Robo1 is a large transmembrane receptor expressed on the axon growth cone. Activation of Robo1 by Slit proteins results in axon repulsion from the midline. Here, the purification and crystallization conditions of first two Ig domains of Robo1 are reported.
Keywords: Roundabout, Slit, mammalian expression
Abstract
Activation of Roundabout 1 (Robo1) by Slit proteins results in axon repulsion from the midline. Robo1 is a large transmembrane receptor expressed on the axon growth cone and the minimal Robo1-binding region required for Slit activation has been mapped to the N-terminal Ig1–2 domains. The cDNA encoding the first two Ig domains of Robo1 has been cloned and the protein has been expressed in HEK293 EBNA-1 mammalian cells. Here, the purification and crystallization conditions of this Robo1 construct are reported. The crystals are orthorhombic, space group P21212, with unit-cell parameters a = 38.8, b = 69.4, c = 103.3 Å and one molecule in the asymmetric unit. X-ray diffraction data have been collected to 2.8 Å resolution on beamline ID29 at the ESRF.
1. Introduction
Bilaterally symmetric nervous systems are divided into left and right mirror images by the midline and communicate via the commissural subset of axons. Developing axons navigate through the embryo by processing a number of different signals in their immediate environment (Tessier-Lavigne & Goodman, 1996 ▶) and the midline provides such target signals for migrating commissural neurons (Garbe & Bashaw, 2004 ▶). Commissural neurons are initially attracted and allowed to cross the midline by activation of the Deleted in Colorectal Cancer (DCC) receptor upon binding the midline-secreted attractant netrin (Stein & Tessier-Lavigne, 2001 ▶). Upon midline crossing, the commissural axons must not recross and this repulsion is mediated by activation of the Roundabout (Robo) receptor upon binding of the midline-secreted repellent Slit (Brose et al., 1999 ▶). The Robo receptors are large multidomain transmembrane receptors expressed on the axonal growth cones (Kidd et al., 1998 ▶). Four Robo genes have been identified in mammals: Robo1, Robo2, Robo3 (or Rig-1) and Robo4 (or Magic Roundabout) (Huminiecki et al., 2002 ▶). All the Robo receptors, with the exception of Robo4, share a common domain architecture containing five immunoglobulin domains (Ig), three fibronectin III (FN3) domains, a transmembrane domain and an intracellular tail. The exception, Robo4, is specifically expressed in the vascular endothelium and contains only two Ig and two FN3 extracellular domains (Park et al., 2003 ▶).
Various combinations of Robo1, Robo2 and Robo3 expression on a given neuron in Drosophila are important for the lateral position these neurons take relative to the midline (Rajagopalan et al., 2000 ▶). While the exact details of this so-called ‘Robo-code’ are still under investigation, it is thought to be mediated by a Slit concentration gradient that spreads out laterally from the midline (Simpson et al., 2000 ▶). A similar, though perhaps more complex, expression pattern of Robo1 and Robo2 has also been observed in vertebrates (Long et al., 2004 ▶). Recent studies have also shown that Robo3 can act as a negative regulator of Slit–Robo signalling (Sabatier et al., 2004 ▶). These data suggest that Robo3 has an important role in sequestering Robo1 by an unknown mechanism, allowing the commissural neurons to cross the midline. This mechanism appears to be quite different to that played by commissureless (comm) in Drosophila, which down-regulates Robo at the intracellular trafficking level to achieve the same effect (Keleman et al., 2002 ▶). The most sequence-conserved region of the Robo receptors is located at the N-terminus and the Slit-binding site on Robo1 has been mapped to the first two Ig domains (Liu et al., 2004 ▶). In order to better understand the molecular basis of this important Slit2–Robo1 ligand–receptor interaction, we decided to undertake a structural study of a minimal Robo1 construct that can interact with Slit2. Here, we describe the successful cloning, purification and crystallization of a Robo1 construct containing the first two Ig domains from Robo1.
2. Materials and methods
2.1. Cloning and mutagenesis
A Robo Ig1–2 construct spanning residues 61–266 was produced by PCR amplification using the full-length cDNA coding for human Robo1 (NP_002932) as a template. Cloning experiments were performed using the following nucleotides (5′ to 3′): R1Ig1F, CAG CAC TAG GGA TCC CTT CGT CAG GAA GAT TTT CCA; R1Ig2R, TCG TCG ATC AGC GGC CGC CTT CAC AAA TGA TGG TCT CTC. The forward primer introduced a BamHI restriction site and the reverse primer introduced a NotI restriction site. The Robo1 PCR product was cloned into a modified pTT3 expression vector (Durocher et al., 2002 ▶) encoding a signal peptide followed by hexahistidine tag. The QuickChange kit (Stratagene) was used to remove a potential glycosylation site and a nonconservative Y238H mutation. One of the two potential asparagine glycosylation sites was mutated to aspartate using the following primers (5′ to 3′): R12N160D_F, CTT GGA GAG GCT GTG AGC CAC GAC GCA TCG CTG GAA GTA G; R12N160D_R, CTA CTT CCA GCG ATG CGT CGT GGC TCA CAG CCT CTC CAA G. The histidine was mutated back to a tyrosine using the following primers (5′ to 3′): R12H238Y_F, GTA AAA GTG ACG CTG GCA AAT ACG TTT GTG TTG GTA CCA ATA TG; R12H238Y_R, CAT ATT GGT ACC AAC ACA AAC GTA TTT GCC AGC GTC ACT TTT AC.
2.2. Protein expression
Human embryonic kidney cells (HEK293) stably expressing Epstein–Barr virus nuclear antigen 1 (EBNA-1) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM high glucose, Sigma) supplemented with l-glutamine, non-essential amino acids (Gibco) and 10% foetal calf serum (FCS, Sigma). Cells were normally grown in standard flasks (Nunclon) at 310 K in a humidified incubator with 5% CO2. The Robo1 Ig1–2 expression plasmid was purified using the Endotoxin-Free Plasmid Mega Kit (Qiagen). Transfections with this vector were performed using polyethyleneimine and were transiently expressed at low serum concentration as described in Durocher et al. (2002 ▶). Briefly, 24 h pre-transfection the cells were diluted to 0.3 × 106 ml−1 in DMEM medium (as above). DNA–PEI complexes (1 µg DNA and 2 µg PEI per millitre of suspension culture) were formed in Optimem medium (Invitrogen), immediately vortexed and then added to the culture medium after a 10 min incubation. Expression medium containing Robo1 Ig1–2 was harvested 120 h after transfection.
2.3. Purification
Cell-free expression medium containing secreted Robo1 Ig1–2 was loaded onto Ni–NTA agarose resin (Qiagen) and extensively washed. The His-tagged Robo1 Ig1–2 was eluted from the resin in 250 mM imidazole, 200 mM NaCl and 50 mM Tris–HCl pH 8.0. The elution fraction was further purified by size-exclusion chromatography (Superdex S200, Pharmacia) using elution buffer containing 200 mM NaCl and 25 mM Tris–HCl pH 8.0. Both gel-filtration and matrix-assisted laser desorption/ionization time-of-flight (MALDI–TOF) mass-spectrometric analysis of purified Robo1 Ig1–2 revealed a homogeneous protein sample. The purified protein was concentrated to 7 mg ml−1 in a spin concentrator (Millipore) and used for crystallization.
2.4. Crystallization and preliminary X-ray data
Initial crystallization screens were carried out using the Cartesian PixSys 4200 (Genomic Solutions) crystallization robot in the EMBL Grenoble high-throughput crystallization facility. The initial crystallization conditions were manually refined using Linbro plates (Hampton Research) and the conventional hanging-drop technique. Robo1 Ig1–2 crystallized at 291 K from 18–20% PEG 3350, 0.2 M potassium thiocyanate and 0.1 M MES pH 6.5. Crystals appeared after 4 d and grew as stacked plates. They could be broken apart into single plates that were suitable for diffraction experiments but were fragile and difficult to work with. Addition of KAuCl4 resulted in much larger and more robust crystals (see §3 for more details). Fine screening of these conditions resulted in an optimized crystallization condition containing 18–20% PEG 3350, 0.2 M potassium thiocyanate, 0.1 M MES pH 6.5 and 2.5 mM KAuCl4. These crystals were transferred to cryogenic conditions containing 22% PEG 3350, 0.2 M potassium thiocyanate, 0.1 M MES pH 6.5, 12.5% glycerol and 2.5 mM KAuCl4 before flash-freezing in liquid nitrogen. An X-ray data set was collected to 2.8 Å resolution on ID29 at the European Synchrotron Radiation Facility (ESRF). All crystals were mounted on SPINE standard pins (Hampton Research) and we exclusively used the EMBL/ESRF/BM14 robotic sample changer (SC3) for crystal screening (Cipriani et al., 2006 ▶). All data were integrated and scaled using the XDS suite (Kabsch, 1993 ▶); a summary of the data statistics is given in Table 1 ▶.
Table 1. Crystal data and data-collection statistics.
Values in parentheses are for the outermost shell of data.
| X-ray source | ID29 |
| Wavelength (Å) | 1.04 |
| Unit-cell parameters | |
| a (Å) | 38.8 |
| b (Å) | 69.4 |
| c (Å) | 103.2 |
| Space group | P21212 |
| No. of molecules in ASU | 1 |
| Matthews coefficient (Å3 Da−1) | 2.9 |
| Solvent content (%) | 57.7 |
| Resolution range (Å) | 30–2.8 (3.0–2.8) |
| Unique reflections | 7042 (1189) |
| Observed reflections [I/σ(I) > 1] | 26114 (3456) |
| Completeness (%) | 96.2 (88.9) |
| Rmerge† | 5.3 (36.6) |
| Mean I/σ(I) | 17.2 (4.0) |
R
merge = 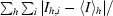
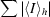 calculated for the whole data set.
calculated for the whole data set.
3. Results
The domain boundary predicted to span the first two Ig domains of Robo1 was designed using the minimal domain boundaries predicted by SMART (Letunic et al., 2006 ▶). Small-scale expression tests confirmed that the Robo1 Ig1–2 construct was present in the medium of the transiently expressing mammalian cells (Fig. 1 ▶ a). A non-conservative mutation from a histidine to a tyrosine at position 238 was subsequently observed on closer examination of the sequenced plasmid. This change was also observed in a duplicate cloning plasmid. Although the Robo1 Ig1–2 is well expressed, we nonetheless decided to correct this nonconservative mutation, as a tyrosine is structurally conserved in related Ig domains and may be important for the stability of the second Robo1 Ig domain. The resulting plasmid was resequenced to ensure that the cDNA encoded for the original Robo1 sequence in the database (NP_002932).
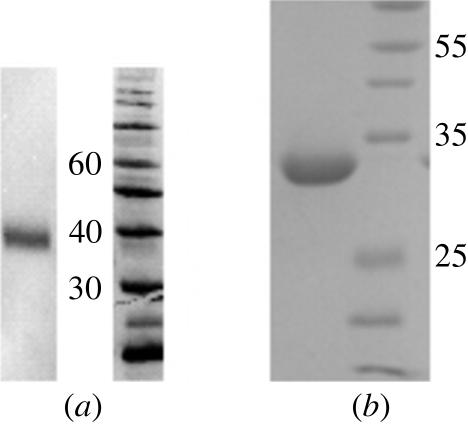
Figure 1.
Western blot and SDS–PAGE (15%) analysis of Robo1. (a) Western blot analysis of media conditioned for 3 d revealed that Robo1 Ig1–2 is well expressed. (b) SDS–PAGE (15%) analysis of purified Robo1 Ig1–2.
The initial tests also revealed that the Robo1 Ig1–2 construct ran at a higher molecular weight than expected and we presumed that this was a consequence of glycosylation (Fig. 1 ▶ a). The most likely glycosylation site was predicted to be Asn160 using the NetNGlyc server (Blom et al., 2004 ▶). This asparagine was mutated to an aspartate in order to improve the chances of crystallization, but the purified protein still ran higher than expected (Fig. 1 ▶ b). Mass-spectroscopic analysis of this mutated Robo1 Ig1–2 protein gives an experimental protein molecular weight of 24.25 kDa, which is close to the predicted molecular weight of 24.3 kDa. It is therefore unlikely that the Robo1 Ig1–2 domains are glycosylated in the native protein. Interestingly, the introduction of both of the mutations significantly boosted the expression level and we presently obtain 20 mg protein from 1 l medium.
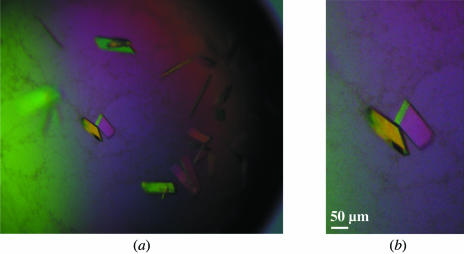
Our initial attempts at solving the structure by molecular replacement were unsuccessful and we decided to attempt heavy-atom derivative soaks for experimental phasing. These experiments were unsuccessful because the crystals obtained were very difficult to work with. We therefore decided to attempt cocrystallization experiments using various heavy-atom solutions. Addition of KAuCl4 greatly improved the crystallization and the resulting single plate-shaped crystals grew to approximate dimensions of 0.03 × 0.5 × 0.1 mm (Fig. 2 ▶). An X-ray data set was collected to 2.8 Å from one of these crystals and an analysis of the systematic absences suggested the space group to be P21212. This corresponds to a Matthews coefficient of 2.9 Å3 Da−1 and a solvent content of 58% with one molecule per asymmetric unit. We are currently trying to calculate experimental phases using the weak gold signal observed in the data.
Figure 2.
Plate-shaped crystals of Robo1 Ig1–2 obtained in the presence of KAuCl4.
Acknowledgments
We gratefully acknowledge the use of the EMBL Grenoble high-throughput crystallization facility and thank the EMBL Grenoble/ESRF Joint Structural Biology Group for access and support at the ESRF beamline ID29. We thank Bernard Dublet for mass-spectrometric analysis and N. Kaldenhoven for his assistance in the early part of this work. We would also like to thank S. Guthrie and A. Naeem (Kings College, London) for the kind gift of full-length Robo1 cDNA. Partial funding for this project was provided by the European Commission Framework 6 Integrated Project ‘SPINE2-Complexes’.
References
- Blom, N., Sicheritz-Ponten, T., Gupta, R., Gammeltoft, S. & Brunak, S. (2004). Proteomics, 4, 1633–1649. [DOI] [PubMed] [Google Scholar]
- Brose, K., Bland, K. S., Wang, K. H., Arnott, D., Henzel, W., Goodman, C. S., Tessier-Lavigne, M. & Kidd, T. (1999). Cell, 96, 795–806. [DOI] [PubMed] [Google Scholar]
- Cipriani, F. et al. (2006). Acta Cryst. D62, 1251–1259. [DOI] [PubMed] [Google Scholar]
- Durocher, Y., Perret, S. & Kamen, A. (2002). Nucleic Acids Res.30, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbe, D. S. & Bashaw, G. J. (2004). Crit. Rev. Biochem. Mol. Biol.39, 319–341. [DOI] [PubMed] [Google Scholar]
- Huminiecki, L., Gorn, M., Suchting, S., Poulsom, R. & Bicknell, R. (2002). Genomics, 79, 547–552. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800. [Google Scholar]
- Keleman, K., Rajagopalan, S., Cleppien, D., Teis, D., Paiha, K., Huber, L. A., Technau, G. M. & Dickson, B. J. (2002). Cell, 110, 415–427. [DOI] [PubMed] [Google Scholar]
- Kidd, T., Brose, K., Mitchell, K. J., Fetter, R. D., Tessier-Lavigne, M., Goodman, C. S. & Tear, G. (1998). Cell, 92, 205–215. [DOI] [PubMed] [Google Scholar]
- Letunic, I., Copley, R. R., Pils, B., Pinkert, S., Schultz, J. & Bork, P. (2006). Nucleic Acids Res.34, D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Z., Patel, K., Schmidt, H., Andrews, W., Pini, A. & Sundaresan, V. (2004). Mol. Cell. Neurosci.26, 232–240. [DOI] [PubMed] [Google Scholar]
- Long, H., Sabatier, C., Ma, L., Plump, A., Yuan, W., Ornitz, D. M., Tamada, A., Murakami, F., Goodman, C. S. & Tessier-Lavigne, M. (2004). Neuron, 42, 213–223. [DOI] [PubMed] [Google Scholar]
- Park, K. W., Morrison, C. M., Sorensen, L. K., Jones, C. A., Rao, Y., Chien, C. B., Wu, J. Y., Urness, L. D. & Li, D. Y. (2003). Dev. Biol.261, 251–267. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, S., Vivancos, V., Nicolas, E. & Dickson, B. J. (2000). Cell, 103, 1033–1045. [DOI] [PubMed] [Google Scholar]
- Sabatier, C., Plump, A. S., Le, M., Brose, K., Tamada, A., Murakami, F., Lee, E. Y. & Tessier-Lavigne, M. (2004). Cell, 117, 157–169. [DOI] [PubMed] [Google Scholar]
- Simpson, J. H., Kidd, T., Bland, K. S. & Goodman, C. S. (2000). Neuron, 28, 753–766. [DOI] [PubMed] [Google Scholar]
- Stein, E. & Tessier-Lavigne, M. (2001). Science, 291, 1928–1938. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne, M. & Goodman, C. S. (1996). Science, 274, 1123–1133. [DOI] [PubMed] [Google Scholar]