Cocrystallization with a peptide, free-interface diffusion crystal chips and crystal dehydration were important in the production of diffraction-quality crystals of the Munc18c protein that helps to regulate membrane fusion.
Keywords: Munc18c, syntaxin4, free-interface diffusion, dehydration
Abstract
The production of diffraction-quality crystals of Munc18c, a protein involved in regulating vesicular exocytosis in mammals, is reported. The diffraction resolution of Munc18c crystals was optimized by (i) cocrystallizing with a peptide fragment of the Munc18c functional binding partner syntaxin4, (ii) using nanolitre free-interface diffusion crystallization-screening chips and microlitre hanging-drop vapour diffusion and (iii) applying a post-crystallization dehydration treatment. Crystals belonging to the cubic space group P213, with unit-cell parameters a = b = c = 170.8 Å, α = β = γ = 90°, were generated that diffract to 3.7 Å resolution on a laboratory X-ray source.
1. Introduction
Munc18c belongs to the Sec1/Munc18 (SM) family of proteins that regulate vesicular exocytosis (Toonen & Verhage, 2003 ▶; Tellam et al., 1995 ▶; Thurmond et al., 1998 ▶). The regulatory effects of SM proteins are mediated through interactions with other exocytotic proteins, principally the soluble N-ethylmaleimide sensitive factor attachment protein receptors (SNAREs; Toonen & Verhage, 2003 ▶; Gallwitz & Jahn, 2003 ▶). Munc18c is a difficult protein to handle; it cannot be expressed in a stable form using bacterial expression systems, although insect-cell expression using recombinant baculovirus does generate protein of suitable stability for crystallization trials. The expression and purification of N-terminally His-tagged Munc18c have been reported in detail previously (Hu et al., 2003 ▶). We found that Munc18c prepared in this manner crystallizes readily, but the diffraction resolution of the 0.2 × 0.2 × 0.2 mm crystals is only ∼10 Å using our laboratory X-ray source (data not shown). We therefore set out to improve the diffraction resolution of Munc18c crystals using a three-pronged approach.
Firstly, we cocrystallized Munc18c with a syntaxin4 peptide. Syntaxin4 is the SNARE binding partner of Munc18c and we showed recently that a peptide corresponding to the N-terminal 29 residues of syntaxin4 is required for the interaction between these two proteins (Latham et al., 2006 ▶). Secondly, our initial crystallization trials were performed using a nanolitre free-interface diffusion screening chip in order to minimize the amount of protein used during the screening phase. The results were then scaled up to microlitre volumes using hanging-drop vapour-diffusion experiments in order to obtain large crystals of the protein–peptide complex. Thirdly, we employed a post-crystallization dehydration treatment in order to improve diffraction quality (Heras et al., 2003 ▶; Heras & Martin, 2005 ▶). Using these three approaches, we generated crystals of dimensions 0.2 × 0.2 × 0.2 mm that diffracted to a resolution of 3.7 Å using a laboratory X-ray source.
2. Methods
2.1. Protein and peptide production
Recombinant N-terminally His-tagged mouse (Mus musculus) Munc18c (73 493 Da, including the six-His tag), residues 1–592, was produced from baculovirus-infected insect cells and purified as described previously (Hu et al., 2003 ▶). A peptide consisting of the N-terminal 29 residues of mouse syntaxin4 (syntaxin41–29, 3392 Da) was chemically synthesized as described previously (Latham et al., 2006 ▶).
2.2. Crystallization
Purified N-terminally tagged Munc18c (in 25 mM HEPES pH 7.0, 150 mM NaCl, 2 mM β-mercaptoethanol) was concentrated to 10 mg ml−1 using an Amicon Ultra-4 centrifugal concentration device with a molecular-weight cutoff of 10 kDa (Millipore, Billerica, MA, USA). Munc18c was mixed with syntaxin41–29 to give a tenfold molar excess of the peptide and the mixture was then incubated for 1–2 h on ice. Cocrystallization trials were set up using free-interface diffusion (Hansen et al., 2002 ▶) Topaz Screening 1.48 chips and Topaz Crystallizer with the Topaz Optimix I screen consisting of 96 conditions (Fluidigm, South San Francisco, CA, USA). After setup, the Topaz chip was stored at 293 K in a temperature-controlled room and examined manually with a Nikon SMZ/U light microscope every 2–3 d. Six conditions that produced small crystals in the Topaz chips were translated to hanging-drop vapour-diffusion format according to the manufacturer’s translation workbook and guide (Fluidigm, South San Francisco, CA, USA). These six crystallization conditions were optimized by screening different types of PEG at different concentrations, pH, protein concentrations and salt concentrations in 24-well plates (pre-greased VDX plates from Hampton Research, San Diego, CA, USA) at 293 K. Hanging drops of 2–4 µl (1–2 µl each of the protein and reservoir solutions, respectively) were equilibrated over 500 µl reservoir solution. The optimized crystallization conditions consisted of a reservoir solution containing 10–13% PEG 3350, 0.2 M magnesium acetate, 0.1 M MES pH 6.5 and 50 mM magnesium chloride. In both the Topaz chip and hanging-drop experiments, crystals were observed after 3 d; the crystals in the hanging-drop experiments grew to full size over a week. It is possible that crystals appeared more quickly in the Topaz chips than in the hanging-drop experiments, but the chips and vapour-diffusion experiments were only checked every 2–3 d. Consequently, crystals were only observed on the third day in both set-ups.
2.3. Diffraction
Crystals were evaluated for X-ray diffraction on a Rigaku FR-E copper rotating-anode generator operating at 45 kV and 45 mA with Osmic Confocal Max-Flux (HiRes2) optics. Reflections were measured using a Rigaku R-AXIS IV++ imaging-plate area detector. One crystal from the Topaz chip experiment and one crystal from the hanging-drop experiment were evaluated at room temperature (293 K) after mounting in quartz capillary tubes (Charles Supper Company, Natick, MA, USA). All other X-ray diffraction analyses were performed at 100 K on crystals harvested with nylon loops (Hampton Research, San Diego, CA, USA) from the hanging-drop experiments. These crystals were cryoprotected by dipping into cryocooling solution for 30 s to 2 min followed by flash-cooling in a nitrogen-gas stream. The cryocooling solution comprised 15% ethylene glycol, 17% PEG 3350, 0.2 M magnesium acetate, 0.1 M MES pH 6.5 and 50 mM magnesium chloride. A CryoCool-LN2 (Cryo Industries, Manchester, NH, USA) was used for cooling crystals. The X-ray diffraction data were integrated, processed and scaled using the program HKL-2000 (Otwinowski & Minor, 1997 ▶).
2.4. Dehydration
For crystal dehydration (Heras et al., 2003 ▶; Heras & Martin, 2005 ▶), crystals were transferred into a buffer comprising 25–30% PEG 3350, 0.2 M magnesium acetate, 0.1 M MES pH 6.5, 50 mM magnesium chloride for 3 h, 1 d or 2 d. After dehydration, the crystals were cryocooled in dehydration buffer also containing 15% ethylene glycol and tested for diffraction quality as above. To confirm the reproducibility of the results shown in Fig. 2 and Table 2, we evaluated three nondehydrated crystals, two crystals after 3 h dehydration, two crystals after 1 d dehydration and two crystals after 2 d dehydration (data not shown).
3. Results and discussion
Initial cocrystallization screening was carried out by free-interface diffusion using Topaz crystallization-screening chips. This allowed 96 conditions to be assessed using just 3 µl of protein, where each well contained 0.75–2.25 nl protein at three protein:precipitant ratios: 1:3, 1:1 and 3:1. After 3 d, crystals were observed in the protein wells of the Topaz chip in six different conditions of the Optimix I screen (Fig. 1 ▶, Table 1 ▶). All six conditions included PEG or PEG monomethyl ether (MME) as a precipitant. A crystal from condition 4 (Table 1 ▶; ∼40 µm in the longest dimension) was retrieved from the chip for diffraction analysis, but no diffraction pattern could be recorded. It was not clear whether the lack of diffraction was a consequence of inherent disorder in the crystal or simply that the crystal was too small for diffraction to be observed using the laboratory X-ray facility. To address this question, larger crystals were required.
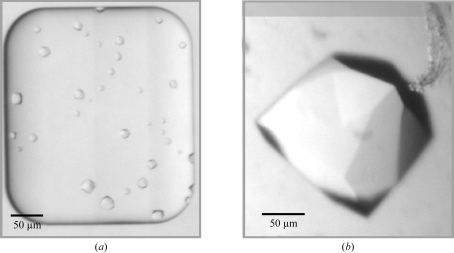
Figure 1.
Munc18c crystallization. (a) Crystals of similar morphology formed in the protein wells of Topaz crystal chips in six related PEG crystallization conditions. Crystals from condition 6 of the Optimix screen (Table 1 ▶) are shown. The crystallization components and approximate dimensions of the crystals for the six conditions are given in Table 1 ▶. The crystal sizes vary from 10 to 40 µm in the longest dimension. (b) Crystallization conditions were translated from free-interface diffusion to hanging-drop vapour diffusion. The crystal shown (longest dimension 200 µm) was grown by hanging-drop vapour diffusion in 12% PEG 3350, 0.2 M magnesium acetate, 0.1 M MES pH 6.5 and 50 mM magnesium chloride.
Table 1. Crystallization conditions.
1–6 refer to conditions producing crystals in the Topaz chip; condition 7 was used to produce crystals using hanging-drop vapour diffusion. The crystals produced from conditions 6 and 7 are shown in Fig. 1 ▶.
| Crystallization conditions | ||||
|---|---|---|---|---|
| Precipitant | Buffer | Salt | Approximate crystal size† (µm) | |
| 1 | 20% PEG MME 1900 | 0.1 M HEPES pH 7.5 | 0.5 M potassium thiocyanate | 30–40 |
| 2 | 20% PEG 20 000 | 0.1 M HEPES pH 7.5 | 0.4 M potassium nitrate | 40 |
| 3 | 20% PEG MME 5000 | — | 0.5 M potassium acetate | 10–20 |
| 4 | 15% PEG 10 000 | 0.1 M sodium acetate pH 4.6 | 0.3 M magnesium formate | 40 |
| 5 | 30% PEG MME 1900 | — | 0.3 M magnesium formate | 30 |
| 6 | 25% PEG 3350 | 0.1 M sodium cacodylate pH 6.5 | 0.6 M magnesium acetate | 20 |
| 7 | 12% PEG 3350 | 0.1 M MES pH 6.5 | 0.2 M magnesium acetate, 50 mM magnesium chloride | 200 |
Longest dimension.
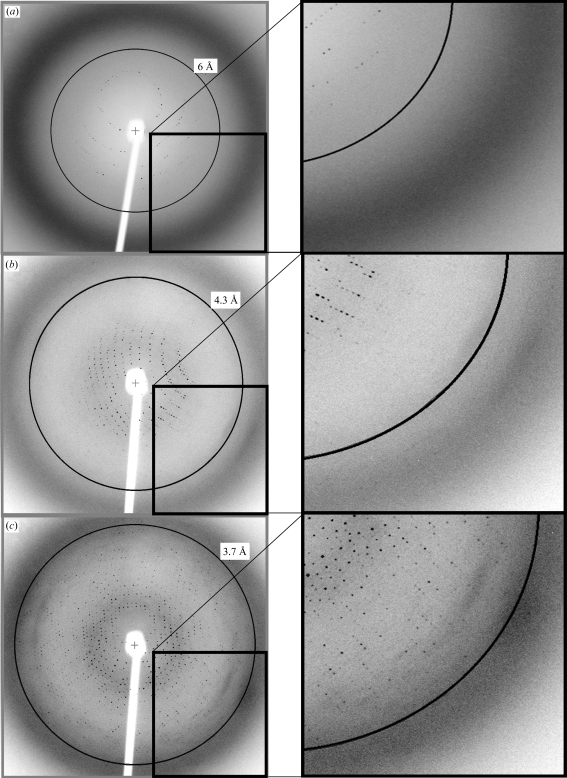
All six successful Optimix I conditions were therefore translated into the larger volume hanging-drop conditions with the aim of growing larger crystals for diffraction analysis. The largest crystals were found with 10–13% PEG 3350, 0.2 M magnesium acetate, 0.1 M MES pH 6.5, 50 mM magnesium chloride, conditions which were related to condition 6 from the free-interface diffusion results. Crystals grew to dimensions of 0.2 × 0.2 × 0.2 mm over a period of 7 d using these conditions. These crystals belong to the cubic space group P213 and diffract X-rays to a resolution of 6 Å at room temperature (Fig. 2 ▶ a; a = b = c = 174.5 Å; mosaicity, 0.2°) and 4.3 Å at 100 K (Fig. 2 ▶ b; a = b = c = 172.5 Å; mosaicity, 0.6°) (Table 2 ▶). The diffraction resolution of the crystals was considerably and reproducibly improved by dehydrating the crystals prior to cryocooling. Dehydration was achieved by placing the crystals in a solution containing a higher concentration of the precipitant (25–30% PEG 3350) than that in which they were grown (10–13% PEG 3350; all other components of the mother liquor remained the same; Heras et al., 2003 ▶, Heras & Martin, 2005 ▶). The diffraction quality was tested for crystals dehydrated for 3 h, 1 d and 2 d. Improvement in diffraction resolution was observed only after the longer periods of 1 or 2 d. Crystals did not show any signs of cracking during dehydration. After cryocooling, the unit-cell volume of the crystals was reduced by 3%. Dehydration reduced the unit-cell volume by a further 3%. After dehydration and cryocooling, the diffraction resolution of the crystals on the in-house X-ray equipment improved to 3.7 Å resolution (Fig. 2 ▶ c; a = b = c = 170.8 Å; mosaicity, 0.4°). The 3.7 Å resolution data set consists of 146 586 independent observations corresponding to 18 037 unique reflections and is 99.9% complete to 3.7 Å with an R merge of 0.079 (0.521 in the highest resolution shell; Table 2 ▶). Assuming the presence of two molecules in the asymmetric unit, the crystal volume per unit molecular weight (V M) is 2.7 Å3 Da−1, with a solvent content of 53%, which is within the normal range for protein crystals (Matthews, 1968 ▶).
Figure 2.
Diffraction images. (a) Room temperature, ∼6 Å resolution. (b) After cryoprotection, 4.3 Å resolution (Table 2 ▶). (c) After cryoprotection and dehydration, 3.7 Å resolution (Table 2 ▶). The right panels show an enlargement of the the same portion of the left panels as indicated.
Table 2. Comparison of representative X-ray data measurements from a cryocooled nondehydrated crystal and from a cryocooled dehydrated crystal.
It was not possible to collect a room-temperature data set for comparison because the crystals deteriorated rapidly in the X-ray beam at this temperature. Values in parentheses are for the highest resolution shell.
| Non-dehydrated | Dehydrated (1 d) | |
|---|---|---|
| Data-collection temperature (K) | 100 | 100 |
| Space group | P213 | P213 |
| Wavelength (Å) | 1.542 | 1.542 |
| Unit-cell parameters | ||
| a = b = c (Å) | 172.5 | 170.8 |
| α = β = γ (°) | 90 | 90 |
| Resolution (Å) | 50–4.30 (4.45–4.30) | 50–3.70 (3.83–3.70) |
| No. observations | 42677 | 146586 |
| No. unique reflections | 11776 | 18037 |
| Rmerge | 0.074 (0.501) | 0.079 (0.521) |
| I/σ(I) | 12.0 (2.9) | 16.4 (5.7) |
| Completeness (%) | 98.2 (98.5) | 99.9 (100.0) |
| Redundancy | 3.7 (3.7) | 8.1 (8.1) |
| Mosaicity (°) | 0.6 | 0.4 |
| Matthews coefficient (Å3 Da−1) | 2.8 | 2.7 |
| Solvent content (%) | 54 | 53 |
| No. of molecules in the ASU | 2 | 2 |
In summary, we grew diffraction-quality crystals of Munc18c by complexing it with an N-terminal peptide from syntaxin4, using free-interface diffusion to screen nanolitre volumes initially, scaling up to hanging-drop vapour diffusion to grow larger crystals and dehydrating the crystals prior to data measurement.
A further improvement in diffraction resolution was achieved recently through the use of synchrotron radiation on the dehydrated crystals, allowing determination of the structure by molecular replacement (Hu et al., 2007 ▶). The structure solution revealed that the crystals contained both Munc18c and the N-terminal peptide, thereby confirming the role of the peptide, in combination with dehydration, in improving the crystal quality.
Acknowledgments
We thank Karl Byriel for assistance with Fig. 2 ▶ and we acknowledge the use of the University of Queensland Macromolecular X-ray Crystallography Facility. We thank Linda Lua, Michelle Christie and the ARC Special Research Centre for Functional and Applied Genomics Protein Expression Facility (The University of Queensland, Australia) for assistance in producing Munc18c. This research was supported by an Australian National Health and Medical Research Council (NHMRC) grant to JLM and DEJ, an NHMRC Senior Research Fellowship to JLM and an NHMRC Senior Principal Research Fellowship to DEJ.
References
- Gallwitz, D. & Jahn, R. (2003). Trends Biochem. Sci.28, 113–116. [DOI] [PubMed] [Google Scholar]
- Hansen, C. L., Skordalakes, E., Berger, J. M. & Quake, S. R. (2002). Proc. Natl Acad. Sci. USA, 99, 16531–16536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heras, B., Edeling, M. A., Byriel, K. A., Jones, A., Raina, S. & Martin, J. L. (2003). Structure, 11, 139–145. [DOI] [PubMed] [Google Scholar]
- Heras, B. & Martin, J. L. (2005). Acta Cryst. D61, 1173–1180. [DOI] [PubMed] [Google Scholar]
- Hu, S.-H., Gee, C. L., Latham, C. F., Rowlinson, S. W., Rova, U., Jones, A., Halliday, J. A., Bryant, N. J., James, D. E. & Martin, J. L. (2003). Protein Expr. Purif.31, 305–310. [DOI] [PubMed] [Google Scholar]
- Hu, S.-H., Latham, C., Gee, C., James, D. & Martin, J. (2007). In the press..
- Latham, C. F., Lopez, J. A., Hu, S.-H., Gee, C. L., Westbury, E., Blair, D. H., Armishaw, C. J., Alewood, P. F., Bryant, N. J., James, D. E. & Martin, J. L. (2006). Traffic, 7, 1408–1419. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Tellam, J. T., McIntosh, S. & James, D. E. (1995). J. Biol. Chem.270, 5857–5863. [DOI] [PubMed] [Google Scholar]
- Thurmond, D. C., Ceresa, B. P., Okada, S., Elmendorf, J. S., Coker, K. & Pessin, J. E. (1998). J. Biol. Chem.273, 33876–33883. [DOI] [PubMed] [Google Scholar]
- Toonen, R. F. & Verhage, M. (2003). Trends Cell Biol.13, 177–186. [DOI] [PubMed] [Google Scholar]