collection. Diffraction data were collected at 100 K on an ADSC Quantum 315 charge-coupled device detector on beamline BL41XU, SPring-8, Japan at a wavelength of 1.00 Å. A diffraction image is shown in Fig. 2 ▶. Diffraction data were indexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). The data-collection statistics are summarized in Table 1 ▶. The relatively low completeness of the data is attributable to radiation damage and to the limited detector area for the outer shell. The crystal belongs to the tetragonal space group P41212 or P43212, with unit-cell parameters a = b = 51.61, c = 256.16 Å. The acceptable range of volume-to-weight ratio (V M) values (Matthews, 1968 ▶) indicates that the crystal contains two protein molecules per asymmetric unit, with a solvent content of 42.1% (V M = 2.12 Å3 Da−1). Although Atg10 is considered to be an E2-like conjugating enzyme, it has little sequence homology to canonical E2 enzymes of reported structure; thus, molecular replacement using other E2 structures as models is not applicable for structure determination of Atg10. Structure determination is in progress using the multiple isomorphous replacement method and/or the multiple anomalous diffraction method.
Figure 2.
A diffraction image of an Atg10 crystal. Circles are labelled with the resolution limit.
Table 1. Summary of crystallographic data.
Values in parentheses are for the highest resolution shell.
| Resolution range (Å) | 50.0–2.30 (2.38–2.30) |
| Observed reflections | 44044 |
| Unique reflections | 14281 |
| Completeness (%) | 86.1 (69.7) |
| Redundancy | 3.1 (1.6) |
| Rmerge(I)† | 0.070 (0.285) |
| I/σ(I) | 16.6 (3.5) |
R
merge(I) = 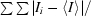
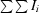 , where I
i is the intensity of the ith observation and 〈I〉 is the mean intensity.
, where I
i is the intensity of the ith observation and 〈I〉 is the mean intensity.
Acknowledgments
The synchrotron-radiation experiments were performed on BL41XU at SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI; Proposal No. 2006B2668). This work was partly supported by Grant-in-Aids for Young Scientists (B) 17790048 and Priority Areas and the National Project on Protein Structural and Functional Analyses from the Ministry of Education, Culture, Sports, Science and Technology, Japan. This work was carried out under the NIBB Cooperative Research Program (4-148).
References
- Ichimura, Y., Kirisako, T., Takao, T., Satomi, Y., Shimonishi, Y., Ishihara, N., Mizushima, N., Tanida, I., Kominami, E., Ohsumi, M., Noda, T. & Ohsumi, Y. (2000). Nature (London), 408, 488–492. [DOI] [PubMed] [Google Scholar]
- Matsushita, M., Suzuki, N. N., Obara, K., Fujioka, Y., Ohsumi, Y. & Inagaki, F. (2007). J. Biol. Chem.282, 6763–6772. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Mizushima, N., Noda, T., Yoshimori, T., Tanaka, Y., Ishii, T., George, M. D., Klionsky, D. J., Ohsumi, M. & Ohsumi, Y. (1998). Nature (London), 395, 395–398. [DOI] [PubMed] [Google Scholar]
- Ohsumi, Y. (2001). Nature Rev. Mol. Cell Biol.2, 211–216. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Shintani, T., Mizushima, N., Ogawa, Y., Matsuura, A., Noda, T. & Ohsumi, Y. (1999). EMBO J.18, 5234–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, N. N., Yoshimoto, K., Fujioka, Y., Ohsumi, Y. & Inagaki, F. (2005). Autophagy, 1, 119–126. [DOI] [PubMed] [Google Scholar]
- Tanida, I., Mizushima, N., Kiyooka, M., Ohsumi, M., Ueno, T., Ohsumi, Y. & Kominami, E. (1999). Mol. Biol. Cell, 10, 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky, A. (1997). Trends Biochem. Sci.22, 383–387. [DOI] [PubMed] [Google Scholar]
- Welchman, R. L., Gordon, C. & Mayer, R. J. (2005). Nature Rev. Mol. Cell Biol.6, 599–609. [DOI] [PubMed]