Diffraction-quality crystals of the apo (1.7 Å) and zinc-bound forms (2.2 Å) of the water-soluble C-terminal domain of the putative zinc transporter CzrB from T. thermophilus have been grown using recombinant production of the protein in E. coli and a combination of vapour-diffusion, batch and seeding crystallogenesis techniques.
Keywords: zinc transporters, CzrB, cation-diffusion facilitator, membrane proteins, zinc binding
Abstract
CzrB is a putative zinc transporter from Thermus thermophilus. The protein is proposed to consist of a hexahelical transmembrane domain with a cytosolic extramembranal C-terminus. The latter 92-residue fragment may be expressed free and may function independently of the full-length integral membrane protein. A 6×His-tagged form of the water-soluble fragment has been overexpressed in Escherichia coli and diffraction-quality crystals of the tagged and tag-free variants have been grown. Preliminary X-ray analyses of tag-free fragment crystals with (2.2 Å resolution) and without zinc ions (1.7 Å resolution) reveal that the former has at least two zinc ions bound per monomer.
1. Introduction
Zinc is considered to be an essential micronutrient despite the fact that it is present in bacterial cells at close to millimolar concentrations. However, whilst the total concentration of this redox-insensitive ion is quite high, essentially none of the million or so atoms in a typical Escherichia coli cell exist free in solution (Outten & O’Halloran, 2001 ▶). This vanishingly low concentration of free zinc represents an interesting challenge for the assorted intracellular and membranal systems concerned with zinc homeostasis. These include proteins and other biomolecules that participate in the sequestration and the import and export of zinc to ensure that it properly serves its vital catalytic, regulatory and structural roles (Hantke, 2005 ▶; Blencowe & Morby, 2003 ▶; Choudhury & Srivastava, 2001 ▶).
One group of proteins that are involved in zinc export belong to the cation-diffusion facilitator (CDF) family (Paulsen & Saier, 1997 ▶). The CDFs are ubiquitous in nature, with representatives in the three taxonomic domains archaea, bacteria and eukaryota. Some examples include FieF in E. coli (Grass et al., 2005 ▶), CzcD in Alcaligenes eutrophus CH34 (since renamed Wautersia metallidurans CH34; Nies et al., 1989 ▶; Munkelt et al., 2004 ▶), Zrc1 in Saccharomyces cerevisiae (Conklin et al., 1992 ▶), AtZat in Arabidopsis thaliana (Bloss et al., 2002 ▶) and ZnTX in mammals (Haney et al., 2005 ▶). In Gram-negative bacteria, CDF proteins move zinc and other metals such as cobalt and manganese from the cytoplasm into the periplasmic space by means of a proton/cation antiporter mechanism (Haney et al., 2005 ▶). Despite the fact that no three-dimensional structure of a representative from this family is yet available, sequence alignment and biochemical and computational analyses reveal common features. These include six membrane-spanning α-helices per monomer, histidine-rich clusters in the N- and C-termini and a tendency to dimerize (Paulsen & Saier, 1997 ▶; Haney et al., 2005 ▶; Spada et al., 2002 ▶).
CzrB (cadmium-zinc resistance protein B) was the first CDF family member to be identified in Thermus thermophilus (Spada et al., 2002 ▶; Kolaj et al., 2006 ▶). As with other CDF transporters, this 291-residue protein has a putative transmembrane hexahelical domain. However, it lacks the hallmark histidine-rich N- and C-termini. Interestingly, it has a 92-amino-acid C-terminus that may form an independent extramembranal cytosolic domain. In support of this, the czrB gene has been shown to contain an ATG consensus sequence at Met200, with a putative ribosome-binding site nearby. This C-terminal domain is proposed to play a role in metal-ion sequestration and transport as a soluble cytoplasmic fragment (sf-CzrB; Spada et al., 2002 ▶). The czrB gene conferred a slightly enhanced tolerance for zinc on E. coli. Surprisingly, both the czrB and sf-czrB genes were found to support the growth of E. coli cultures to higher densities during bacteriophage and recombinant protein production (Spada et al., 2002 ▶). This suggested an ill-defined chaperone-like function for the corresponding gene products.
Here, we describe the production, purification, crystallization and preliminary X-ray analysis of the soluble C-terminal fragment of CzrB in its apo and zinc-loaded forms.
2. Materials and methods
2.1. Cloning and expression
A DNA fragment that encodes the C-terminal 92-residue soluble fragment of CzrB was cloned as a NotI–PstI insert into the pIVEX2.4d plasmid (Roche Diagnostics, Basel, Switzerland). The soluble fragment incorporated a proteolytically cleavable 6×His tag at its N-terminus. The expressed protein consisted of 111 residues with the sequence MSGSHHHHHHSSGIEGR*GRMDEGLPPEEVERIRAFLQERIRGRALEVHDLKTRRAGPRSFLEFHLVVRGDTPVEEAHRLCDELERALAQAFPGLQATIHVEPEGERKRTNP. It is referred to hereafter as 6×His-sf-CzrB. Factor Xa protease cleaves at the C-terminal side of the sequence IEGR (shown in bold in the 6×His-sf-CzrB sequence) and was used to produce the tag-free water-soluble fragment sf-CzrB.
In preparation for overexpression, the recombinant plasmid was transformed into E. coli BL21 (DE3) cells (Invitrogen, Carlsbad, USA) by electroporation. The induction temperature (297–310 K), length of induction (3–6 h) and inducer concentration (0.1–1 mM IPTG) were varied in the initial analyses in order to optimize the yield of soluble protein in the E. coli cytoplasm. Detection of the protein was carried out by SDS–PAGE and by immunoblotting the N-terminal 6×His tag according to standard procedures (Hu et al., 2005 ▶). Subsequently, E. coli BL21 (DE3) cells containing the pIVEX2.4d-czrB plasmid were grown overnight at 310 K and with agitation at 240 rev min−1 (C24 Incubator/Shaker, New Brunswick Scientific, Edison, USA) in 40 ml Luria–Bertani (LB) broth containing 100 µg ml−1 ampicillin (American Bioanalytical, Natick, USA). The overnight culture was used to inoculate 2 l of the same medium, followed by growth at 310 K and 240 rev min−1 agitation until an OD600 of 0.5 was reached (Agilent 8453 UV–Vis spectrometer, Agilent Technologies, Santa Clara, USA). Expression of the CzrB cytoplasmic domain was induced by addition of isopropyl β-d-thiogalactopyranoside (IPTG; American Bioanalytical) to a final concentration of 0.5 mM and protein production was allowed to proceed at 298 K and a frequency of agitation of 160 rev min−1 for 4 h. Cells were harvested by centrifugation at 8000g (Sorvall RC6, Kendro Laboratory Equipment, Asheville, USA) for 10 min at 277 K. The moist cell pellet (8.5 g) was stored at 193 K.
2.2. Protein purification
The frozen cell pellet was thawed on ice for 15 min and resuspended in 5 ml cold (277 K) buffer (50 mM Tris–HCl pH 8.0, 300 mM NaCl, 10 mM imidazole) per gram of wet cell pellet. 5 mg lysozyme (American Bioanalytical) per gram of wet cell pellet and Triton X-100 to 0.1%(v/v) were added to the suspension and the mixture was stirred for 1 h on ice. Cells were disrupted by sonication (Sonicator 3000, Misonix Inc., Farmingdale, USA) on ice with three 30 W pulses of 30 s duration at 2 min intervals. DNase I (750 U; Lot 1339311, Invitrogen, Carlsbad, USA) and RNase A (0.5 mg; Lot 124101816, Qiagen, Valencia, USA) were added and the mixture was stirred for another hour on ice. Cell debris was removed by centrifugation (Sorvall RC6) for 30 min at 47 800g and 277 K. The supernatant (40 ml) was applied onto a 1 ml bed-volume homemade gravity Ni–NTA column (1 cm diameter, Ni–NTA agarose; Lot 11230934, Qiagen) which was pre-equilibrated with 25 ml 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 20 mM imidazole. The protein was eluted by sequentially passing over the column five 15 ml volumes of buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 100–500 mM imidazole) in steps of 100 mM imidazole. The fractions eluting in 100, 200 and 300 mM imidazole all contained the protein and were combined. To remove the imidazole, the protein-containing sample was dialyzed (dialysis tubing, 3500 MWCO; Lot 3223607, Spectra/Por, Rancho Dominguez, USA) thrice for 4 h at 277 K against 500 ml 20 mM Tris–HCl pH 8.0. Following dialysis, the protein was concentrated by ultrafiltration (Amicon Ultra-4, 5000 MWCO) to 10–30 mg ml−1 and used in crystallization trials.
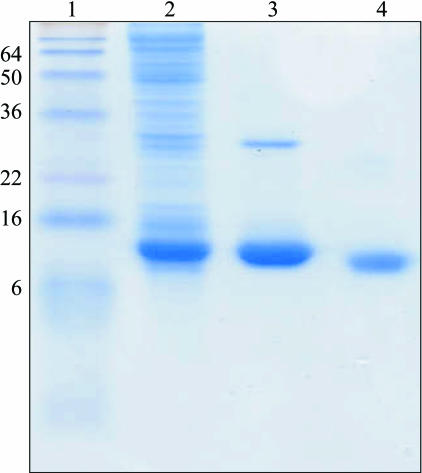
In order to remove the 6×His tag, the dialyzed protein was concentrated to 5 ml by ultrafiltration (Amicon Ultra-4, 5000 MWCO). It was then washed four times by ultrafiltration (Amicon Ultra-4, 5000 MWCO) with 10 ml protease buffer consisting of 50 mM Tris–HCl pH 6.5, 100 mM NaCl and 5 mM CaCl2. Factor Xa protease (Novagen, Lot N67059-2, EMD Chemicals, Inc., San Diego, USA) was added to the 6×His-sf-CzrB (1 U per 3.125 mg 6×His-sf-CzrB per millilitre) and incubated overnight with gentle stirring at 293 K. Precipitate was removed by centrifugation (Sorvall RC6, Kendro Laboratory Equipment) for 30 min at 47 800g and 277 K. The cleavage enzyme was removed by adding factor Xa removal resin (Qiagen; Lot 12194722; 50 µl resin binds 4 U factor Xa). After incubation for 10 min at 293 K with gentle shaking, the resin was pelleted at 1000g for 5 min at 293 K. The supernatant was decanted and its pH was adjusted from 6.5 to 8.0 by the addition of 1 M Tris–HCl pH 8.0. The mixture was subsequently applied onto a gravity Ni–NTA column as above. The flowthrough containing the cleaved protein (sf-CzrB) was collected and the cleaved 6×His tag and uncleaved 6×His-sf-CzrB protein were retained on the column. The sf-CzrB was concentrated from a volume of 15 ml to 1.5 ml by ultrafiltration (Amicon Ultra-4, 5000 MWCO) and applied onto a gel-filtration column (120–124 ml volume, HiLoad 16/60, Superdex 75, preparative grade, Amersham Biosciences, GE, Piscataway, USA) equilibrated with 20 mM Tris–HCl pH 8.0 to remove any remaining impurities. The His-tag-free protein was concentrated by ultrafiltration (Amicon Ultra-4, 5000 MWCO) to 10–30 mg ml−1 for use in crystallization trials. The yield of sf-CzrB ranged from 25 to 35 mg per litre of primary culture. Protein concentration was determined by the Bradford assay (Bradford, 1976 ▶) using bovine serum albumin (albumin standard; 2 mg ml−1 in 0.9% aqueous NaCl solution containing sodium azide; Lot GH97262, Pierce, Rockford, USA) and lyophilized salt-free sf-CzrB as standards. Both standards gave a similar absorbance response at 595 nm in the range up to 0.25 mg ml−1 protein. To evaluate progress during purification and 6×His-tag removal, protein composition was monitored by SDS–PAGE (Fig. 1 ▶).
Figure 1.
SDS–PAGE of sf-CzrB at various stages in its production, purification and 6×His-tag removal. Lane 1, molecular-weight standard markers (molecular weights are shown on the left in kDa). Lane 2, soluble protein prepared from E. coli BL21 (DE3) cells 4 h post induction with IPTG. Lane 3, 6×His-sf-CzrB following Ni–NTA column purification. Lane 4, sf-CzrB following gel-permeation chromatography.
2.3. Crystallization
2.3.1. 6×His-sf-CzrB
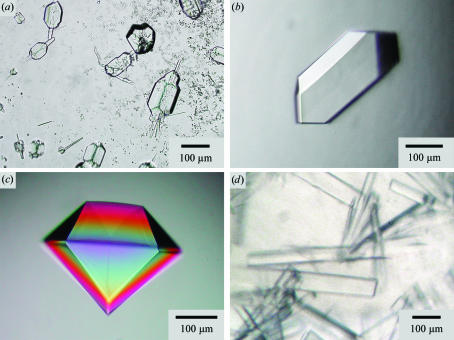
Initial crystallization trials were performed using the 6×His-tagged protein in 20 mM Tris–HCl pH 8.0. Hanging drops were set up in VDXm plates (Hampton Research, Aliso Viejo, USA) against Crystal Screens I and II (Lots 04169940 and 05039921, Hampton Research) using 18 mm silanized circular glass cover slides at 293 and 277 K (1 µl protein solution at 12 mg ml−1 was added to 1 µl precipitant over a reservoir containing 400 µl precipitant solution). Crystal hits were detected in 4 M sodium formate, 0.1 M sodium acetate pH 4.6 at 293 K. These conditions were optimized to 3–3.5 M sodium formate, 0.1 M sodium acetate pH 4.6 at 293 K. The best crystals measured 40 × 40 × 150 µm after 4 d, but typically had visual growth defects (Fig. 2 ▶ a). Seeding was used in order to improve crystal quality. For this purpose, a crystal was removed from the crystallization drop (above) and washed in 5 µl precipitant solution. It was then transferred to a 10 µl drop of precipitant solution and the crystal was crushed with a spatula. The suspension was transferred into an Eppendorf tube, diluted 1:10(v:v) with precipitant solution and stored at 293 K. To set up seeding drops, 1 µl of a protein solution at or below 6 mg ml−1 was mixed with 1 µl precipitant solution and was incubated over a reservoir containing 400 µl precipitant solution. The drop was equilibrated for 2 d at 293 K and 0.1 µl of the 1:10 diluted seeding suspension was added to the drop. Following this protocol, crystals free of visual defects grew to maximum dimensions of 30 × 75 × 150 µm within a week of seeding (Fig. 2 ▶ b). Prior to cryocooling, crystals were transferred into and allowed to sit for a few seconds in a drop containing the reservoir ingredients at their original concentrations (3–3.5 M sodium formate, 0.1 M sodium acetate pH 4.6) that had been adjusted to 10%(v/v) glycerol. They were then plunged directly into liquid nitrogen.
Figure 2.
Crystals of sf-CzrB with and without the 6×His tag grown with and without added zinc. (a) 6×His-sf-CzrB. (b) 6×His-sf-CzrB crystals produced by seeding. (c) sf-CzrB. (d) Cocrystals of sf-CzrB and Zn2+. The image in (c) was recorded using polarized light.
2.3.2. sf-CzrB
Initial crystallization conditions for the His-tag-free protein were found by screening against precipitant solutions from Crystal Screens I and II (Lots 04169940 and 05039921, Hampton Research). Hanging-drop crystallization was performed in VDXm Plates (Hampton Research); drops consisting of 1 µl protein solution (15 mg ml−1 in 20 mM Tris–HCl pH 8.0) and 1 µl precipitant solution were suspended above a reservoir containing 300 µl precipitant solution. The wells were covered with siliconized glass slides from Hampton Research. Two conditions [Crystal Screen I No. 20, 0.1 M sodium acetate pH 4.6, 0.2 M ammonium sulfate, 25%(w/v) polyethylene glycol 4000 (PEG 4000), and Crystal Screen II No. 13, 0.1 M sodium acetate pH 4.6, 0.2 M ammonium sulfate, 30%(w/v) polyethylene glycol monomethyl ether 2000 (PEG MME 2000)] gave rise to crystalline material. They were optimized with a view to growing crystals of sufficient size and diffracting power by adjusting the concentrations in the range 0.1–0.2 M ammonium sulfate, 10–22.5% PEG 4000 and 12.5–25% PEG 2000 MME. Crystals usually grew within one week to average dimensions of 150 × 150 × 300 µm. The best diffracting crystals were produced under the following conditions: 0.1 M sodium acetate pH 4.6, 0.2 M ammonium sulfate, 15%(w/v) PEG 4000 and 0.1 M sodium acetate pH 4.6, 0.2 M ammonium sulfate, 17.5%(w/v) PEG MME 2000 (Fig. 2 ▶ c).
Cryoconditions were obtained by replacing the precipitant solution in the reservoir over which crystals grew by either 0.1 M sodium acetate pH 4.6, 0.2 M ammonium sulfate and 25.5%(w/v) PEG 2000 MME or 0.1 M sodium acetate pH 4.6, 0.2 M ammonium sulfate and 25.5%(w/v) PEG 4000 depending on the initial condition. Drops were allowed to adjust to the new conditions for 2 d at 293 K, at which point the crystals were harvested and plunged directly into liquid nitrogen.
In preparation for small-angle X-ray scattering (SAXS) measurements, 2.5 µl of a 1.0 M aqueous zinc chloride solution was added to 100 µl sf-CzrB (28 mg ml−1) in 50 mM Tris buffer pH 7.48. The initial turbidity produced by the zinc chloride disappeared upon gentle shaking at 277 K and the clear solution was used for SAXS measurements, which will be reported separately. After 5 d storage at 277 K, a shower of protein crystals formed in an unused portion of the SAXS sample (∼70 µl stored in a sealed 500 µl tube). Well formed crystals, which reached maximum dimensions of 50 × 50 × 300 µm (Fig. 2 ▶ d), were harvested from the batch, mounted in immersion oil (Hampton Research HR3-611, type A) and flash-cooled directly in the cryostream immediately prior to data collection.
2.4. X-ray data collection
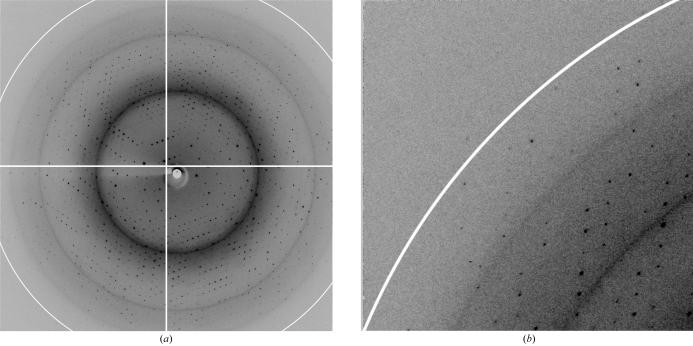
Diffraction measurements were performed on the F1 and F2 beamlines at the Cornell High-Energy Synchrotron Source (CHESS, Ithaca, NY). On F1, a 100 µm diameter 0.9124 Å beam was used together with an ADSC Quantum-4 detector positioned 180 mm from the sample. For data collection at F2, a 20 µm diameter beam produced by single-bounce capillary optics was used. Data sets at F2 were collected at 1.2818 Å using a Quantum-210 detector 134 mm from the sample. Larger crystals from cleaved and uncleaved protein (>300 µm) suffered from high mosaicity and disorder (spots were streaky or elongated in one direction and/or had associated diffuse scattering). The best crystals of the His-tag-free protein had maximum dimensions of 100–200 µm and diffracted to 1.7 Å resolution (Fig. 3 ▶). For comparison, the best diffraction observed with His-tagged protein crystals was to 2.7 Å. Data reduction and scaling were performed with HKL-2000 (Otwinowski & Minor, 1997 ▶). Data-collection statistics are shown in Table 1 ▶.
Figure 3.
(a) X-ray diffraction recorded from a crystal of sf-CzrB. An expanded view of the upper left quadrant of (a) is shown in (b). The white circle identifies diffraction at 1.7 Å.
Table 1. Unit-cell characteristics and X-ray data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Sample | 6×His-sf-CzrB | sf-CzrB | sf-CzrB + Zn2+ |
|---|---|---|---|
| CHESS Station | F1 | F1 | F2 (microbeam) |
| Temperature (K) | 100 | 100 | 100 |
| Space group | P3221 | P3212 | C2221 |
| Unit-cell parameters (Å, °) | a = b = 109.1, c = 118.1, α = β = 90, γ = 120 | a = b = 60.33, c = 92.14, α = β = 90, γ = 120 | a = 50.78, b = 74.60, c = 43.96, α = β = γ = 90 |
| Mosaicity (°) | 0.6 | 0.3 | 0.5 |
| Resolution limits (Å) | 50–2.7 (2.8–2.7) | 50–1.70 (1.76–1.70) | 30–2.2 (2.32–2.20) |
| Total reflections | 210595 | 270944 | 30714 |
| Unique reflections | 22816 | 21179 | 4408 |
| Rejected reflections (%) | 0.08 | 0.05 | 0.17 |
| Redundancy | 9.2 (9.4) | 12.8 (6.2) | 7 (7.2) |
| Completeness (%) | 100 (100) | 98.9 (92.4) | 98.7 (97.8) |
| I/σ(I) | 33.5 (3.9) | 54.8 (4.4) | 19.8 (8.8) |
| Rsym (%) | 7.0 (65.9 | 4.2 (39.3) | 10.1 (24.2) |
| Rp.i.m.† (%) | 4.2 (31.6) | 1.9 (23.3) | 5.9 (14.1) |
| Rr.i.m.‡ (%) | 12.5 (96.5) | 6.9 (61.7) | 11.8 (28.1) |
| Wilson B factor (Å2) | 80.0 | 25.7 | 19.8 |
3. Results and discussion
The cytosolic water-soluble C-terminal domain of the membrane protein CzrB has been overexpressed and a highly purified form of the fragment has been produced (Fig. 1 ▶, lane 4). Crystals of the His-tagged variant had visible defects (Fig. 2 ▶ a) and diffracted poorly. Two approaches were taken to improve crystal quality. The first involved seeding and yielded optically perfect crystals (Fig. 2 ▶ b). However, the diffraction quality did not improve. The second approach, in which the N-terminal His tag was removed, produced crystals that diffracted to 1.7 Å. Interestingly, both forms of the protein crystallized in the same trigonal space group but with different unit-cell parameters (Table 1 ▶). The Matthews coefficient (Matthews, 1968 ▶; Kantardjieff & Rupp, 2003 ▶) is consistent with eight or nine 6×His-sf-CzrB monomers per asymmetric unit and an estimated solvent content of 48% or 42%, respectively. In contrast, the His-tag-free form contains only two sf-CzrB monomers per asymmetric unit, with an estimated solvent content of 48.2%.
Attempts were made to prepare heavy-atom derivatives of the His-tagged and tag-free protein crystals. The first involved soaking crystals in 1 mM solutions of heavy-atom compounds [zinc sulfate, zinc chloride, potassium hexachloroplatinate(IV), potassium tetrachloroplatinate, sodium hexachloroiridate(III) hydrate, iridium(III) chlorohydrate and methylmercury(III) chloride] for 24 h at 293 K. However, no significant heavy-atom signal was detected by X-ray diffraction. A second approach, involving cocrystallization in the presence of zinc chloride, proved successful with sf-CzrB (Fig. 2 ▶ d; Table 1 ▶). Crystals grew in space group C2221 and diffracted to 2.2 Å. Matthews coefficient calculations suggest that each asymmetric unit contains a single monomer with an estimated solvent content of 39.8%. Patterson maps revealed at least two bound zinc ions per monomer.
Acknowledgments
The authors thank M. Szebenyi (CHESS, Cornell University) for processing the zinc/sf-CzrB data. Support was provided by grants from Science Foundation Ireland (02-IN1-B266), the National Institutes of Health (GM61070 and GM75915) and the National Science Foundation (IIS-0308078) to MC and by Enterprise Ireland grants SC/2001/432 and CFTD/04/106 to GW. This work is based on research conducted at the Cornell High Energy Synchrotron Source (CHESS), which is supported by the National Science Foundation under award DMR 0225180, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, supported by award RR-01646 from the National Institutes of Health through its National Center for Research Resources.
References
- Blencowe, D. K. & Morby, A. P. (2003). FEMS Microbiol. Rev.27, 291–311. [DOI] [PubMed] [Google Scholar]
- Bloss, T., Clemens, S. & Nies, D. H. (2002). Planta, 214, 783–791. [DOI] [PubMed] [Google Scholar]
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed] [Google Scholar]
- Choudhury, R. & Srivastava, S. (2001). Curr. Sci.81, 768–775.
- Conklin, D. S., McMaster, J. A., Culbertson, M. R. & Kung, C. (1992). J. Mol. Cell. Biol.12, 3678–3688. [DOI] [PMC free article] [PubMed]
- Grass, G., Otto, M., Fricke, B., Haney, C. J., Rensing, C., Nies, H. D. & Munkelt, D. (2005). Arch. Microbiol.183, 9–18. [DOI] [PubMed] [Google Scholar]
- Haney, C. J., Grass, G., Franke, S. & Rensing, C. (2005). J. Ind. Microbiol. Biotechnol.32, 215–226. [DOI] [PubMed] [Google Scholar]
- Hantke, K. (2005). Curr. Opin. Microbiol.8, 196–202. [DOI] [PubMed] [Google Scholar]
- Hu, X., O’Dwyer, R. & Wall, J. G. (2005). J. Biotechnol.120, 38–45. [DOI] [PubMed] [Google Scholar]
- Kantardjieff, K. A. & Rupp, B. (2003). Protein Sci.12, 1865–1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaj, O., Li, H., Burrowes, E., Moore, J., Caffrey, M. & Wall, J. G. (2006). Microb. Cell Fact.5, S21.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Munkelt, D., Grass, G. & Nies, D. H. (2004). J. Bacteriol.186, 8036–8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies, D. H., Nies, A., Chu, L. & Silver, S. (1989). Proc. Natl. Acad. Sci. USA, 86, 7351–7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Outten, C. E. & O’Halloran, T. V. (2001). Science, 292, 2488–2492. [DOI] [PubMed] [Google Scholar]
- Paulsen, I. T. & Saier, M. H. (1997). J. Membr. Biol.156, 99–103. [DOI] [PubMed] [Google Scholar]
- Spada, S., Pembroke, J. T. & Wall, J. G. (2002). Extremophiles, 6, 301–308. [DOI] [PubMed] [Google Scholar]
- Weiss, M. S. (2001). J. Appl. Cryst.34, 130–135. [Google Scholar]