An α/β-type small, acid-soluble spore protein (SASP) from Bacillus subtilis, a major source of DNA protection against damaging effects in spores, was crystallized in a functionally relevant complex with a double-stranded DNA. This report provides insights into initial characterization of the complex and its structure elucidation.
Keywords: small acid-soluble spore protein, spore resistance, DNA, Bacillus subtilis
Abstract
An engineered variant of an α/β-type small acid-soluble spore protein (SASP) from Bacillus subtilis was crystallized in a complex with a ten-base-pair double-stranded DNA by the hanging-drop vapor-diffusion method using ammonium sulfate as a precipitating agent. Crystals grew at 281 K using sodium cacodylate buffer pH 5.5 and these crystals diffracted X-rays to beyond 2.4 Å resolution using synchrotron radiation. The crystallized complex contains two or three SASP molecules bound to one DNA molecule. The crystals belong to the hexagonal space group P6122 or P6522, with unit-cell parameters a = b = 87.0, c = 145.4 Å, α = β = 90.0, γ = 120.0°. Diffraction data were 96.6% complete to 2.4 Å resolution, with an R sym of 8.5%. Structure solution by the multiwavelength/single-wavelength anomalous dispersion method using isomorphous crystals of selenomethionine-labeled protein is in progress.
1. Introduction
The DNA in the spores of Bacillus species is extremely resistant to a variety of damaging agents including heat, UV radiation and many genotoxic chemicals (Setlow, 1999 ▶, 2006 ▶). A major reason for the resistance of the DNA in the spores is the saturation of the DNA with a group of nonspecific DNA-binding proteins, the small acid-soluble spore proteins (SASPs) of the α/β-type (Driks & Setlow, 1999 ▶; Setlow, 1999 ▶, 2006 ▶, 2007 ▶). The α/β-type SASPs have 59–72 residues and their primary sequence is highly conserved in Bacillus species, although they show no obvious homology to any other type of protein or any known DNA-binding motifs. Binding of α/β-type SASPs to DNA has significant effects on the structure of both the DNA and the protein components of the complex, as the DNA appears to adopt an A-like helical conformation, although not the classical A-helix, while the protein becomes highly α-helical (Hayes et al., 2000 ▶; Setlow, 2006 ▶, 2007 ▶). Studies on the interaction of α/β-type SASPs with DNA in vitro have shown that (i) only double-stranded (ds) DNAs are bound, (ii) GC-rich DNAs are bound most tightly, in particular polydG·polydC, (iii) the affinity of α/β-type SASPs for DNA increases as the pH is lowered below 7, (iv) there is one α/β-type SASP per approximately four base pairs (bp) of DNA in the complex, (v) ds oligonucleotides as small as decamers are bound and (vi) binding of α/β-type SASPs to DNA is cooperative and important contacts between adjacent protein molecules bound to DNA have been identified (Hayes et al., 2000 ▶; Hayes & Setlow, 1998 ▶; Kosman & Setlow, 2003 ▶; Setlow et al., 1992 ▶; Setlow, 2003 ▶). Relatively low-resolution information on the structure of the α/β-type SASP–DNA complex has also been obtained by both electron microscopy and cryo-electron microscopy (Frenkiel-Krispin et al., 2004 ▶; Griffith et al., 1994 ▶).
Although some information about the complex formed between α/β-type SASP and DNA is available as noted above, a high-resolution structure of the complex has not been determined. Consequently, it is unclear how α/β-type SASP binding modifies DNA properties such that the spore DNA acquires resistance to so many different agents. Consequently, in this work we report the crystallization and preliminary structural characterization of the complex between a 10 bp ds oligonucleotide and an engineered α/β-type SASP that binds extremely tightly to DNA.
2. Experimental procedures
2.1. Production of α/β-type SASP and DNA and complex formation
The B. subtilis α/β-type SASP SspCΔN11-D13K-C3, a 64-residue protein with a molecular weight of 6797.7 Da, was chosen for this work since this protein had been engineered to have tighter binding to DNA than any native α/β-type SASP (Kosman & Setlow, 2003 ▶) and it was felt that tight binding of the protein to DNA would be likely to be essential for the crystallization of a complex. This protein was overexpressed in Escherichia coli BL21(DE3) from plasmid pET11d carrying sspC ΔN11-D13K-C3 and extracted and purified as described in Kosman & Setlow (2003 ▶). The purified protein was dialyzed at 279 K against 10 mM sodium phosphate buffer pH 6 in Spectra/Por tubing (3500 Da moelcular-weight cutoff) and had a final concentration of ∼250 µM as determined by a dye-binding assay that was calibrated for this α/β-type SASP by amino-acid analysis (Bradford, 1976 ▶; Kosman & Setlow, 2003 ▶). SspCΔN11-D13K-C3 contains three methionine residues, although the N-terminal Met is removed post-translationally. The protein was labeled with selenomethionine (SeMet) by overexpression from plasmid pET11d carrying sspC ΔN11-D13K-C3 in E. coli B834 (Novagen). The cells were grown at 310 K in M9 minimal medium supplemented with SeMet as described by Doublié (1997 ▶), SspCΔN11-D13K-C3 overexpression was induced by addition of isopropyl β-d-thiogalactoside to 0.5 mM at an OD600nm of ∼1, growth was continued for a further 2 h, cells were harvested by centrifugation, the pellet was washed with cold 0.15 M NaCl and the final cell pellet was frozen and lyophilized. The dry cells were ruptured and the SeMet protein was extracted and purified as described for the Met-containing protein (Kosman & Setlow, 2003 ▶), except that dithiothreitol (DTT) was present at 10 mM in all solutions used in the extraction and purification to minimize SeMet oxidation.
The DNA used for SASP binding was a ds oligonucleotide, MW 6686.4 Da, generated by annealing two 11-nucleotide oligonucleotides, 5′-G10A-3′ and 5′-C10A-3′, that formed ten GC base pairs with single 3′-overhangs to prevent strand slippage. The oligonucleotides, which had been purified by high-pressure liquid chromatography, were obtained from the Midland Certified Reagent Company, Midland, TX, USA and annealed as suggested by the supplier in 10 mM Tris–HCl pH 7.5, 0.1 M NaCl, 1 mM ethylenediamine tetraacetic acid (EDTA), giving a ds DNA concentration of ∼100 µM.
The α/β-type SASP–DNA complex was prepared at room temperature by slowly adding purified protein to the DNA to a molar protein:DNA ratio of ∼3:1; the mixture was allowed to sit for an additional 1 h at room temperature and was then dialyzed at 279 K in Spectra/Por 3 tubing against filtered 10 mM sodium phosphate pH 6. Previous work suggested that two to three α/β-type SASP molecules would bind to the ds DNA used (Hayes et al., 2000 ▶, 2001 ▶; Setlow et al., 1992 ▶). However, we used a 3:1 ratio of protein:DNA to ensure that the DNA was saturated with protein. Formation of the α/β-type SASP–DNA complex was assessed by circular-dichroism (CD) spectroscopy (Hayes et al., 2000 ▶; Kosman & Setlow, 2003 ▶) and this indicated that the α/β-type SASP had bound to the DNA. The complex was then concentrated to ∼2.4 mM total protein by centrifugation in Centricon YM-3 concentrators (Millipore Corporation) in 10 mM sodium phosphate buffer pH 6.0 (also used for crystallization; see below).
2.2. Crystallization
The α/β-type SASP–DNA complex was crystallized at 281 K (ECHOtherm 40 chilling incubator, Torrey Pines Scientific LLC, San Marcos, CA, USA) by the hanging-drop vapor-diffusion method using 24-well Linbro culture plates (McPherson, 1999 ▶; Scott et al., 1995 ▶). Preliminary crystallization conditions were determined using a screen of several common precipitating agents incorporated in the Natrix crystallization screen for the crystallization of nucleic acids and protein–nucleic acid complexes (Hampton Research). The hanging drops contained equal volumes of 1–2 µl protein solution and 1–2 µl reservoir solution. The reservoir volume was 500 µl. The initial and final crystallizations were performed using the same setup.
2.3. X-ray diffraction, data collection and analysis
For X-ray diffraction studies, crystals were cryoprotected using ethylene glycol and flash-frozen in liquid nitrogen. X-ray data were collected using synchrotron radiation on beamline 8.2.2 at the Advanced Light Source, Lawrence Berkeley National Laboratory. The X-ray diffraction data for the crystals of the complex were collected using rotation (oscillation) photography utilizing an ADSC Q315 3 × 3 CCD array detector. The collected data were analyzed, indexed, integrated and scaled using the HKL-2000 software package (Otwinowski & Minor, 1997 ▶). The unit cells and space groups of highest symmetry were determined by examining the diffraction pattern, its symmetry and systematic absences and the statistics of the native diffraction data.
2.4. Other methods
UV absorption was measured using a Shimadzu BioSpec 1601 spectrophotometer at room temperature using 50 µl quartz cuvettes. All UV spectra were determined at least in triplicate.
3. Results and discussion
3.1. Preparation of the α/β-type SASP–DNA complex
The SASP chosen for crystallization studies was a variant of the minor B. subtilis α/β-type SASP, SspCΔN11-D13K-C3, that had been engineered to bind very tightly to DNA (Kosman & Setlow, 2003 ▶). The binding of this protein to DNA is the tightest of any α/β-type SASP tested, but this protein has the same effect on DNA properties in vitro as other wild-type α/β-type SASPs (Kosman & Setlow, 2003 ▶). This protein also lacks 11 N-terminal residues present in the wild-type parental protein that do not appear to be involved in DNA binding but that might interfere with crystal formation (Hayes & Setlow, 2001 ▶; Kosman & Setlow, 2003 ▶). The binding affinity of this protein for DNA increases significantly as the pH is lowered below 7, which is consistent with the observation that the pH in the dormant spore is 6.3–6.5 but rises to 7.5–7.8 when spores germinate and α/β-type SASP must dissociate from DNA (Kosman & Setlow, 2003 ▶; Magill et al., 1994 ▶, 1996 ▶; Setlow & Setlow, 1980 ▶).
The ds oligonucleotide chosen for complex formation with the SspC variant was a 10 bp oligodG·oligodC with single 3′-A overhangs to prevent strand slippage. A GC-rich oligonucleotide was chosen because of the much tighter binding of α/β-type SASP to GC-rich DNAs, in particular homopolymeric dG·dC regions, and the 10 bp length was chosen based on previous work showing that this was as small as would give tight binding to α/β-type SASP (Hayes et al., 2000 ▶, 2001 ▶; Kosman & Setlow, 2003 ▶).
3.2. Crystallization and crystal analysis
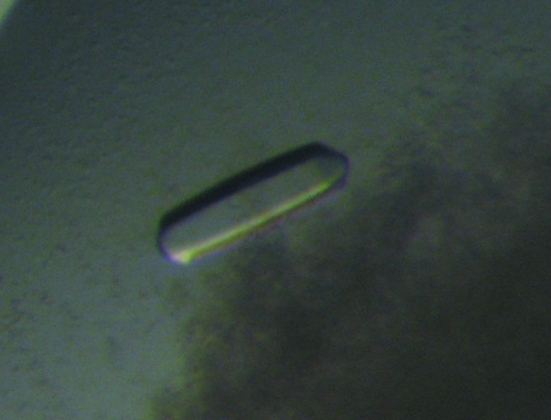
Initially, small hexagonal bipyramidal crystals grew using 10 mM magnesium sulfate, 50 mM sodium cacodylate buffer pH 6.5 and 2.0 M ammonium sulfate as the precipitating agent. After refining the composition of the crystallization solution, the optimal condition for growing crystals proved to be 10 mM magnesium sulfate, 50 mM sodium cacodylate buffer pH 5.5, 1.4 M ammonium sulfate, with 10 mM sodium bromide, 100 mM taurine, 5% polyvinylpyrrolidone, 5 mM spermine (Fig. 1 ▶). The latter four compounds were instrumental in growing larger crystals that grew to approximate dimensions of 0.15 × 0.15 × 0.30 mm in several weeks.
Figure 1.
Crystal of the methionyl α/β-type SASP–DNA complex.
The crystallization of the SeMet protein–DNA complex was performed under conditions similar to those given above, but using a lower concentration of ammonium sulfate (1.3 M) and adding EDTA to 10–20 mM and DTT to 1.5–2.0 mM. Both EDTA and DTT were important for SeMet protein-crystal growth. Macroseeding was employed to obtain larger crystals, but these crystals were still smaller than those formed using the Met protein. Both Met and SeMet protein crystals were cryoprotected using ethylene glycol and flash-frozen in liquid nitrogen at 93 K.
3.3. Diffraction data collection
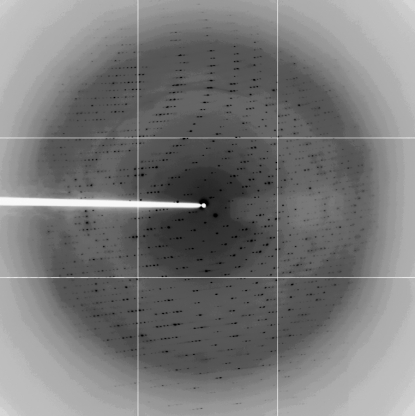
One cryoprotected and frozen crystal of the Met protein was used to collect a full data set to 2.4 Å (Fig. 2 ▶, Table 1 ▶), although decay of crystals in the X-ray beam was observed. Analysis of the diffraction data using the autoindexing procedure of DENZO (Otwinowski & Minor, 1997 ▶) indicated that the crystals belong to the hexagonal space group P6122 or P6522, with unit-cell parameters a = b = 87.0, c = 145.4 Å, α = β = 90.0, γ = 120.0°. A crystal volume per unit of protein molecular weight, V M, of 4.0 or 2.9 Å3 Da−1 is consistent with the presence of one molecule of the complex in the asymmetric unit. For the V M of 4.0 Å3 Da−1, there are two SASP molecules and one DNA molecule in a complex (MW 20 282 Da); a V M of 2.9 Å3 Da−1 corresponds to three SASP molecules in the same complex (MW 27 079 Da). The solvent content is 68 or 59% for two or three SASP molecules in the complex, respectively (Matthews, 1968 ▶).
Figure 2.
Diffraction image from a crystal of the methionyl α/β-type SASP–DNA complex. The edge of the detector is at ∼3.0 Å resolution.
Table 1. Statistics of the native and three-wavelength MAD X-ray diffraction data for the α/β-type SASP–DNA complex.
X-ray data for the complex with the Met protein were acquired at 103 K using a synchrotron source at 1.1 Å and a CCD detector. The SeMet protein complex data were obtained at three wavelengths: inflection, peak and high-energy remote. Values in parentheses are for the highest resolution shells.
| SeMet complex | ||||
|---|---|---|---|---|
| Data set | Native complex | Peak | Inflection | High remote |
| Wavelength (Å) | 1.1 | 0.9800 | 0.9798 | 0.9500 |
| Resolution (Å) | 50.0–2.40 (2.59–2.40) | 50.0–3.50 (3.63–3.50) | 50.0–3.50 (3.63–3.50) | 50.0–3.50 (3.63–3.50) |
| Total reflections | 247290 (15962) | 45089 (3451) | 43708 (3389) | 48922 (3922) |
| Unique reflections | 13171 (1177) | 4412 (426) | 4418 (429) | 4426 (431) |
| Redundancy | 18.8 (12.9) | 10.2 (8.1) | 9.9 (7.9) | 11.0 (9.1) |
| Completeness (%) | 96.6 (81.0) | 99.0 (99.5) | 99.0 (99.5) | 98.9 |
| I/σ(I) | 31.6 (3.0) | 9.5 (2.5) | 8.5 (2.0) | 10.2 (2.4) |
| Rsym† (%) | 8.5 (73.6) | 25.3 (85.4) | 26.5 (85.8) | 24.7 (93.0) |
R
sym = 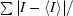
 , where I is the intensity of an observation of a multiply observed reflection.
, where I is the intensity of an observation of a multiply observed reflection.
Data images were processed with HKL-2000 (Otwinowski & Minor, 1997 ▶). The overall merging R factor, completeness and I/σ(I) for all the data between 50.0 and 2.4 Å resolution are 8.5%, 96.6% and 31.6, respectively (Table 1 ▶). The crystals of the complex with the SeMet protein were utilized to collect three-wavelength anomalous dispersion diffraction data at the Se edge to 3.5 Å resolution (Table 1 ▶). X-ray fluorescence was used to detect and confirm the presence of Se in these crystals as well as to determine the peak and inflection points for the Se K edge for the multiwavelength anomalous dispersion (MAD) data collection. The presence of Se in these crystals was unambiguously confirmed. The poorer quality of the MAD data compared with those for the Met protein–DNA complex in a large part reflects the smaller size of the crystals of the SeMet protein compared with those of the Met protein.
3.4. Initial structure solution
Utilizing the automated methodology of SOLVE (Terwilliger & Berendzen, 1999 ▶) with the MAD diffraction data, four to six Se atoms were identified in the asymmetric unit of the crystal and these were used to obtain initial phases with an initial figure of merit (FOM) of 0.35 at 3.5 Å. Further density modification with maximum-likelihood methods employed in RESOLVE (Terwilliger, 2002 ▶) resulted in phases with an FOM of 0.52 at 3.5 Å. The automated residue-fitting procedures of RESOLVE also yielded coordinates for 82 protein residues (Terwilliger, 2003 ▶). Inspection of graphics using O (Jones et al., 1991 ▶) suggested that these protein residues correspond to the two to three SASP molecules expected to be bound to the ds oligonucleotide in the complex. Further manual building of the model is in process.
3.5. Confirmation of complex formation
CD spectroscopy of the protein–DNA mixture after completion of protein addition indicated that the protein had bound to the DNA and further that the amount of protein bound was approximately what would be expected for two to three proteins bound to the DNA used (see §2). The crystals formed were presumably of the complex and not of any of the individual components present in the crystallization mixture, the DNA or the protein. However, this was confirmed in several ways. The presence of α/β-type SASP in the crystals was confirmed from the fluorescence data obtained during diffraction experiments for collection of the MAD data. The presence of DNA in the crystals was confirmed by taking several crystals from the crystallization drops, washing out remaining mother liquor by multiple transfer of crystals into the crystallization reservoir solution, dissolving the crystals in water and measuring the UV spectrum of the dissolved crystals in a 50 µl cuvette. This UV spectrum was essentially identical to that of the α/β-type SASP–DNA mixture used for crystallization (data not shown).
Acknowledgments
We thank Dr James E. Littlejohn for assistance and discussion and Dr Ki Seog Lee for providing a picture of the crystal. The diffraction data were collected at the Berkeley Center for Structural Biology, Advanced Light Source, Lawrence Berkeley National Laboratory using beamline 8.2.2. The authors thank the staff of this beamline for help and assistance. This study was supported by a DARPA contract (DAAD19-03-C-0051 to MJJ and PS) and a grant from the NIH (GM 19698 to PS and MJJ).
References
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed] [Google Scholar]
- Doublié, S. (1997). Methods Enzymol.276, 523–530. [PubMed] [Google Scholar]
- Driks, A. & Setlow, P. (1999). Prokaryotic Development, edited by Y. V. Brun & L. J. Shimkets, pp. 191–218. Washington DC: American Society for Microbiology.
- Frenkiel-Krispin, D., Sack, R., Englander, J., Shimoni, E., Eisenstein, M., Bullitt, E., Horowitz-Scherer, R., Hayes, C. S., Setlow, P., Minsky, A. & Wolf, S. G. (2004). J. Bacteriol.186, 3525–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith, J., Makhov, A., Santiago-Lara, L. & Setlow, P. (1994). Proc. Natl Acad. Sci. USA, 91, 8224–8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes, C. S., Alarcon-Hernandez, E. & Setlow, P. (2001). J. Biol. Chem.276, 2267–2275. [DOI] [PubMed] [Google Scholar]
- Hayes, C. S., Peng, Z. Y. & Setlow, P. (2000). J. Biol. Chem.275, 35040–35050. [DOI] [PubMed] [Google Scholar]
- Hayes, C. S. & Setlow, P. (1998). J. Biol. Chem.273, 17326–17332. [DOI] [PubMed] [Google Scholar]
- Hayes, C. S. & Setlow, P. (2001). J. Bacteriol.183, 2662–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T. A., Zou, J.-Y., Cowan, S. W. & Kjeldgaard, M. (1991). Acta Cryst. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kosman, J. & Setlow, P. (2003). J. Bacteriol.185, 6095–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson, A. (1999). Crystallization of Biological Macromolecules. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
- Magill, N. G., Cowan, A. E., Koppel, D. E. & Setlow, P. (1994). J. Bacteriol.176, 2252–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magill, N. G., Cowan, A. E., Leyva-Vazquez, M. A., Brown, M., Koppel, D. E. & Setlow, P. (1996). J. Bacteriol.178, 2204–2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Scott, W. G., Finch, J. T., Grenfell, R., Fogg, J., Smith, T., Gait, M. J. & Klug, A. (1995). J. Mol. Biol.250, 327–332. [DOI] [PubMed] [Google Scholar]
- Setlow, B. & Setlow, P. (1980). Proc. Natl Acad. Sci. USA, 77, 2474–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow, B., Sun, D. & Setlow, P. (1992). J. Bacteriol.174, 2312–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow, P. (1999). Bacterial Stress Responses, edited by G. Storz & R. Hennge-Aronis, pp. 217–230. Washington DC: American Society for Microbiology.
- Setlow, P. (2003). Curr. Opin. Microbiol.6, 550–556. [DOI] [PubMed] [Google Scholar]
- Setlow, P. (2006). J. Appl. Microbiol.101, 514–525. [DOI] [PubMed] [Google Scholar]
- Setlow, P. (2007). Trends Microbiol.15, 172–180. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T. C. (2002). Acta Cryst. D58, 1937–1940. [DOI] [PubMed] [Google Scholar]
- Terwilliger, T. C. (2003). Acta Cryst. D59, 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger, T. C. & Berendzen, J. (1999). Acta Cryst. D55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]