The first crystal of a coxsackievirus RNA-dependent RNA polymerase is reported.
Keywords: coxsackievirus B3, RNA-dependent RNA polymerase
Abstract
The Picornaviridae virus family contains a large number of human pathogens such as poliovirus, hepatitis A virus and rhinoviruses. Amongst the viruses belonging to the genus Enterovirus, several serotypes of coxsackievirus coexist for which neither vaccine nor therapy is available. Coxsackievirus B3 is involved in the development of acute myocarditis and dilated cardiomyopathy and is thought to be an important cause of sudden death in young adults. Here, the first crystal of a coxsackievirus RNA-dependent RNA polymerase is reported. Standard crystallization methods yielded crystals that were poorly suited to X-ray diffraction studies, with one axis being completely disordered. Crystallization was improved by testing crystallization solutions from commercial screens as additives. This approach yielded crystals that diffracted to 2.1 Å resolution and that were suitable for structure determination.
1. Introduction
Coxsackieviruses belong to the Enterovirus genus of the Picornaviridae family. They are grouped into two serogroups, A and B, encompassing 24 and six serotypes, respectively (for a review, see Pallansch & Roos, 2001 ▶). Viruses of serogroup B are responsible for the most severe clinical syndromes, such as pancreatitis, myocarditis, encephalitis and aseptic meningitis. It has been known for several decades that coxsackievirus B3 (CVB3) is the most widespread source of human viral myocarditis, which can eventually progress to dilated cardiomyopathy (Woodruff, 1980 ▶) and is associated with sudden death in young infants. In addition, other reports have suggested that CVB3 can induce damage to and/or death of human pancreatic islet β-cells and can thus be implicated in the development of type I juvenile or insulin-dependent diabetes mellitus (Foulis et al., 1990 ▶; Kukreja & Maclaren, 2000 ▶; Roivainen et al., 1998 ▶; Tauriainen et al., 2003 ▶). To date, the mechanisms leading to these varying pathologies still remain unclear or unknown. They could either be dependent on the CVB3 strain or be linked to genetic, environmental and/or immunological factors.
The positive single-stranded RNA genome of the Picornaviridae virus family contains one open reading frame encoding for one polyprotein that is subsequently processed by viral proteinases (2Apro, 3Cpro and 3CDpro) to give rise to mature viral proteins during and after the translation process (Bedard & Semler, 2004 ▶; Racaniello, 2001 ▶). The structural P1 region of the polyprotein generates the four capsid proteins (VP1, VP2, VP3 and VP4). The C-terminal part of the polyprotein contains the nonstructural regions (P2 and P3), which are processed by the 3Cpro and 3CDpro proteases into functional proteins. The viral RNA replication is catalyzed by the RNA-dependent RNA polymerase (RdRp), also known as protein 3Dpol. Replication involves the synthesis of a complementary negative-strand RNA intermediate. The initiation of RNA replication requires essential cis-acting replication elements located at the 5′ and 3′ untranslated regions (UTRs; Racaniello, 2001 ▶; Xiang et al., 1995 ▶) and the 2C-coding region (van Ooij et al., 2006 ▶). Additionally, RNA synthesis is initiated by uridylylation of VPg (viral protein genome-linked or protein 3B), which serves as a primer for positive- and negative-strand synthesis.
The crystallographic structure of poliovirus 3Dpol was the first RdRp structure to be solved, although with an important part of the protein disordered (Hansen et al., 1997 ▶). Recently, the complete structure has been reported (Thompson & Peersen, 2004 ▶). This study showed why a precise N-terminus is important for RdRp activity. CVB3 3Dpol shares 73.4% sequence identity with poliovirus 3Dpol; it is therefore likely that the same applies to CBV3 3Dlpol. We thus produced CBV3 3Dpol with a C-terminal His tag in Escherichia coli. The protein was purified to homogeneity. The crystallization of CVB3 3Dpol was optimized to yield crystals that diffracted to 2.1 Å resolution and were suitable for structure determination.
2. Protein expression and purification
The gene coding for CVB3 RdRp was amplified from cDNA (CVB3 strain, Nancy GenBank accession No. gi:323421) using two primers (forward primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCGAAGGAGATAGAACCATGGGTGAAATAGAATTTATTGAGAGC-3′; reverse primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAGTGATGGTGATGGTGATGAAAGGAGTCCAACCACTTCCTG-3′) containing the attB sites of the Gateway recombination system (Invitrogen) and spanning amino acids 1724–2185 of the corresponding polyprotein. A six-histidine tag coding sequence is attached directly to the 3′ end of the construct. The resulting PCR product was cloned into the pDest14 plasmid, giving pExp1464HC, using Gateway technology. Appropriate protein-expression levels were subsequently obtained using E. coli strain Rosetta(pLysS) (Novagen). Transformed cells were grown in LB medium at 310 K. When the OD600nm reached 0.6, expression was induced with 500 µM IPTG. Expression took place at 298 K for 5 h. Cells from 1 l culture (∼4.5 g) were harvested and resuspended in 35 ml lysis buffer consisting of 50 mM Tris–HCl pH 9.0, 500 mM NaCl, 10 mM imidazole, 0.1% Triton X-100 (Sigma), 5%(v/v) glycerol and stored at 253 K. To a thawed bacterial suspension, lysozyme was added to a final concentration of 1 mg ml−1, DNase I to 10 µg ml−1, β-mercaptoethanol to 10 mM, phenylmethylsulfonyl fluoride (PMSF) to 1 mM and protease-inhibitor cocktail (Sigma) to 50 µl g−1. The cells were lysed for 30 min at 277 K and then sonicated. The soluble fraction was recovered by centrifugation for 30 min at 25 000g.
Two steps of purification led to apparent protein homogeneity. Firstly, immobilized metal-ion affinity chromatography (IMAC) was performed. The soluble fraction was incubated in batch with 3 ml Chelating Sepharose Flast Flow resin (GE Healthcare) loaded with Ni2+. After incubation for 30 min at 277 K and one washing step with 50 mM Tris buffer pH 8.5 containing 500 mM NaCl, 10 mM imidazole, 10 mM β-mercaptoethanol, 1 mM PMSF and a second step using the same buffer with 1 M NaCl, the protein was eluted with 50 mM Tris pH 8.5, 500 mM NaCl and 150 mM imidazole. The protein was then dialyzed into 20 mM Tris pH 9.0, 300 mM NaCl, 15% glycerol and 0.5 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP–HCl, Sigma). The optimal conditions for protein solubility and stability had previously been determined by microdialysis by screening different buffers covering a pH range between 6.0 and 9.0, different ionic strength conditions (0, 150 and 300 mM NaCl) and glycerol (0–20%) and TCEP (0 and 0.5 mM) concentrations as described previously (Geerlof et al., 2006 ▶). The second purification step consisted of size-exclusion chromatography using a Superdex 200 column (GE Healthcare) in 20 mM Tris pH 9.0, 300 mM NaCl, 15% glycerol, 0.5 mM TCEP. The protein purification typically yields up to 4 mg of homogenous protein per litre of culture. The protein was homogeneous as judged by SDS–PAGE analysis and was used for crystallization trials. The protein was concentrated to 15 mg ml−1 using Centricon Plus-20 (Millipore) concentrators (5000 kDa molecular-weight cutoff). Aliquots were flash-frozen in liquid nitrogen and stored at 193 K for subsequent use.
3. Crystallization
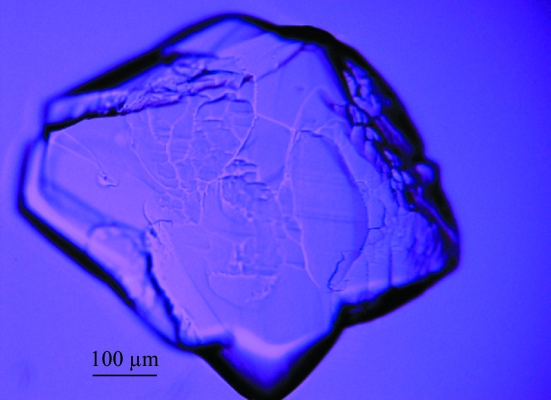
Initial crystallization trials were performed in 96-well Greiner plates using a nanolitre-drop crystallization robot (Sulzenbacher et al., 2002 ▶) and the commercial screens Crystal Screens I and II (Hampton Research), Wizard Screens 1 and 2 (Emerald Biostructures), Structure Screens 1 and 2 and Stura Footprint Screen (Molecular Dimensions Ltd). Protein solution (15 mg ml−1 in 20 mM Tris pH 9.0, 300 mM NaCl, 15% glycerol, 0.5 mM TCEP) droplets of 300, 200 and 100 nl were mixed with 100 nl precipitant solution; the volume of the well buffer was 150 µl. Small crystals appeared within 1–2 d in condition No. 33 of Wizard Screen 1 (Emerald Biostructures), which contained 2.0 M (NH4)2SO4, 100 mM CAPS pH 10.5, 200 mM Li2SO4. After optimization, the size of the crystals was increased using the hanging-drop technique by mixing the protein solution and crystallization solution [2.0–2.3 M (NH4)2SO4, 100 mM CAPS pH 9.8–10.8, 200 mM Li2SO4] in ratios of 3:1, 2.5:1 and 2:1 to give a final drop size of 3–6 µl. The crystals grew in 1–2 d and reached maximum dimensions of 400 × 400 × 350 µm (Fig. 1 ▶). All these crystals displayed many defects such as cracks and/or satellites, as shown in Fig. 1 ▶, but diffracted to 2.5 Å resolution. In addition, their diffraction patterns revealed large trails of reflections in one direction, indicating that one axis was completely disordered (Fig. 2 ▶). All common strategies to improve the crystals (addition of frequently used additives and detergents, protein ligands, salts, PEGs, alcohols and other small organic compounds and changes of buffer and pH) failed. To overcome this apparent dead end, we mixed the previously obtained crystallization condition with standard conditions from several commercial crystallization screens, such as MPD Grid and Natrix Screens (Hampton Research), Emerald Cryo I and II (Emerald BioSystems), Structure Screens 1 and 2 (Molecular Dimensions), Nextal Anions, Cations and MPDs Suites (Qiagen). The reservoirs of 96-well Greiner plates were filled with crystallization solution containing 2.0 M (NH4)2SO4, 100 mM CAPS pH 9.8–10.8, 200 mM Li2SO4 mixed with these commercial screen conditions in a 2:1 volume ratio.
Figure 1.
Initial crystals of CVB3 3Dpol. The crystals have approximate dimensions of 400 × 400 × 350 µm. They show flaws and/or satellites and the diffraction experiments revealed a disordered axis.
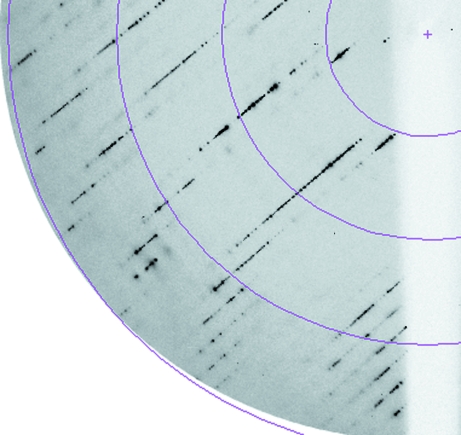
Figure 2.
The typical diffraction pattern of crystals presented in Fig. 1 ▶ reveals crystal disorder along an axis. The crystal was exposed for 3 s duration with a Δϕ of 0.2°. The distance between the crystal and the detector was fixed at 215 mm. From low to high resolution the four rings correspond to 11, 5.5, 3.7 and 2.8 Å resolution, respectively.
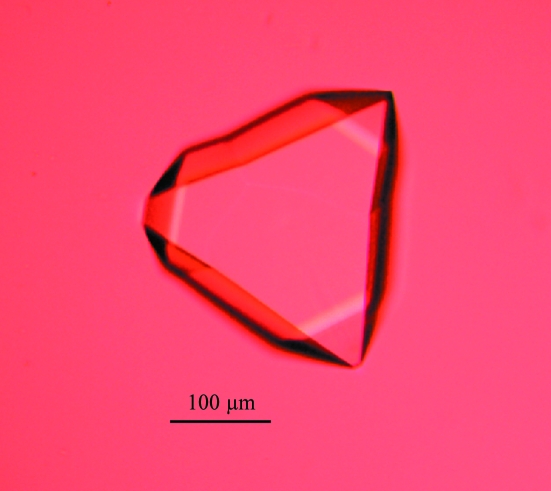
Small prismatic crystals grew in 4–6 d in two crystallization conditions (Nos. 1 and 2) of the Nextal Cations Suite. The final composition of the mother liquor was obtained by mixing two parts 2.0 M (NH4)2SO4, 100 mM CAPS pH 10, 200 mM Li2SO4 with one part 1.75 or 3.5 M NH4Cl, 100 mM sodium acetate pH 4.6 by volume. In order to optimize this condition, one solution was kept unchanged while both the precipitant and the pH of the other were varied. Suitable crystals for X-ray diffraction studies were obtained using two parts 2.0 M (NH4)2SO4, 100 mM CAPS pH 10, 200 mM Li2SO4 and one part 1.5–2 M NH4Cl, 100 mM sodium acetate pH 4.6–5.2 by volume (Fig. 3 ▶). The diffraction patterns exhibits well scattered diffraction spots, indicating a strong positive effect on crystal packing of the solution containing 1.5–2 M NH4Cl, 100 mM sodium acetate pH 4.6–5.2 (Fig. 4 ▶).
Figure 3.
Optimized crystals of CVB3 3Dpol with approximate dimensions 300 × 250 × 250 µm. They grew within 3–6 d.
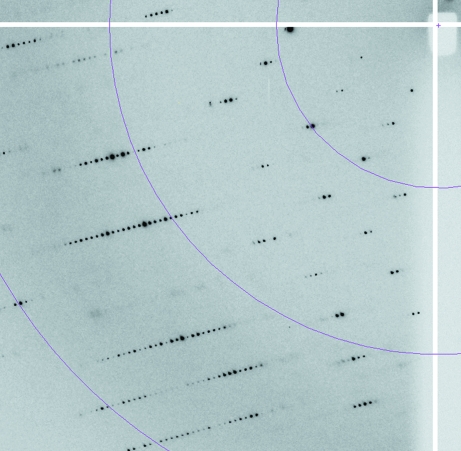
Figure 4.
Enlargement of a diffraction frame of an optimized crystal. The crystal was exposed for 2 s using the same oscillation range as for Fig. 2 ▶ (Δϕ = 0.2°). The distance between the crystal and the detector was again fixed at 215 mm. The three resolution rings from low to high resolution correspond to 11, 5.5 and 3.7 Å, respectively.
4. Preliminary X-ray analysis
Glycerol was added to the mother liquor to a final concentration of 27%. Crystals were soaked for 30 s in this cryoprotectant solution and subsequently flash-frozen in a gaseous nitrogen stream at 110 K. A native diffraction data set was collected at European Synchrotron Radiation Facility (ESRF, Grenoble, France) beamline ID14-3 using a MAR 165 CCD detector. The diffraction frames were indexed and integrated using MOSFLM (Leslie, 2006 ▶) and scaled with SCALA (Collaborative Computational Project, Number 4, 1994 ▶).
CVB3 3Dpol crystallized in space group P41212 or enantiomorph, with unit-cell parameters a = b = 74.38, c = 285.81 Å. With one molecule per asymmetric unit, V M is 3.7 Å3 Da−1, corresponding to a solvent content of 67%. One crystal was found to diffract to 1.7 Å resolution. However, the length of the c axis and the crystal orientation in the beam lead to overlaps between 2.1 and 1.7 Å. A strong native data set was recorded to 2.1 Å resolution with an R merge of 3.8% and a completeness of 90.6%. Data-collection statistics are summarized in Table 1 ▶. Structure determination by the molecular-replacement method using the structure of poliovirus 3Dpol has been initiated.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| X-ray source | ID14-3, ESRF |
| Wavelength (Å) | 0.9330 |
| Space group | P41212 or enantiomorph |
| Unit-cell parameters (Å) | a = b = 74.38, c = 285.81 |
| Resolution (Å) | 2.1 (2.21–2.1) |
| Unique data | 43457 |
| Observations | 236464 |
| Redundancy | 5.4 (3.7) |
| Completeness (%) | 90.6 (90.6) |
| I/σ(I) | 12.5 (7.7) |
| Rmerge† (%) | 3.8 (9.3) |
R
merge = 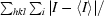
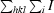 .
.
5. Conclusion
We have established the expression, purification and crystallization of CVB3 3Dpol. The diffraction data from the first crystals revealed a disordered axis and were unsuitable for X-ray structure determination. Among various attempts to improve crystal quality, our results suggest that the combination of a promising condition with all possible conditions from other screens is a very easy and helpful method when common strategies fail. For this purpose, we used a 2:1 volume ratio mixture of the first determined crystallization condition and various conditions from commercial kits, resulting in well ordered crystals that were suitable for X-ray analysis.
Acknowledgments
The ESRF is greatly acknowledged for beam time allocation. We especially wish to thank local contacts at ID14-3 beamline for their kind support. This work was supported by the SPINE project (QLRT-2001-00988) and subsequently by the VIZIER integrated project (LSHG-CT-2004-511960) of the European Union Sixth Framework Programme (FP6).
References
- Bedard, K. M. & Semler, B. L. (2004). Microbes Infect.6, 702–713. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Foulis, A. K., Farquharson, M. A., Cameron, S. O., McGill, M., Schonke, H. & Kandolf, R. (1990). Diabetologia, 33, 290–298. [DOI] [PubMed] [Google Scholar]
- Geerlof, A. et al. (2006). Acta Cryst. D62, 1125–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen, J. L., Long, A. M. & Schultz, S. C. (1997). Structure, 5, 1109–1122. [DOI] [PubMed] [Google Scholar]
- Kukreja, A. & Maclaren, N. K. (2000). Cell Mol. Life Sci.57, 534–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed] [Google Scholar]
- Ooij, M. J. van, Vogt, D. A., Paul, A., Castro, C., Kuijpers, J., van Kuppeveld, F. J., Cameron, C. E., Wimmer, E., Andino, R. & Melchers, W. J. (2006). J. Gen. Virol.87, 103–113. [DOI] [PubMed] [Google Scholar]
- Pallansch, M. A. & Roos, R. P. (2001). Fields Virology, 4th ed., edited by D. M. Knipe & P. M. Howley, Vol. 1, pp. 723–775. Philadelphia, USA: Lippicott Williams & Wilkins.
- Racaniello, V. (2001). Fields Virology, 4th ed., edited by D. M. Knipe & P. M. Howley, Vol. 1, pp. 685–722. Philadelphia, USA: Lippicott Williams & Wilkins.
- Roivainen, M., Knip, M., Hyoty, H., Kulmala, P., Hiltunen, M., Vahasalo, P., Hovi, T. & Akerblom, H. K. (1998). J. Med. Virol.56, 74–78. [DOI] [PubMed] [Google Scholar]
- Sulzenbacher, G. et al. (2002). Acta Cryst. D58, 2109–2115. [DOI] [PubMed] [Google Scholar]
- Tauriainen, S., Salminen, K. & Hyoty, H. (2003). Ann. NY Acad. Sci.1005, 13–22. [DOI] [PubMed] [Google Scholar]
- Thompson, A. A. & Peersen, O. B. (2004). EMBO J.23, 3462–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff, J. F. (1980). Am. J. Pathol.101, 425–484. [PMC free article] [PubMed] [Google Scholar]
- Xiang, W., Harris, K. S., Alexander, L. & Wimmer, E. (1995). J. Virol.69, 3658–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]