A preliminary crystallographic study of cysteine synthase, a major enzyme in the cysteine-biosynthesis pathway, from the amoebic pathogen E. histolytica.
Keywords: cysteine synthase, Entamoeba histolytica
Abstract
Entamoeba histolytica, the causative agent of human amoebiasis, is essentially anaerobic, requiring a small amount of oxygen for growth. It cannot tolerate the higher concentration of oxygen present in human tissues or blood. However, during tissue invasion it is exposed to a higher level of oxygen, leading to oxygen stress. Cysteine, which is a vital thiol in E. histolytica, plays an essential role in its oxygen-defence mechanisms. The major route of cysteine biosynthesis in this parasite is the condensation of O-acetylserine with sulfide by the de novo cysteine-biosynthetic pathway, which involves cysteine synthase (EhCS) as a key enzyme. In this study, EhCS was cloned, expressed in Escherichia coli and purified by affinity and size-exclusion chromatography. The purified protein was crystallized in space group P41 with two molecules per asymmetric unit and a complete data set was collected to a resolution of 1.86 Å. A molecular-replacement solution was obtained using the Salmonella typhimurium O-acetylserine sulfhydrylase structure as a probe and had a correlation coefficient of 37.7% and an R factor of 48.8%.
1. Introduction
The cysteine-biosynthetic pathway contributes significantly to the incorporation of inorganic sulfur into organic compounds. In bacteria and plants, l-cysteine is the precursor of most sulfur-containing metabolites, including methionine and glutathione. Once the extracellularly transported sulfate enters the cell, it is activated and then reduced to form sulfide. Sulfide reacts with O-acetylserine, which is produced from serine and acetyl-CoA by serine acetyltransferase. This final reaction, which forms l-cysteine by transfer of the alanyl group of O-acetylserine to the sulfide, is catalyzed by cysteine synthase [CS; O-acetyl-l-serine (thiol)-lyase; EC 4.2.99.8]. Studies have shown that this pyroxidal phosphate-dependent enzyme, which is more commonly referred to as O-acetylserine sulfhydrylase (OASS), is ubiquitously present in various bacteria (Byrne et al., 1988 ▶; Ogasawara et al., 1994 ▶) and also in plants (Hell et al., 1994 ▶; Noji et al., 1994 ▶; Romer et al., 1992 ▶; Saito et al., 1992 ▶).
Entamoeba histolytica is an enteric protozoan parasite that causes amoebic colitis and extraintestinal abscesses (e.g. hepatic, pulmonary and cerebral; World Health Organization, 1995 ▶) in approximately 50 million inhabitants of the areas in which it is endemic. This organism is an amitochondriate and its basic metabolic pathways involve several proteins that contain low-midpoint redox potential iron–sulfur centres coordinated by cysteines. E. histolytica trophozoites preferably survive in the colonic lumen, which is specifically an anaerobic environment. However, they are subjected to high oxygen stress during tissue invasion, metastasis and extraintestinal propagation. Interestingly, despite being anaerobes, E. histolytica trophozoites have been shown to take up oxygen and also to tolerate oxygen pressure (Weinbach et al., 1976 ▶). In the host they invade aerobic tissues, but they require a reduced medium for growth in vitro, indicating that E. histolytica must be capable of defensive mechanisms to deal with reactive oxygen species in the host. The mechanisms of its antioxidative defences are poorly understood. Superoxide dismutase is present, but catalase and the glutathione system involved in the antioxidative defences of other organisms, including its host, are lacking in this organism (Hell et al., 1994 ▶). Cysteine was shown to be the major thiol in E. histolytica (Nozaki et al., 1998 ▶) and plays a vital role in the survival of this organism, including its antioxidative defences and the matrix adhesion, elongation, motility and growth in vitro of this glutathione-deficient organism (Romer et al., 1992 ▶; Saito et al., 1992 ▶). The major route of cysteine biosynthesis in this parasite is the condensation of O-acetylserine with sulfide by the de novo cysteine-biosynthetic pathway involving the key enzyme cysteine synthase (EhCS).
The E. histolytica genome encodes two genes for EhCS which only differ from one another by three nucleotides, consequently changing two amino acids in the sequence. Phylogenetic analysis of various CS sequences shows that EhCS does not belong to any existing families of the cysteine synthase superfamily (Nozaki et al., 1998 ▶). There is an extra N-terminal insertion that is unique to EhCS compared with CSs from other bacteria and plants. Because of the interesting characteristics attributed to this organism as well as to EhCS, we are focusing on structural studies of cysteine synthase from E. histolytica, which may be further exploitable for drug design.
2. Experimental procedure
2.1. Cloning of EhCS
The cysteine synthase (GenBank accession No. 2346963) coding sequence was amplified by PCR from genomic DNA of E. histolytica strain HM1.IMSS at the trophozoite stage (with the generous support of Professor Alok Bhattacharya, School of Life Sciences, JNU) and subcloned in the TA (Fermentas) vector between NcoI and XhoI flanking sites. Subsequently, the gene (digested at NcoI and XhoI) was cloned in pET-28a vector with a C-terminal His6 tag (Novagen). The PCR reaction was performed using the following forward and reverse oligonucleotides primers: CS_FP, 5′-CATGCCATGGAACAAATAAGTATTAGCTCTC-3′, and CS_RP, 5′-CCGCTCGAGTTCATTCAATAATGAATCAAG-3′, respectively.
2.2. Overexpression and purification
pET28a-EhCS was transformed into Escherichia coli strain BLR (DE3) cells, which were grown in LB media supplemented with 50 µg ml−1 kanamycin at 310 K. When the OD600 reached 0.5, overexpression of EhCS was induced by the introduction of 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG) into the culture and incubation at 303 K for an additional 3 h. The cells were harvested by centrifugation at 6000g for 5 min at 277 K, suspended in buffer A (50 mM Tris–HCl pH 8.0, 200 mM NaCl, 100 µM PMSF) and lysed with 1 mg ml−1 lysozyme and 0.1% Triton-X. After sonication on ice, a clear supernatant was obtained by centrifugation at 15 000 rev min−1 for 15 min at 277 K.
The first chromatographic step involved the selection of His-tagged protein on an Ni–NTA column (Sigma–Aldrich) pre-equilibrated with buffer A (lysis buffer) and 5 mM imidazole. The column was washed with five column volumes of buffer B (50 mM Tris–HCl pH 8.0, 200 mM NaCl, 30 mM imidazole) at 277 K. The bound protein was eluted as 1 ml fractions with buffer B containing 300 mM imidazole. The fractions with optimum absorbance at 280 nm were checked for homogeneity on SDS–PAGE (Laemmli, 1970 ▶), pooled and subjected to 40% ammonium sulfate precipitation. The precipitant was dissolved in 0.5 ml buffer C (100 mM Tris–HCl pH 8.0, 50 mM NaCl, 10 mM β-mercaptoethanol).
The final purification step involved gel filtration on a HiLoad Superdex 75G 16/60 column (Amersham Biosciences). The column was pre-equilibrated with buffer A (50 mM Tris, 150 mM NaCl, 10 mM β-mercaptoethanol pH 8.0). The purity of the protein was assessed on SDS–PAGE and the purified protein was concentrated using YM3K Centricon tubes (Amicon) to a final concentration of 15 mg ml−1 as estimated by the Bradford method (Bradford, 1976 ▶).
2.3. Crystallization
Purified EhCS was concentrated to a final concentration of 15 mg ml−1 in 50 mM Tris–HCl pH 8.0 buffer containing 150 mM NaCl for crystallization trials. Crystallization trials were carried out at 289 and 277 K using the hanging-drop method by mixing equal volumes (3 µl) of protein and reservoir solutions. The drops were equilibrated against 500 µl of the same precipitant solution. Several crystallization conditions were tested with PEG, ammonium sulfate and sodium malonate.
2.4. Collection and processing of diffraction data
A single crystal was separated from the drop using a cryoloop (Hampton Research). The crystal was then flash-cooled to 100 K in a nitrogen-gas stream. Prior to flash-freezing, the crystal was soaked sequentially for 15 s in reservoir solution supplemented with 5, 10, 15 and 20% glycerol as a cryoprotectant. Diffraction data were collected on a MAR imaging plate using a Rigaku MicroMax-007 X-ray generator with X-ray optics to focus the beam (International Center of Genetic Engineering and Biotechnology, New Delhi). The data were indexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶).
2.5. Structure solution
Sequence-similarity searches using the BlastP algorithm (Altschul et al., 1997 ▶) with EhCS against the Protein Data Bank (http://www.rcsb.org) selected several cysteine synthase structures. EhCS has a sequence identity of 47% to Salmonella typhimurium CS (PDB code 1oas; Burkhard et al., 1998 ▶), 46% to Arabidopsis CS (PDB code 1z7w; Bonner et al., 2005 ▶) and 39% to E. coli CS (PDB code 2bht; Claus et al., 2005 ▶). Molecular replacement was attempted using various structures of S. typhimurium CS in various conformations (PDB codes 1oas, 1fcj and 1d6s), as the S. typhimurium CS exhibited the highest sequence identity.
3. Results and discussion
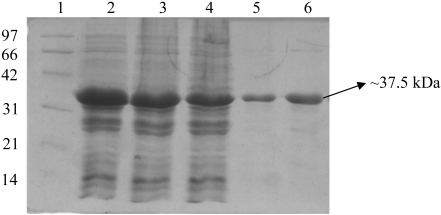
Cysteine synthase from E. histolytica was expressed in BLR (DE3) cells, resulting in approximately 100% soluble protein. The recombinant protein was purified by affinity and size-exclusion chromatography. The sample was 99% pure as estimated by SDS–PAGE (Fig. 1 ▶). The retention time of the protein on the size-exclusion column indicated that the protein is a dimer of ∼75 kDa (data not shown).
Figure 1.
SDS–PAGE showing fractions purified by Ni–NTA and gel-filtration chromatography. Proteins were separated on 12% SDS–PAGE and stained with Coomassie Brilliant Blue. Lane 1, molecular-weight markers (kDa); lanes 2–4, Ni–NTA purified fractions; lanes 5–6, gel-filtration purified fractions.
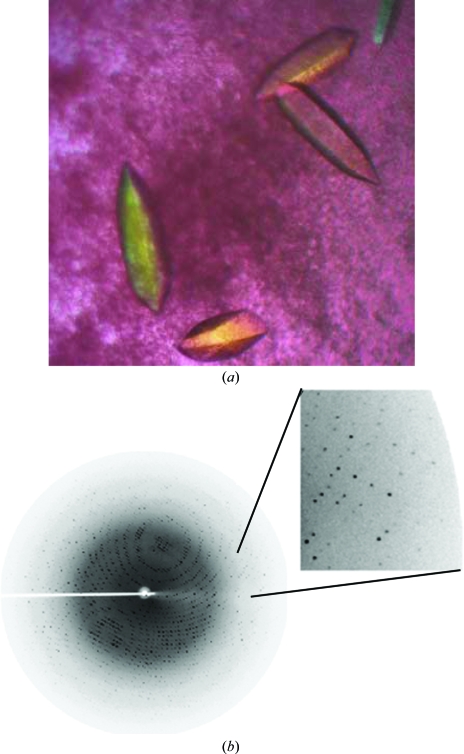
Many drops containing ammonium sulfate as a precipitant at 289 K developed phase separation in 1 or 2 d. Optimization of these conditions with ammonium sulfate resulted in the production of good-quality single crystals (ranging in size from 0.3 × 0.2 × 0.2 to 0.5 × 0.2 × 0.2 mm) using 2.5 M ammonium sulfate as precipitant in 100 mM Tris pH 7.0. Elongated hexagonal shaped crystals of EhCS were obtained in 20 d at 289 K (Fig. 2 ▶ a).
Figure 2.
(a) Single elongated hexagonal shaped crystals of EhCS obtained along with protein precipitate at 289 K using the hanging-drop vapour-diffusion method. (b) Diffraction pattern of EhCS crystals to 1.86 Å resolution. The data were collected using a Rigaku MicroMax-007 generator and a MAR imaging plate. The imaging plate was adjusted to a distance of 150 mm and the crystals were exposed for 90 s per frame. Diffraction spots were observed to the edges of the image plate, as shown in the enlargement.
The crystals belong to the tetragonal space group P41, with unit-cell parameters a = 80.3, b = 80.3, c = 112.2 Å. A complete X-ray diffraction data set of about sevenfold redundancy was collected to 1.86 Å resolution (Fig. 2 ▶ b). The volume of the asymmetric unit allows the presence of a dimer, giving a Matthews volume (V M) of 2.5 Å3 Da−1 and a solvent content of 50.7% (Matthews, 1968 ▶). This is consistent with the solution obtained by molecular replacement and allowed us to assign the space group as P41 (in contrast to P43). Details of the data-collection statistics are reported in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.54 |
| Space group | P41 |
| Unit-cell parameters (Å) | |
| a | 80.31 |
| b | 80.31 |
| c | 112.21 |
| Resolution range (Å) | 50–1.86 (1.93–1.86) |
| Rmerge† (%) | 4.3 (33.6) |
| Completeness (%) | 99.8 (98.1) |
| Total No. of observations | 392808 |
| No. of unique observations | 59571 |
| Redundancy | 6.6 (5.9) |
| Average I/σ(I) | 16.2 (5.55) |
| Crystal mosaicity (°) | 0.4 |
R
merge = 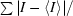
 .
.
The best solution was obtained using the S. typhimurium CS structure (PDB code 1oas; Burkhard et al., 1998 ▶) as a model and resulted in a correlation coefficient of 37.7% and an R factor of 48.8% when data to 4 Å resolution were used. Rigid-body refinement using CNS (Brünger et al., 1998 ▶) and three rigid bodies (residues 1–44, 45–148 and 149–315) resulted in a model with an R factor of 47.6% to 2.5 Å resolution. Further refinement and model building are in progress.
Acknowledgments
The authors thank Dr Amit Sharma, ICGEB (International Center for Genetic Engineering and Biotechnology), New Delhi for allowing them to use the rotating-anode generator and imaging-plate system for data collection and Dr M. Yogavel for his help during data collection. CK thanks ICMR, RJ thanks UGC and TK thanks UGC for fellowships. NA thanks the Department of Science and Technology, Government of India for funding. We thank Professor Alok Bhattacharya for his support. Part of the study was supported by grants from CSIR and DBT, Government of India to Professor Alok Bhattacharya, School of Life Sciences, JNU.
References
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res.25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner, E. R., Cahoon, R. E., Knapke, S. M. & Jez, J. M. (2005). J. Biol. Chem.280, 38803–38813. [DOI] [PubMed] [Google Scholar]
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T. & Warren, G. L. (1998). Acta Cryst. D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burkhard, P., Jagannatha Rao, G. S., Hohenester, E., Schnackerz, K. D., Cook, P. F. & Jansonius, J. N. (1998). J. Mol. Biol.283, 121–133. [DOI] [PubMed] [Google Scholar]
- Byrne, C. R., Monroe, R. S., Ward, K. A. & Kredich, N. M. (1988). J. Bacteriol.170, 3150–3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claus, M. T., Zocher, G. E., Maier, T. H. P. & Schulz, G. E. (2005). Biochemistry, 44, 8620–8626. [DOI] [PubMed] [Google Scholar]
- Hell, R., Bork, C., Bogdanova, N., Frolov, I. & Hauschild, R. (1994). FEBS Lett.351, 57–62. [DOI] [PubMed]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Nozaki, T., Asai, T., Kobayashi, S., Ikegami, F., Noji, M., Saito, K. & Takeuchi, T. (1998). Mol. Biochem. Parasitol.97, 33–44. [DOI] [PubMed] [Google Scholar]
- Noji, M., Murakoshi, I. & Saito, K. (1994). Mol. Gen. Genet.244, 57–66. [DOI] [PubMed] [Google Scholar]
- Ogasawara, N., Nakai, S. & Yoshikawa, H. (1994). DNA Res.1, 1–14. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Romer, S., d’Harlingue, A., Camara, B., Schantz, R. & Kuntz, M. (1992). J. Biol. Chem.267, 17966–17970. [PubMed] [Google Scholar]
- Saito, K., Miura, N., Yamazaki, M., Hirano, H. & Murakoshi, I. (1992). Proc. Natl Acad. Sci. USA, 89, 8078–8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbach, E. C., Diamond, L. S., Claggett, C. E. & Kon, H. (1976). J. Parasitol.62, 127–128. [PubMed] [Google Scholar]
- World Health Organization (1995). The World Health Report 1995: Bridging the Gaps, p. 28. Geneva: World Health Organization.