Est25, a ketoprofen-specific hormone-sensitive lipase from a metagenomic library, was crystallized and diffraction data were collected to 1.49 Å resolution.
Keywords: Est25, ketoprofen-specific hormone-sensitive lipase
Abstract
Ketoprofen, a nonsteroidal anti-inflammatory drug, inhibits the synthesis of prostaglandin. A novel hydrolase (Est25) with high ketoprofen specificity has previously been identified using a metagenomic library from environmental samples. Recombinant Est25 protein with a histidine tag at the N-terminus was expressed in Escherichia coli and purified in a homogenous form. Est25 was crystallized from 2.4 M sodium malonate pH 7.0 and X-ray diffraction data were collected to 1.49 Å using synchrotron radiation. The crystals belong to the monoclinic space group C2, with unit-cell parameters a = 197.8, b = 95.2, c = 99.4 Å, β = 97.1°.
1. Introduction
Ketoprofen [(R,S)-2-(3-benzoylphenyl)propionic acid; C16H14O3], a nonsteroidal anti-inflammatory drug, inhibits the synthesis of prostaglandin (Sohn & Krotz, 2006 ▶; Turini & DuBois, 2002 ▶). Specifically, (S)-ketoprofen has attracted great academic and industrial interest owing to its anti-inflammatory and analgesic effects (Mazieres, 2005 ▶). Therefore, many methods with limited application have been employed to prepare large quantities of (S)-ketoprofen with high purity (Mullangi et al., 2003 ▶). Recently, ketoprofen ester hydrolysis catalyzed by hydrolytic enzymes such as Candina rugosa lipase and Carica papaya lipase has been used for this purpose (Zhu et al., 2005 ▶; Ng & Tsai, 2005 ▶). In previous studies, we searched a metagenomic library to discover new catalytic enzymes with high ketoprofen specificities and a novel hydrolase (Est25) from environmental samples was characterized (Yoon et al., 2007 ▶; Kim et al., 2006 ▶; Ahn et al., 2006 ▶). Est25 is optimally active at pH 7.0 and 298 K and has a catalytic triad consisting of Ser201, Asp303 and His333 residues.
Interestingly, the primary sequence of Est25 shows similarities to members of the hormone-sensitive lipase (HSL) family such as brefeldin A esterase (BFAE, 48% identity) from Bacillus subtilis, a carboxylesterase (AFEST, 33%) from Archaeoglobus fulgidus, heroin esterase (HerE, 29%) from Rhodococcus sp. and a carboxyesterase (EST2, 30%) from Alicyclobacillus acidocaldarius. The HSL family is a class of enzymes that mobilize fatty acids for energy metabolism in adipose tissue (Yeaman, 2004 ▶; Osterlund, 2001 ▶). Although the members of the HSL family share an α/β-hydrolase fold, each structure has its own arrangement of α-helices and β-sheets (Larsen et al., 2002 ▶; De Simone et al., 2001 ▶; Wei et al., 1999 ▶). Therefore, the crystal structure of Est25 is essential to understand the enzyme’s catalytic mechanism and substrate specificities for (RS)-ketoprofen at the molecular level. In addition, the Est25 crystal structure is expected to provide structural and functional insights into the HSL family. In the present study, the expression, crystallization and preliminary crystallographic studies of Est25 are reported.
2. Experimental procedures
2.1. Protein expression and purification
Molecular cloning of Est25 from uncultured microorganisms has been described previously (Streit & Schmitz, 2004 ▶; Kim et al., 2006 ▶). For purification of the Est25 protein, Est25 cDNA was inserted into the T5 promoter-driven expression vector pQE30. The forward and reverse primers were 5′-CAGGATCCatgagcaacgcaacactaaaccaa-3′ (BamHI) and 5′-GCAAGCTTttatgctcgagaataagcgaaggcc-3′ (HindIII), respectively. Overexpression of Est25 with an MRGSHHHHHHGS tag at the N-terminus was performed in Escherichia coli XL1-Blue strain according to a standard protocol (Qiagen, USA). Briefly, transformed E. coli cells were grown at 310 K in LB medium containing 100 µg ml−1 ampicillin to an OD600 of ∼0.5. Protein expression was induced by adding 0.5 mM isopropyl β-d-1-thiogalactoside (IPTG). After 3 h induction, the cells were harvested by centrifugation, resuspended and sonicated in 50 mM sodium phosphate buffer pH 8.0 containing 10 mM imidazole and 300 mM NaCl in a chilled water bath. In some cases, harvested cells were stored at 253 K for several days. After centrifugation at 12 000 rev min−1 for the removal of cell debris, the supernatants were applied onto an Ni–NTA column (Qiagen, USA) equilibrated with 10 mM imidazole and the His-tagged protein was purified according to the manufacturer’s instructions. SDS–PAGE analysis indicated that the purity of the Est25 protein was >98%. The protein concentration was determined using an extinction coefficient at 280 nm calculated from the amino-acid sequence. The purified protein was concentrated to 10 mg ml−1 in phosphate-buffered saline (140 mM sodium chloride, 2.7 mM potassium chloride, 10 mM phosphate pH 7.4) using an Amicon Ultrapure-15 centrifugal filter (30 kDa MWCO, Millipore, USA). The purified protein with a His tag at the N-terminus was used for crystallization without further cleavage.
2.2. Crystallization
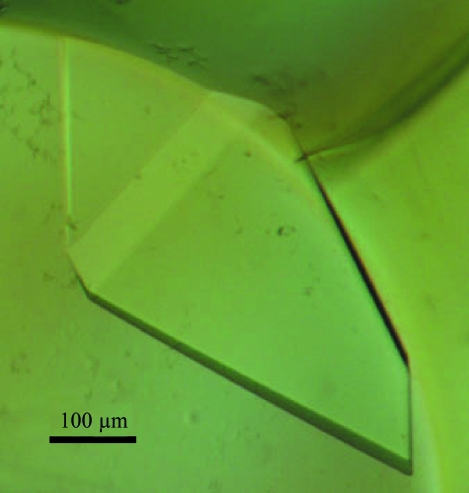
Initial crystallization was performed using the microbatch crystallization method (Chayen et al., 1990 ▶) under a thin layer of Al’s oil using Crystal, Index, Natrix (Hampton Research, CA, USA), Wizard and Cryo screening kits (Jena Bioscience, Germany) at 295 K. Each drop was prepared by mixing 1 µl reservoir solution and 1 µl protein solution and was covered with oil using Nunc MiniTrays (Nalge Nunc International, USA). Crystallization conditions for diffraction experiments were further optimized by changing various parameters. Diffraction-quality crystals were obtained using 2.4 M sodium malonate pH 7.0 (Fig. 1 ▶).
Figure 1.
A crystal of Est25 grown in a solution containing 2.4 M sodium malonate. Crystal dimensions were 0.4 × 0.3 × 0.1 mm.
2.3. X-ray data collection and processing
Crystals were transferred to cryosolvent consisting of the original precipitant containing an additional 30%(v/v) glycerol just before being flash-frozen in a cold nitrogen stream. Initial X-ray diffraction experiments were performed at beamline 4A of the Pohang Accelerator Laboratory, South Korea and at beamline NW12A of the Photon Factory, Japan. X-ray diffraction data were collected at 100 K using an ADSC Quantum 210 detector at beamline NW12, Photon Factory, Japan and were indexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). The wavelength of the synchrotron radiation was 1.00 Å.
3. Results and discussion
Est25, a ketoprofen-specific hydrolase identified from a metagenomic library, was overexpressed, purified and crystallized for structural analysis. Biophysical analyses have demonstrated that Est25 exists as a dimer under physiological conditions (Kim et al., 2006 ▶). Of a number of initial crystals obtained under various conditions, only the plate-shaped crystals obtained using screen No. 27 of Index Screen (2.4 M sodium malonate pH 7.0) showed reasonable diffraction. However, these crystals had cracks and the diffraction pattern did not appear to be homogeneous. Although efforts such as changing the concentration of protein or precipitant and using detergents failed to improve the crystal quality, the replacement of Al’s oil by paraffin oil improved the diffraction quality and the cracks disappeared. Finally, diffraction-quality crystals were grown using 2.4 M sodium malonate pH 7.0 and reached dimensions of 0.4 × 0.3 × 0.1 mm within 2 d (Fig. 1 ▶). X-ray diffraction data were collected to 1.49 Å at 100 K using synchrotron radiation with an R merge (on intensity) of 5.0%. The merged data set is 96.2% complete to 1.49 Å (Table 1 ▶). The Est25 crystals belong to the monoclinic space group C2, with unit-cell parameters a = 197.8, b = 95.2, c = 99.4 Å, α = γ = 90.0, β = 97.1°. Based on a molecular weight of 32 kDa for the Est25 monomer, the crystal-packing parameter (V M) was 3.62 Å3 Da−1 with a solvent content of 65.8% for two dimers in the asymmetric unit. In the case of three dimers in the asymmetric unit, V M and the solvent content would be 2.40 Å3 Da−1 and 48.7%, respectively. The data-collection and processing statistics for Est25 are summarized in Table 1 ▶. To obtain phase information, molecular replacement was attempted using brefeldin A esterase (BFAE; PDB code 1jkm) as a template. However, this was unsuccessful. Multi-wavelength anomalous dispersion methods as well as further molecular-replacement experiments using other template structures will be performed to determine the crystal structure. It is expected that the crystal structure of Est25 will provide new molecular insights into the substrate specificity of Est25 for (RS)-ketoprofen as well as the HSL family.
Table 1. Data-collection and processing statistics.
Values in parentheses are for the highest resolution shell.
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 197.8, b = 95.2, c = 99.4, β = 97.1 |
| Wavelength (Å) | 1.0000 |
| Resolution (Å) | 50–1.49 (1.65–1.59) |
| Unique reflections | 285917 (26733) |
| Completeness (%) | 96.2 (90.1) |
| Redundancy | 3.5 (3.3) |
| Rmerge† (%) | 5.0 (31.6) |
| 〈I/σ(I)〉 | 27.8 (3.6) |
R
merge = 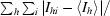
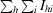 .
.
Acknowledgments
This work was supported by the Korean Government (MOEHRD-KRF-2004-D00235; YR) and by the 21C Frontier Functional Proteomics Program (FPR06B2-120), the Ubiquitome Research Program (M10533010001-05N3301-00100) and the National Research Laboratory Program (NRL-2006-02287) of the Korean Ministry of Science and Technology (to KKK). This work was also supported by the ‘GRRC’ project of Gyeonggi Provincial Government, South Korea.
References
- Ahn, M., Kim, S., Kang, M., Ryu, Y. & Kim, T. D. (2006). Biochem. Biophys. Res. Commun.346, 1142–1149. [DOI] [PubMed] [Google Scholar]
- Chayen, N. E., Shaw Stewart, P. D., Maeder, D. L. & Blow, D. M. (1990). J. Appl. Cryst.23, 297–302. [Google Scholar]
- De Simone, G., Menchise, V., Manco, G., Mandrich, L., Sorrentino, N., Lang, D., Rossi, M. & Pedone, C. (2001). J. Mol. Biol.314, 507–518. [DOI] [PubMed] [Google Scholar]
- Kim, Y. J., Choi, G. S., Kim, S. B., Yoon, G. S., Kim, Y. S. & Ryu, Y. W. (2006). Protein Expr. Purif.45, 315–323. [DOI] [PubMed] [Google Scholar]
- Larsen, N. A., Turner, J. M., Stevens, J., Rosser, S. J., Basran, A., Lerner, R. A., Bruce, N. C. & Wilson, I. A. (2002). Nature Struct. Biol.9, 17–21. [DOI] [PubMed] [Google Scholar]
- Mazieres, B. (2005). Drugs R D, 6, 337–344. [DOI] [PubMed] [Google Scholar]
- Mullangi, R., Yao, M. & Srinivas, N. R. (2003). Biomed. Chromatogr.17, 423–434. [DOI] [PubMed] [Google Scholar]
- Ng, I. S. & Tsai, S. W. (2005). Biotechnol. Bioeng.91, 106–113. [DOI] [PubMed] [Google Scholar]
- Osterlund, T. (2001). Eur. J. Biochem.268, 1899–1907. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Sohn, H. Y. & Krotz, F. (2006). Curr. Drug Targets, 7, 1275–1284. [DOI] [PubMed] [Google Scholar]
- Streit, W. R. & Schmitz, R. A. (2004). Curr. Opin. Microbiol.7, 492–498. [DOI] [PubMed] [Google Scholar]
- Turini, M. E. & DuBois, R. N. (2002). Annu. Rev. Med.53, 35–57. [DOI] [PubMed] [Google Scholar]
- Wei, Y., Contreras, J. A., Sheffield, P., Osterlund, T., Derewenda, U., Kneusel, R. E., Matern, U., Holm, C. & Derewenda, Z. S. (1999). Nature Struct. Biol.6, 340–345. [DOI] [PubMed] [Google Scholar]
- Yeaman, S. J. (2004). Biochem. J.379, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon, S., Kim, S., Ryu, Y. & Kim, T. D. (2007). Int. J. Biol. Macromol.41, 1–7. [DOI] [PubMed] [Google Scholar]
- Zhu, S., Wu, Y. & Yu, Z. (2005). J. Biotechnol.116, 397–401. [DOI] [PubMed] [Google Scholar]