Native ACTIBIND was successfully crystallized and it was shown that the interaction between ACTIBIND and actin is in a molar ratio of 1:2, with a binding constant of 16.17 × 104 M −1.
Keywords: RNase T2, ACTIBIND, anticarcinogens, antiangiogens
Abstract
ACTIBIND is a T2 RNase extracellular glycoprotein produced by the mould Aspergillus niger B1 (CMI CC 324626) that possesses anticarcinogenic and antiangiogenic activities. ACTIBIND was found to be an actin-binding protein that interacts with rabbit muscle actin in a 1:2 molar ratio (ACTIBIND:actin) with a binding constant of 16.17 × 104 M −1. Autoclave-treated ACTIBIND (EI-ACTIBIND) lost its RNase activity, but its actin-binding ability was conserved. ACTIBIND crystals were grown using 20% PEG 3350, 0.2 M ammonium dihydrogen phosphate solution at room temperature (293 K). One to four single crystals appeared in each droplet within a few days and grew to approximate dimensions of 0.5 × 0.5 × 0.5 mm after about two weeks. Diffraction studies of these crystals at low temperature (100 K) indicated that they belong to the P3121 space group, with unit-cell parameters a = 78, b = 78, c = 104 Å.
1. Introduction
Ribonucleases (RNases) are ubiquitous and efficient enzymes that hydrolyze RNA to 3′-mononucleotides via 2′,3′-cyclic nucleotides. RNases have been classified into three major families, RNase A, RNase T1 and RNase T2, based on their molecular weight (11–14, 12 and 20–40 kDa, respectively) and specificity (towards pyrimidine bases, guanine bases or nonspecific bases, respectively) (Deshpande & Shankar, 2002 ▶).
In addition to their ability to degrade RNA, certain RNases display a variety of biological activities. From the RNase A family, human angiogenin and eosinophil cationic protein (ECP) exhibit angiogenic and cytostatic activities, respectively (Maeda et al., 2002 ▶; Schein, 1997 ▶; Strydom, 1998 ▶). Also from the RNase A family, bovine seminal RNase has antispermatogenic, transplanted bone-marrow stimulating, immunosuppressive and antitumour activities (Antignani et al., 2001 ▶; Matousek, 2001 ▶). Onconase isolated from northern leopard frog (Rana pipiens) oocytes and early embryos has been shown to possess antitumour properties and is currently being investigated in phase III trials for use in cancer therapy (Lee et al., 2000 ▶; Lin et al., 1994 ▶).
Members of the T2 RNase family are widely distributed in living organisms, ranging from viruses to mammals (Irie & Ohgi, 2001 ▶). Some T2 RNases are capable of digesting extracellular polyribonucleotides, thereby accelerating phosphate uptake, and others protect against possible pathogens (Irie & Ohgi, 2001 ▶; Nurnberger et al., 1990 ▶). In some plants, specific T2 RNases encoded by the S-locus are responsible for rejecting self-pollen, thus preventing self-fertilization (Clarke & Newbigin, 1993 ▶; Silva & Goring, 1997 ▶). In the human genome, deletion of a region of chromosome 6 (6q27) is associated with several malignancies, e.g. ovarian (Cooke et al., 1996 ▶; Saito et al., 1992 ▶), breast (Theile et al., 1996 ▶; Tibiletti et al., 2000 ▶) and colon/rectal cancer (Honchel et al., 1996 ▶). This region contains the tumour suppressor gene RNASET2, which encodes the putative RNASET2 protein that shares high amino-acid homology with the RNase T2 family (Acquati et al., 2001 ▶; Trubia et al., 1997 ▶).
ACTIBIND is an RNase T2, an extracellular glycoprotein produced by the mould Aspergillus niger B1 (CMI CC 324626). ACTIBIND was purified by two-step anion-exchange chromatography (Roiz et al., 2000 ▶). Our previous studies have shown that ACTIBIND contains 32 and 36 kDa isoforms, both of which share a common 29 kDa protein moiety (Roiz et al., 1995 ▶, 2000 ▶; Roiz & Shoseyov, 1995 ▶). Owing to its ability to bind actin and to interfere with the cytoskeletal network structure, ACTIBIND inhibits cell extension and cell migration in cancer cells as well as in endothelial cells. In mice ACTIBIND has been shown to inhibit HT-29 xenograft tumour development and in rats it has been shown to exert preventive and therapeutic effects on dimethylhydrazine-induced colonic tumours (Roiz et al., 2006 ▶). Similar effects were obtained using enzymatically inactive ACTIBIND (EI-ACTIBIND), indicating that these antitumorigenic and antiangiogenic activities are not related to its RNase activity (Roiz et al., 2006 ▶).
The crystal structure of ACTIBIND has not yet been determined; however, the crystal structures of four members of the RNase T2 family are known: RNase Rh from the mould Rhizopus niveus (Kurihara et al., 1992 ▶, 1996 ▶), RNase LE from cultured tomato cells (Tanaka et al., 2000 ▶), RNase MC1 from bitter gourd seeds (Nakagawa et al., 1999 ▶) and S-RNase from pear (Matsuura et al., 2001 ▶). Comparison of the crystal structures of T2 RNases revealed that they belong to the α+β-type class of proteins, with six α-helices and seven β-strands. The overall structures of the four RNases are similar except for some differences in the exposed loop regions. Their core includes the conserved active site that is responsible for degrading RNA.
Here, we present quantified binding of actin to ACTIBIND as well as the preliminary crystallographic characterization of ACTIBIND crystals. Efforts to cocrystallize ACTIBIND with actin in order to identify the domain(s) responsible for its anticarcinogenic and antiangiogenic activities are in progress.
2. Experimental
All materials and other chemicals such as buffers and reagents for this work were obtained from Sigma–Aldrich Company and were of extra pure grade.
2.1. Actin-binding assay
The binding of ACTIBIND to actin was investigated by cross-linking followed by gel electrophoresis. The assay was carried out as described previously (Hu et al., 1993 ▶; Prochniewicz & Yanagida, 1991 ▶; Van Dijk et al., 2000 ▶). Globular actin (G-actin; 10 µg, Sigma–Aldrich Co., St Louis, MO, USA) was mixed with ACTIBIND (20 µg) in 15 µl buffer containing 2 mM Tris–HCl pH 8.0, 0.2 mM CaCl2 and 0.2 mM ATP. EI-ACTIBIND was obtained after autoclaving (393 K, 120 kPa) for 30 min, after which its RNase activity was undetectable. BSA was used as a control. Each protein mixture was incubated for 30 min at room temperature and 1-[3-(dimethylamino)propyl]-3-ethyl-carbodiimide methiodide (EDC-methiodide, Sigma) was then added to a final concentration of 10 mM. After another 30 min incubation at room temperature, the cross-linked complex was run on 12.5% Tricine SDS–PAGE. Proteins were visualized by Coomassie Blue staining.
2.2. Scatchard analysis of ACTIBIND
The ability of ACTIBIND to bind actin was quantified by a modified procedure of Simm et al. (1987 ▶). Rabbit muscle G-actin was polymerized to form filamentous actin (F-actin) in buffer containing 10 mM Tris pH 8, 0.1 mM ATP, 0.2 mM CaCl2, 0.1 M KCl, 2 mM MgCl2 for 30 min at room temperature. 50 µl samples containing between 33 and 1 µM ACTIBIND were incubated overnight (277 K) in the presence or absence of 30 µM F-actin. As a control, the same concentrations of ACTIBIND were incubated with buffer alone. The samples were centrifuged (40 min, 15 000g) and the RNase activity of the supernatant was determined as described below. The concentration of bound ACTIBIND (Rb) was calculated as the difference between the total ACTIBIND concentration and the concentration of free ACTIBIND (Rf).
RNase activity was determined as described previously (Roiz et al., 2000 ▶). 10 µl samples were added to 490 µl ice-cold buffer containing 4 mg ml−1 yeast RNA (Sigma, St Louis, MO, USA). Half of the mixture was immediately transferred to another tube containing 50 µl 0.75%(w/v) uranyl sulfate in 25%(w/v) perchloric acid (stop mixture) for use as a blank. The remainder was incubated for 10 min and 50 µl stop mixture was then added. Following centrifugation at 15 000g for 5 min, the supernatant was diluted 20-fold with distilled water and the absorbance was determined at 260 nm. One unit of RNase activity was determined as the amount of enzyme releasing soluble nucleotides at a rate of one A 260nm per minute.
2.3. Crystallization
Crystallization experiments were set up using the hanging-drop vapour-diffusion method with siliconized cover slips and Linbro 24-well tissue-culture plates. In these experiments, droplets ranging in size from 5 to 10 µl prepared by mixing equal volumes of protein solution and reservoir solution were equilibrated against 1.0 ml reservoir solution at room temperature (293 K). The protein solution was prepared by dissolving 17 mg lyophilized and purified ACTIBIND in 0.5 ml distilled water (Roiz et al., 2000 ▶).
2.4. Data collection
A native data set was collected at the Wolfson Center for Structural Biology at the Hebrew University of Jerusalem. Diffraction data from ACTIBIND crystals were collected at low temperature (100 K) using an R-AXIS IV electronic area detector and a Rigaku RU-200 HB generator. Determination of unit-cell parameters, crystal orientation and integration of reflection intensities was performed using HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results
3.1. SDS–PAGE analysis of ACTIBIND
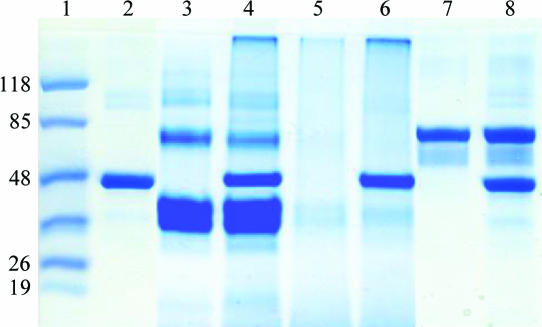
Coomassie Blue staining of the SDS–PAGE revealed that when ACTIBIND was mixed with actin a band at a high molecular weight that hardly penetrated the running gel appeared, suggesting that a high-molecular-weight complex was formed between ACTIBIND and actin (Fig. 1 ▶, lane 4). Similarly to the native ACTIBIND, the same high-molecular-weight complex was also formed when EI-ACTIBIND (Fig. 1 ▶, lane 5) was mixed with actin and stained with Coomassie Blue (Fig. 1 ▶, lane 6). No complex was observed when actin was mixed with BSA (Fig. 1 ▶, lane 8). In lane 2, the faint high-molecular-weight band is a result of self-aggregation of actin, as reported previously (Bonafe & Chaussepied, 1995 ▶; Van Dijk et al., 1999 ▶). In lane 3, in addition to the major ACTIBIND band, faint high-molecular-weight bands appeared on the SDS–PAGE. These bands are a result of self-aggregation of ACTIBIND, as confirmed by Western blot analysis with anti-ACTIBIND (data not shown).
Figure 1.
SDS–PAGE analysis. 10 µg actin was mixed with 20 µg ACTIBIND, EI-ACTIBIND and BSA as control. Following incubation for 30 min at room temperature, the mixture was cross-linked for 30 min with EDC-methiodide and analyzed by 12.5% Tricine SDS–PAGE. Lane 1, molecular-weight markers (kDa); lane 2, actin; lane 3, ACTIBIND; lane 4, ACTIBIND and actin; lane 5, EI-ACTIBIND; lane 6, EI-ACTIBIND and actin; lane 7, BSA; lane 8, BSA and actin.
3.2. Scatchard analysis of ACTIBIND
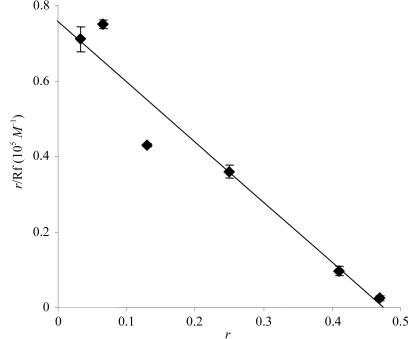
Preliminary in vitro experiments indicated that addition of ACTIBIND (or EI-ACTIBIND) to F-actin resulted in precipitation of RNase–actin complexes (not shown), indicating an interaction between ACTIBIND and actin. Scatchard plot analysis was used to quantify this interaction (Fig. 2 ▶). A regression line intercepting the ordinate at 7.6 × 104 M −1 and at the abscissa at 0.47 (R = 0.95) indicated that the ACTIBIND:actin molar ratio was 1:2, with a binding constant of 16.17 × 104 M −1 (and K d = 6 µM). A similar binding constant of 9.2 × 104 M −1 was measured for bovine seminal RNase bound to actin (Simm et al., 1987 ▶).
Figure 2.
Scatchard plot of ACTIBIND–actin binding. The actin concentration (A) was 30 µM. Rf, free ACTIBIND concentration (µM). The bound ACTIBIND concentration, Rb (µM), was calculated as the difference between the total concentration of ACTIBIND and Rf; r is defined as r = Rb/A.
3.3. Preliminary crystallographic characterization
Crystallization experiments were first carried out by the fast screening method using Hampton Research Crystal Screens I and II. Initial screening resulted in small crystals that were not suitable for data collection in some of the solutions. However, experiments using the hanging-drop method and Wizard 1 and 2 screens (Emerald Biostructures) resulted in crystals with dimensions of 0.4 × 0.4 × 0.3 mm (condition No. 28 of Wizard 1). These crystals were obtained using 0.1 M HEPES pH 7.5, 20% PEG 3K, 0.2 M NaCl as the reservoir solution. Preliminary X-ray characterization indicated that they diffracted to about 8 Å. Efforts to improve the quality of the crystals using a detergent screen (such as the Detergent Screen from Hampton Research) as well as a wide variety of additives (such as the 96-condition Hampton Research Additive Screen) were unsuccessful.
Crystals that were suitable for data collection were finally obtained using the PEG/Ion Screen from Hampton Research (condition No. 43; 20% PEG 3350, 0.2 M ammonium dihydrogen phosphate). Between one and four crystals appeared in the droplet after 3 d and grew to maximal dimensions of 0.5 × 0.5 × 0.5 mm within two weeks (Fig. 3 ▶). The crystallization conditions, which are very similar to the Wizard 1 conditions but do not include any buffer solution, resulted in crystals that diffracted to about 2.0 Å. The ACTIBIND crystals belong to space group P3121, with unit-cell parameters a = 78, b = 78, c = 104 Å. The Matthews coefficient (V M) for these crystals is 2.52 Å3 Da−1, indicating the presence of a monomer in the asymmetric unit. A summary of data collection and statistics is given in Table 1 ▶. This data set will be used to determine the crystal structure of ACTIBIND using the molecular-replacement method with the crystal structure of RNase Rh (PDB code 1bol), another member of the RNase T2 family, as a starting model.
Figure 3.
A typical ACTIBIND crystal of dimensions 0.4 × 0.4 × 0.4 mm.
Table 1. Summary of data collection and statistics for ACTIBIND crystals.
Values in parentheses are for the highest resolution shell.
| Unit-cell parameters (Å) | a = b = 78, c = 104 |
| Space group | P3121 |
| Resolution (Å) | 50.0–2.07 (2.14–2.07) |
| No. of observations | 40948 |
| Unique reflections | 23021 |
| Rmerge† (%) | 7.6 (38.4) |
| Completeness (%) | 98.7 (96) |
| Average redundancy | 23.2 |
| Average I/σ(I) | 17 (7.5) |
R
merge = 
 . Taken from Weiss (2001 ▶).
. Taken from Weiss (2001 ▶).
To summarize, we have succeeded in crystallizing native ACTIBIND. We showed that the interaction between ACTIBIND and actin is in a molar ratio of 1:2, with a binding constant of 16.17 × 104 M −1.
References
- Acquati, F., Nucci, C., Bianchi, M. G., Gorletta, T. & Taramelli, R. (2001). Methods Mol. Biol.160, 87–101. [DOI] [PubMed] [Google Scholar]
- Antignani, A., Naddeo, M., Cubellis, M. V., Russo, A. & D’Alessio, G. (2001). Biochemistry, 40, 3492–3496. [DOI] [PubMed] [Google Scholar]
- Bonafe, N. & Chaussepied, P. (1995). Biophys. J.68, 35S–43S. [PMC free article] [PubMed]
- Clarke, A. E. & Newbigin, E. (1993). Annu. Rev. Genet.27, 257–279. [DOI] [PubMed] [Google Scholar]
- Cooke, I. E., Shelling, A. N., Le Meuth, V. G., Charnock, M. L. & Ganesan, T. S. (1996). Genes Chromosomes Cancer, 15, 223–233. [DOI] [PubMed] [Google Scholar]
- Deshpande, R. A. & Shankar, V. (2002). Crit. Rev. Microbiol.28, 79–122. [DOI] [PubMed] [Google Scholar]
- Honchel, R., McDonnell, S., Schaid, D. J. & Thibodeau, S. N. (1996). Cancer Res.56, 145–149. [PubMed] [Google Scholar]
- Hu, G. F., Strydom, D. J., Fett, J. W., Riordan, J. F. & Vallee, B. L. (1993). Proc. Natl Acad. Sci. USA, 90, 1217–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie, M. & Ohgi, K. (2001). Methods Enzymol.341, 42–55. [DOI] [PubMed] [Google Scholar]
- Kurihara, H., Mitsui, Y., Ohgi, K., Irie, M., Mizuno, H. & Nakamura, K. T. (1992). FEBS Lett.306, 189–192. [DOI] [PubMed] [Google Scholar]
- Kurihara, H., Nonaka, T., Mitsui, Y., Ohgi, K., Irie, M. & Nakamura, K. T. (1996). J. Mol. Biol.255, 310–320. [DOI] [PubMed] [Google Scholar]
- Lee, I., Lee, Y. H., Mikulski, S. M., Lee, J., Covone, K. & Shogen, K. (2000). J. Surg. Oncol.73, 164–171. [DOI] [PubMed] [Google Scholar]
- Lin, J. J., Newton, D. L., Mikulski, S. M., Kung, H. F., Youle, R. J. & Rybak, S. M. (1994). Biochem. Biophys. Res. Commun.204, 156–162. [DOI] [PubMed] [Google Scholar]
- Maeda, T., Kitazoe, M., Tada, H., de Llorens, R., Salomon, D. S., Ueda, M., Yamada, H. & Seno, M. (2002). Eur. J. Biochem.269, 307–316. [DOI] [PubMed] [Google Scholar]
- Matousek, J. (2001). Comput. Biochem. Physiol. C Toxicol. Pharmacol.129, 175–191. [DOI] [PubMed]
- Matsuura, T., Sakai, H., Unno, M., Ida, K., Sato, M., Sakiyama, F. & Norioka, S. (2001). J. Biol. Chem.276, 45261–45269. [DOI] [PubMed] [Google Scholar]
- Nakagawa, A., Tanaka, I., Sakai, R., Nakashima, T., Funatsu, G. & Kimura, M. (1999). Biochim. Biophys. Acta, 1433, 253–260. [DOI] [PubMed] [Google Scholar]
- Nurnberger, T., Abel, S., Jost, W. & Glund, K. (1990). Plant Physiol.92, 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Prochniewicz, E. & Yanagida, T. (1991). Adv. Biophys.27, 207–211. [DOI] [PubMed] [Google Scholar]
- Roiz, L., Goren, R. & Shoseyov, O. (1995). Physiol. Plant.94, 585–590.
- Roiz, L., Ozeri, U., Goren, R. & Shoseyov, O. (2000). J. Am. Soc. Hortic. Sci.125, 9–14.
- Roiz, L. & Shoseyov, O. (1995). Int. J. Plant Sci.156, 37–41.
- Roiz, L., Smirnoff, P., Bar-Eli, M., Schwartz, B. & Shoseyov, O. (2006). Cancer, 106, 2295–2308. [DOI] [PubMed] [Google Scholar]
- Saito, S., Saito, H., Koi, S., Sagae, S., Kudo, R., Saito, J., Noda, K. & Nakamura, Y. (1992). Cancer Res.52, 5815–5817. [PubMed] [Google Scholar]
- Schein, C. H. (1997). Nature Biotechnol.15, 529–536. [DOI] [PubMed]
- Silva, N. F. & Goring, D. R. (1997). Cell. Mol. Life Sci.58, 1988–2007. [DOI] [PMC free article] [PubMed]
- Simm, F., Kreitsch, C. & Isenberg, G. (1987). Eur. J. Biochem.166, 49–54. [DOI] [PubMed] [Google Scholar]
- Strydom, D. J. (1998). Cell. Mol. Life Sci.54, 811–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, N., Arai, J., Inokuchi, N., Koyama, T., Ohgi, K., Irie, M. & Nakamura, K. T. (2000). J. Mol. Biol.298, 859–873. [DOI] [PubMed] [Google Scholar]
- Theile, M., Seitz, S., Arnold, W., Jandrig, B., Frege, R., Schlag, P. M., Haensch, W., Guski, H., Winzer, K. J., Barrett, J. C. & Scherneck, S. (1996). Oncogene, 13, 677–685. [PubMed] [Google Scholar]
- Tibiletti, M. G., Sessa, F., Bernasconi, B., Cerutti, R., Broggi, B., Furlan, D., Acquati, F., Bianchi, M., Russo, A., Capella, C. & Taramelli, R. (2000). Clin. Cancer Res.6, 1422–1431. [PubMed] [Google Scholar]
- Trubia, M., Sessa, L. & Taramelli, R. (1997). Genomics, 42, 342–344. [DOI] [PubMed] [Google Scholar]
- Van Dijk, J., Celine, F., Barman, T. & Chaussepied, P. (2000). Biophys. J.78, 3093–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk, J., Furch, M., Derancourt, J., Batra, R., Knetsch, M. L., Manstein, D. J. & Chaussepied, P. (1999). Eur. J. Biochem.260, 672–683. [DOI] [PubMed] [Google Scholar]
- Weiss, M. S. (2001). J. Appl. Cryst.34, 130–135. [Google Scholar]