The NAD(P)H:ferredoxin oxidoreductase in carbazole 1,9a-dioxygenase from Janthinobacterium sp. J3 was crystallized and diffraction data were collected to 2.60 Å resolution.
Keywords: angular dioxygenases, NAD(P)H:ferredoxin oxidoreductases, Rieske nonhaem iron oxygenase system, electron transfer, carbazole
Abstract
Carbazole 1,9a-dioxygenase (CARDO), which consists of an oxygenase component (CARDO-O) and the electron-transport components ferredoxin (CARDO-F) and ferredoxin reductase (CARDO-R), catalyzes dihydroxylation at the C1 and C9a positions of carbazole. CARDO-R was crystallized at 277 K using the hanging-drop vapour-diffusion method with the precipitant PEG 8000. Two crystal types (types I and II) were obtained. The type I crystal diffracted to a maximum resolution of 2.80 Å and belonged to space group P42212, with unit-cell parameters a = b = 158.7, c = 81.4 Å. The type II crystal was obtained in drops from which type I crystals had been removed; it diffracted to 2.60 Å resolution and belonged to the same space group, with unit-cell parameters a = b = 161.8, c = 79.5 Å.
1. Introduction
Rieske nonhaem iron oxygenase systems (ROSs) are the initial catalysts in the degradation pathways of various aromatic compounds, including dioxins, polychlorinated biphenyls and crude-oil components such as polycyclic aromatic hydrocarbons and heteroaromatics, which are of serious environmental concern (Wittich, 1998 ▶; Bressler & Fedorak, 2000 ▶; Nojiri & Omori, 2002 ▶; Habe & Omori, 2003 ▶; Furukawa et al., 2004 ▶). With very few exceptions, ROSs catalyze the incorporation of both O atoms of molecular dioxygen at tandem C atoms of an aromatic ring as two hydroxyl groups in the cis configuration. This dioxygenation is known as lateral dioxygenation or cis-dihydroxylation (Mason & Cammack, 1992 ▶). On the other hand, in the initial dioxygenation of several heteroaromatics such as carbazole and dioxins one carbon that is bonded to the heteroatom and its adjacent carbon in the aromatic ring are both hydroxylated. This reaction, called angular dioxygenation, is catalyzed by a limited number of ROSs, which are called angular dioxygenases (Nojiri & Omori, 2002 ▶). The terminal oxygenase components of ROSs invariably consist of an iron–sulfur protein with a Rieske-type [2Fe–2S] cluster and a mononuclear iron. The electron-transport chain consists of either one or two separate proteins: reductase alone or ferredoxin and reductase in combination (Mason & Cammack, 1992 ▶). ROSs catalyze the dioxygenation of aromatics using dioxygen and two electrons. The electrons, which are originally derived from NAD(P)H, are transferred through the electron-transport proteins. ROSs have been classified into three major groups based on the number of constituent components and the nature of the redox centre (Batie et al., 1991 ▶; Ferraro et al., 2005 ▶). Class I ROSs consist of reductase and oxygenase components, with the reductases containing both flavin and a plant-type [2Fe–2S] cluster, and are further divided into two subclasses by the type of flavin, FMN (class IA) or FAD (class IB). Both class II and III ROSs contain a ferredoxin component in addition to the reductase and oxygenase components. Class II is further divided into classes IIA and IIB, which use putidaredoxin-type and Rieske-type ferredoxins, respectively, to mediate electron transfer between the reductase and oxygenase. Class II reductases contain FAD as the only cofactor, whereas class III reductases contain FAD and a plant-type [2Fe–2S] cluster.
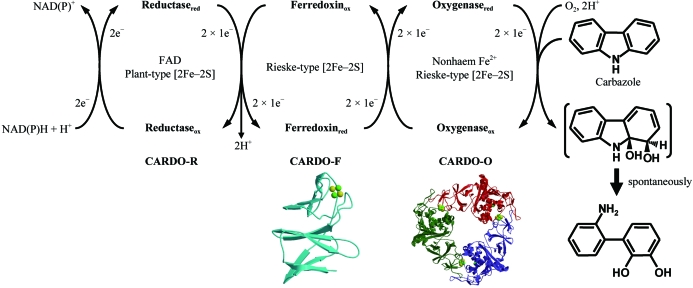
We have investigated the enzymatic function of carbazole 1,9a-dioxygenase (CARDO) from various bacteria (Nojiri & Omori, 2002 ▶; Inoue et al., 2004 ▶, 2005 ▶). All CARDOs consist of three components: a terminal oxygenase CARDO-O, a ferredoxin CARDO-F and a ferredoxin reductase CARDO-R, which are encoded by carAa, carAc (or fdx) and carAd (or fdr) genes, respectively (Fig. 1 ▶). The CARDOs from Pseudomonas resinovorans CA10 (CARDOCA10), Janthinobacterium sp. J3 (CARDOJ3), Sphingomonas sp. KA1 (CARDOKA1) and Nocardioides aromaticivorans IC177 (CARDOIC177) are grouped into classes III, III, IIA and IIB, respectively (Sato et al., 1997 ▶; Inoue et al., 2004 ▶, 2006 ▶; Urata et al., 2006 ▶), indicating that these CARDOs have diverse types of electron-transfer components (CARDO-F and CARDO-R). Therefore, CARDO is an excellent model system for studying structure–function relationships and the mechanisms of inter-component electron transfer in ROSs.
Figure 1.
Components, functions and structures solved for the clas III CARDO system. The proposed electron-transfer reactions and the conversion of carbazole to 2′-aminobiphenyl-2,3-diol are illustrated. The subscripts ‘ox’ and ‘red’ indicate oxidized and reduced states of the CARDO components, respectively. The crystal structures of CARDO-OJ3 (PDB code 1ww9) and CARDO-FCA10 (1vck) are shown.
Recently, we determined the crystal structures of CARDO-F from P. resinovorans CA10 (CARDO-FCA10; Nam et al., 2005 ▶), CARDO-O from Janthinobacterium sp. J3 (CARDO-OJ3; Nojiri et al., 2005 ▶; Fig. 1 ▶) and the electron-transfer complex between CARDO-OJ3 and CARDO-FCA10 (Ashikawa et al., 2005 ▶, 2006 ▶). Class III ROS reductases, including CARDO-RCA10 and CARDO-RJ3, contain a plant-type [2Fe–2S] cluster and FAD and their structures have not yet been determined. ROS reductases are classified into two separate structural families: ferredoxin-NADP reductases (FNR) and glutathione reductases (GR). The structures of two FNR-family ROS reductases, phthalate dioxygenase reductase (PDO-R; class IA) from P. cepacia PHK (Correll et al., 1992 ▶) and benzoate dioxygenase reductase (BZDO-R; class IB) from Acinetobacter baylyi ADP1 (Karlsson et al., 2002 ▶), have been determined. The structure of biphenyl dioxygenase reductase from Pseudomonas sp. KKS102, which belongs to the GR family, has also been determined (class IIB; Senda et al., 2000 ▶). Although the amino-acid sequences of class III CARDO-Rs appear to align well with that of BZDO-R (23–24% amino-acid sequence identity), there may be significant structural differences between class III CARDO-Rs and BZDO-R as the reductases have different redox partners (a ferredoxin component and an oxygenase component, respectively). Structural and functional studies on class III CARDO-Rs could enrich our knowledge and further our understanding of the mechanisms of electron transfer between ROS components. For this purpose, we report the crystallization of and preliminary X-ray diffraction studies on a class III CARDO-R (both CARDO-RCA10 and CARDO-RJ3 were composed of 329 amino acids with a molecular weight of approximately 36 kDa).
2. Protein expression and purification
Although we tried to crystallize the C-terminally His-tagged form of CARDO-RCA10 (accession No. NP_758573) that was used in a previous purification and characterization study (Nam et al., 2002 ▶), crystals of CARDO-RCA10 could not be obtained. Therefore, in the present study, we used the N-terminally His-tagged form of CARDO-RJ3 (accession No. BAC56748; 95% amino-acid sequence identity to the sequence of CARDO-RCA10). The carAd gene was amplified by PCR from plasmid pBJ3001 (Inoue et al., 2004 ▶) using the primers 5′-GGAATTCCATATGCACCACCACCACCACCACTACCAACTCAAAATTG-3′ and 5′-CCGGAGCTCTTAGAAAAATGCGTCAAAATGAATTTGATCGCGTGGAACACC-3′. The PCR product was ligated into pT7Blue T-vector (Novagen). The nucleotide sequence of the insert was checked against the original sequence and the plasmid was digested with NdeI and SacI. The resultant 1.0 kbp NdeI–SacI fragment was inserted into the overexpression vector pET-26b(+) (Novagen; designated pEJ3NAd). pEJ3NAd contains the genes for the full-length CARDO-RJ3 with a 6×His tag at the N-terminus.
Escherichia coli BL21 (DE3) (Novagen) harbouring pEJ3NAd was grown in LB medium (Sambrook & Russell, 2001 ▶) or SB medium (Nam et al., 2002 ▶) supplemented with kanamycin (50 µg ml−1) at 310 K for initial incubation and at 303 K for induction of protein expression with shaking at 120 rev min−1. When the optical density at 600 nm reached approximately 0.5, 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG) was added. After a 15 h incubation with IPTG, the cells were harvested by centrifugation at 5000g and washed with TG buffer (Nam et al., 2002 ▶). Appropriate E. coli cells were suspended in TI buffer (20 mM Tris–HCl pH 7.5, 30 mM imidazole, 0.5 M NaCl). The crude cell extract prepared by sonication and subsequent centrifugation at 25 000g was applied onto a HiTrap Chelating HP column (GE Healthcare) connected to an ÄKTA FPLC instrument (GE Healthcare) according to the manufacturer’s recommendation. All purification procedures were carried out at 277 K. CARDO-RJ3 was eluted with TI buffer containing 300 mM imidazole. The fractions that contained CARDO-RJ3, as confirmed by SDS–PAGE, were then pooled and concentrated by ultrafiltration using Centriprep-10 (Millipore) and Vivaspin 20 membranes (10 000 MWCO; Sartorius). The resultant preparation was further purified by gel-filtration chromatography (GFC) using a Superdex 200 prep-grade column (26 × 600 mm; GE Healthcare) and GFC buffer [20 mM Tris–HCl pH 7.5, 0.2 M NaCl, 10%(v/v) glycerol]. The purified CARDO-RJ3 was almost homogeneous on SDS–PAGE and gel-filtration analysis showed that CARDO-RJ3 was monomeric, as was CARDO-RCA10 (Nam et al., 2002 ▶; data not shown). Prior to crystallization, the purified CARDO-RJ3 was confirmed to have retained electron-transfer ability [from NAD(P)H to CARDO-FCA10] by the detection of CARDO activity in a component-reconstitution system (data not shown). For the crystallization experiments, fractions that contained CARDO-RJ3 were concentrated and buffer-exchanged with 50 mM Tris–HCl buffer pH 7.5 containing 10%(v/v) glycerol. Protein concentrations were estimated using a protein-assay kit (Bio-Rad; Bradford, 1976 ▶) with bovine serum albumin as a standard.
3. Crystallization
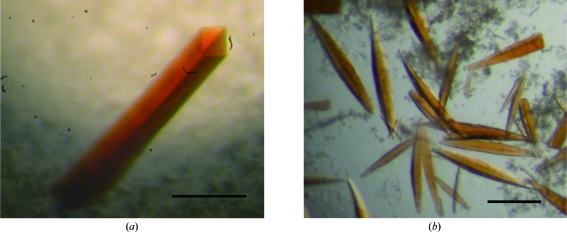
The protein concentration was adjusted to 5–25 mg ml−1 in 50 mM Tris–HCl buffer pH 7.5 containing 10%(v/v) glycerol. Crystallization was performed using the hanging-drop vapour-diffusion method with VDX Plates (Hampton Research) at 277 K. Drops containing 2 µl protein solution and 2 µl mother liquor were equilibrated against 700 µl reservoir solution. Initial crystallization conditions were screened using Crystal Screens I and II, Crystal Screen Lite, Crystal Screen Cryo, Grid Screens, Index, SaltRx (Hampton Research), Wizard I and II (Jena Bioscience) and JBScreen (Emerald Biostructures). Unique crystals were obtained using a third of the concentration of Crystal Screen I condition No. 46 [0.067 M calcium acetate, 6%(w/v) PEG 8000, 0.033 M sodium cacodylate pH 6.5] in the reservoir and protein solution at a concentration of 10–25 mg ml−1. After optimizing the crystallization parameters and protein concentration, rod-shaped crystals (type I crystals) appeared within 2–3 d and grew to approximate maximum dimensions of 0.1 × 0.1 × 0.4 mm after one week using a protein concentration of 18–22 mg ml−1 (Fig. 2 ▶ a). Type I crystals were formed using 0.16 mM calcium acetate or magnesium acetate, 8–12%(w/v) PEG 8000 and 0.2 M sodium iodide in 0.08 M MES pH 6.5–6.7. Type II crystals were often observed after several weeks in drops from which type I crystals grown using magnesium acetate as a salt had been removed and grew to maximum dimensions of approximately 0.05 × 0.05 × 0.4 mm (Fig. 2 ▶ b).
Figure 2.
Photographs of CARDO-RJ3 crystals. The scales bars indicate 0.2 mm. (a) Type I crystal grown in 0.16 mM calcium acetate, 8%(w/v) PEG 8000, 0.2 M sodium iodide and 0.08 M MES pH 6.5. (b) Type II crystals grown in drops from which type I crystals had been removed [0.16 mM magnesium acetate, 12%(w/v) PEG 8000, 0.2 M sodium iodide in 0.08 M MES pH 6.6].
4. X-ray analysis
Type I crystals were first transferred into cryoprotectant solution [reservoir solution to which glycerol had been added to a final concentration of 15–20%(v/v)] and then mounted in a nylon loop and flash-cooled in a nitrogen stream at 100 K. Type II crystals were directly flash-cooled in a nitrogen stream at 100 K. The diffraction experiments were conducted at beamline AR-NW12, Photon Factory, Tsukuba, Japan. Diffraction data were gathered at a wavelength of 1.0 Å using a Quantum 210 CCD X-ray detector (ADSC). For the type I crystal, a data set was collected using a single crystal with oscillation steps of 0.5° over a range of 120° with 5 s exposure per frame. The type II crystal data set was collected with oscillation steps of 0.5° over a range of 90° with a 10 s exposure per frame. All diffraction images were indexed, integrated and scaled using the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶).
The type I crystal diffracted to a maximum resolution of 2.80 Å and belonged to space group P42212, with unit-cell parameters a = b = 158.7, c = 81.4 Å. The type II crystal diffracted to 2.60 Å resolution and belonged to the same space group, with unit-cell parameters a = b = 161.8, c = 79.5 Å. The collected data and processing statistics are summarized in Table 1 ▶. Initial analysis of the crystal solvent content of type I and type II crystals using the Matthews coefficient (Matthews, 1968 ▶) suggested that the asymmetric unit contains three CARDO-RJ3 molecules with 47.0 and 47.8% solvent content, which led to acceptable packing densities (V M) of 2.32 and 2.35 Å3 Da−1, respectively. Although the molecular-replacement method was attempted using the structure of BZDO-R from A. baylyi ADP1 (23% amino-acid sequence identity; Karlsson et al., 2002 ▶) as a search model, it did not give sufficient phases for structure determination; we are now preparing selenomethionine-substituted CARDO-RJ3 crystals for phase determination using the MAD technique.
Table 1. Crystal parameters and data-collection statistics.
The data sets were collected at beamline AR-NW12 at the Photon Factory, Tsukuba, Japan. Values in parentheses are for the highest resolution shell.
| Crystal type | Type I | Type II |
|---|---|---|
| Wavelength (Å) | 1.0 | 1.0 |
| Space group | P42212 | P42212 |
| Unit-cell parameters (Å, °) | a = b = 158.7, c = 81.4, α = β = γ = 90.0 | a = b = 161.8, c = 79.5, α = β = γ = 90.0 |
| Resolution range (Å) | 50.00–2.80 (2.90–2.80) | 50.00–2.60 (2.69–2.60) |
| Total No. of reflections | 232454 | 235698 |
| No. of unique reflections | 25971 (2539) | 33087 (3258) |
| Completeness (%) | 99.5 (99.9) | 99.9 (100.0) |
| Average I/σ(I) | 46.3 (6.5) | 40.0 (4.1) |
| Rsym† (%) | 8.9 (39.5) | 6.4 (41.7) |
| Multiplicity | 9.0 (9.4) | 7.1 (6.9) |
R
sym = 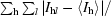
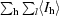 , where I
l is the lth observation of reflection h and 〈I
h〉 is the weighted average intensity for all observations l of reflection h.
, where I
l is the lth observation of reflection h and 〈I
h〉 is the weighted average intensity for all observations l of reflection h.
Several reports have indicated that whereas the reductase components of ROSs can be replaced by unrelated or other ROS reductases, the ferredoxin components cannot be replaced (Subramanian et al., 1981 ▶, 1985 ▶; Haigler & Gibson, 1990 ▶; Fukuda et al., 1994 ▶; Yu et al., 2007 ▶). In the class III CARDO system, CARDO-R can be replaced by an unrelated reductase such as ferredoxin reductase from spinach, but CARDO-F is indispensable for electron transfer to CARDO-O, suggesting that there is a specific interaction between CARDO-O and CARDO-F but not between CARDO-R and CARDO-F (Nam et al., 2002 ▶). The structure of the complex between CARDO-O and CARDO-F provided important information for understanding their specific interactions and the electron transport between the components (Ashikawa et al., 2006 ▶). The forthcoming structure of CARDO-RJ3 will provide insight into the structural basis of the nonspecific interaction between ferredoxin and reductase in the CARDO system, which is a common feature of various ROSs.
Acknowledgments
The authors thank Dr Atsuko Yamashita of RIKEN Harima Institute for helpful advice. Part of this work was supported by a Grant-in-Aid for Scientific Research (17380052 to HN) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and by the Institute for Bioinformatics Research Development, Japan Science Technology Agency (BIRD-JST). The use of synchrotron radiation in this work was approved by the Photon Factory Advisory Committee and the High Energy Accelerator Research Organization (KEK; proposal Nos. 2005G060 and 2006G171) and the Japan Synchrotron Radiation Research Institute (JASRI; proposal No. 2005A0671-NL1-np).
References
- Ashikawa, Y., Fujimoto, Z., Noguchi, H., Habe, H., Omori, T., Yamane, H. & Nojiri, H. (2005). Acta Cryst. F61, 577–580. [DOI] [PMC free article] [PubMed]
- Ashikawa, Y., Fujimoto, Z., Noguchi, H., Habe, H., Omori, T., Yamane, H. & Nojiri, H. (2006). Structure, 14, 1779–1789. [DOI] [PubMed] [Google Scholar]
- Batie, C. J., Ballou, D. P. & Correll, C. C. (1991). Chemistry and Biochemistry of Flavoenzymes, Vol. 3, edited by F. Muller, pp. 543–556. Boca Raton: CRC Press.
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed] [Google Scholar]
- Bressler, D. C. & Fedorak, P. M. (2000). Can. J. Microbiol.46, 397–409. [PubMed] [Google Scholar]
- Correll, C. C., Batie, C. J., Ballou, D. P. & Ludwig, M. L. (1992). Science, 258, 1604–1610. [DOI] [PubMed] [Google Scholar]
- Ferraro, D. J., Gakhar, L. & Ramaswamy, S. (2005). Biochem. Biophys. Res. Commun.338, 175–190. [DOI] [PubMed] [Google Scholar]
- Fukuda, M., Yasukouchi, Y., Kikuchi, Y., Nagata, Y., Kimbara, K., Horiuchi, H., Takagi, M. & Yano, K. (1994). Biochem. Biophys. Res. Commun.202, 850–856. [DOI] [PubMed] [Google Scholar]
- Furukawa, K., Suenaga, H. & Goto, M. (2004). J. Bacteriol.186, 5189–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habe, H. & Omori, T. (2003). Biosci. Biotechnol. Biochem.67, 225–243. [DOI] [PubMed] [Google Scholar]
- Haigler, B. E. & Gibson, D. T. (1990). J. Bacteriol.172, 465–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, K., Habe, H., Yamane, H. & Nojiri, H. (2006). Appl. Environ. Microbiol.72, 3321–3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, K., Habe, H., Yamane, H., Omori, T. & Nojiri, H. (2005). FEMS Microbiol. Lett.245, 145–153. [DOI] [PubMed] [Google Scholar]
- Inoue, K., Widada, J., Nakai, S., Endoh, T., Urata, M., Ashikawa, Y., Shintani, M., Saiki, Y., Yoshida, T., Habe, H., Omori, T. & Nojiri, H. (2004). Biosci. Biotechnol. Biochem.68, 1467–1480. [DOI] [PubMed] [Google Scholar]
- Karlsson, A., Beharry, Z. M., Eby, D. M., Coulter, E. D., Neidle, E. L., Kurtz, D. M. Jr, Eklund, H. & Ramaswamy, S. (2002). J. Mol. Biol.318, 261–272. [DOI] [PubMed] [Google Scholar]
- Mason, J. R. & Cammack, R. (1992). Annu. Rev. Microbiol.46, 277–305. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Nam, J.-W., Noguchi, H., Fujimoto, Z., Mizuno, H., Ashikawa, Y., Abo, M., Fushinobu, S., Kobashi, K., Wakagi, T., Iwata, K., Yoshida, T., Habe, H., Yamane, H., Omori, T. & Nojiri, H. (2005). Proteins, 58, 779–789. [DOI] [PubMed] [Google Scholar]
- Nam, J.-W., Nojiri, H., Noguchi, H., Uchimura, H., Yoshida, T., Habe, H., Yamane, H. & Omori, T. (2002). Appl. Environ. Microbiol.68, 5882–5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojiri, H., Ashikawa, Y., Noguchi, H., Nam, J.-W., Urata, M., Fujimoto, Z., Yoshida, T., Habe, H. & Omori, T. (2005). J. Mol. Biol.351, 355–370. [DOI] [PubMed] [Google Scholar]
- Nojiri, H. & Omori, T. (2002). Biosci. Biotechnol. Biochem.66, 2001–2016. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Sambrook, J. & Russell, D. W. (2001). Molecular Cloning. A Laboratory Manual, 3rd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
- Sato, S., Nam, J.-W., Kasuga, K., Nojiri, H., Yamane, H. & Omori, T. (1997). J. Bacteriol.179, 4850–4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senda, T., Yamada, T., Sakurai, N., Kubota, M., Nishizaki, T., Masai, E., Fukuda, M. & Mitsuidagger, Y. (2000). J. Mol. Biol.304, 397–410. [DOI] [PubMed] [Google Scholar]
- Subramanian, V., Liu, T.-N., Yeh, W.-K., Narro, M. & Gibson, D. T. (1981). J. Biol. Chem.256, 2723–2730. [PubMed] [Google Scholar]
- Subramanian, V., Liu, T.-N., Yeh, W.-K., Serdar, C. M., Wakett, L. P. & Gibson, D. T. (1985). J. Biol. Chem.260, 2355–2363. [PubMed] [Google Scholar]
- Urata, M., Uchimura, H., Noguchi, H., Sakaguchi, T., Takemura, T., Eto, K., Habe, H., Omori, T., Yamane, H. & Nojiri, H. (2006). Appl. Environ. Microbiol.72, 3206–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittich, R.-M. (1998). Appl. Microbiol. Biotechnol.49, 489–499. [DOI] [PubMed] [Google Scholar]
- Yu, C. L., Liu, W., Ferraro, D. J., Brown, E. N., Parales, J. V., Ramaswamy, S., Zylstra, G. J., Gibson, D. T. & Parales, R. E. (2007). J. Ind. Microbiol. Biotechnol.34, 311–324. [DOI] [PubMed] [Google Scholar]