By heating a highly amyloidogenic mutant of the human plasma protein transthyretin at 328 K for 48 h, diffraction-quality crystals could be reproducibly produced. The procedure precipitated ∼40% of the protein, but rendered what remained in solution more homogenous.
Keywords: TTR, transthyretin, amyloid, heating
Abstract
The use of high temperatures in the purification procedures of heat-stable proteins is a well established technique. Recently, rapid pre-heat treatment of protein samples prior to crystallization trials was described as a final polishing step to improve the diffraction properties of crystals [Pusey et al. (2005 ▶), Prog. Biophys. Mol. Biol. 88, 359–386]. The present study demonstrates that extended high-temperature incubation (328 K for 48 h) of the highly amyloidogenic transthyretin mutant TTR G53S/E54D/L55S successfully removes heterogeneities and allows the reproducible growth of well diffracting crystals. Heat treatment might be applied as an optimization method to other cases in which the protein/biomolecule fails to form diffracting crystals.
1. Introduction
Self-assembly and deposition of proteins into amyloid fibrils and plaques are phenomena that have currently been linked to around 20 different human diseases (Pepys, 2006 ▶). Amyloids are long unbranched fibrils that typically have a diameter of between 50 and 130 Å and display a characteristic cross-β pattern formed by an extended β-helical structure (for reviews, see Ghiso & Frangione, 2002 ▶; Kelly, 1998 ▶; Pepys, 2006 ▶; Rochet & Lansbury, 2000 ▶). Structural studies of amyloids are hampered by their size and heterogeneous nature.
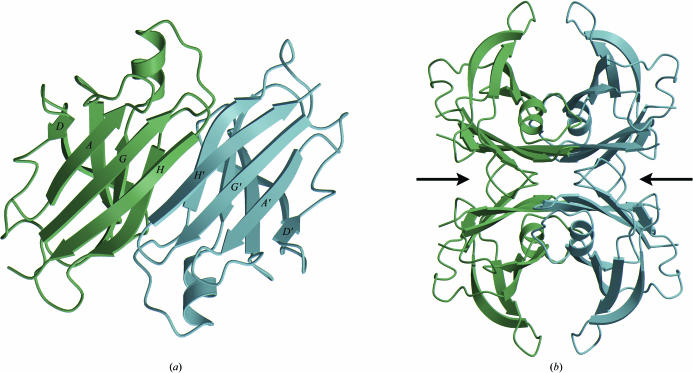
Transthyretin (TTR) is a homotetrameric protein that is involved in thyroid-hormone transport in humans. It is implicated in several types of amyloid diseases in which the misfolding of the wild-type protein as well as a number of substitution mutants causes the protein molecules to aggregate into amyloids. The normally folded tetrameric structure of human TTR has been described at high resolution with and without its natural ligand thyroxine (Fig. 1 ▶; Blake et al., 1978 ▶; Hamilton et al., 1993 ▶; Hörnberg et al., 2000 ▶).
Figure 1.
The structure of the native transthyretin tetramer (PDB code 1f41; Hörnberg et al., 2000 ▶). (a) The TTR dimer structure. Each monomer of 127 amino acids forms a β-barrel formed by two β-sheets comprising β-strands D-A-G-H and C-B-E-F. Dimerization occurs predominantly through an intermolecular main-chain interaction between the H β-strands of each monomer so that a continuous eight-stranded β-sheet is formed. (b) The tetramer is generated through hydrophobic interactions between the AB and GH loops, creating a central hydrophobic channel with two hormone-binding sites, indicated by the two arrows.
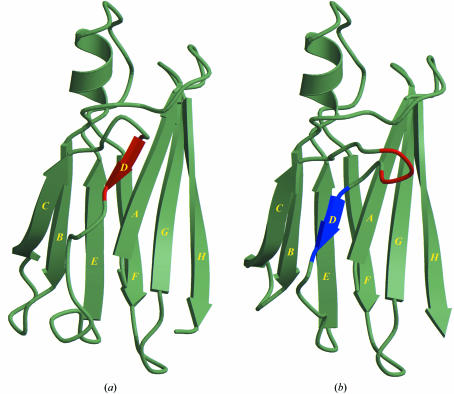
None of the structures of naturally occurring TTR mutants solved by X-ray crystallography display structural features that give a deeper insight into the events responsible for amyloid assembly (Hörnberg et al., 2000 ▶). A substitution mutant TTR G53S/E54D/L55S has been constructed that proved to be highly prone to aggregation into fibril-like structures in vitro (Goldsteins et al., 1997 ▶). Despite a high degree of aggregation, one batch of TTR G53S/E54D/L55S protein could be successfully crystallized in our laboratory. The structure was solved at 2.3 Å and displayed features that were suggested to be of relevance for fibril formation (PDB code 1g1o; Eneqvist et al., 2000 ▶). A dramatic shift in the positioning of β-strand D was observed, whereas the location of the C strand remained unchanged (Fig. 2 ▶). This so-called ‘β-slipped’ conformation of the D strand allowed new crystal-contact interactions with neighbouring molecules and it was suggested that other amyloidogenic mutants of TTR could undergo a similar β-strand register shift as that observed in the engineered triple mutant (Eneqvist et al., 2000 ▶).
Figure 2.
The β-slip. (a) Monomer A from the structure of native transthyretin (PDB code 1f41; Hörnberg et al., 2000 ▶). The D strand and the position of the three residues Gly53-Glu54-Leu55 are shown in red. (b) Monomer B from the structure of the triple mutant TTR G53S/E54D/L55S (PDB code 1g1o; Eneqvist et al., 2000 ▶). A three-residue shift of β-strand D places the mutation sites in the new extended CD loop.
To elucidate the significance of the β-slip model for amyloid formation, further structural and biochemical studies of the triple mutant are required. Unfortunately, until now we have not been able to reproduce crystals of the triple mutant, despite extensive optimization of both protein-expression and protein-purification protocols. The problem of crystallizing the triple mutant is probably a consequence of the high degree of heterogeneity as observed on native PAGE and size-exclusion chromatography (Eneqvist et al., 2000 ▶). By lowering the protein-expression temperature from 310 to 293 K, the formation of inclusion bodies could be prevented. Crystals could be obtained from the purified soluble material; however, these did not diffract.
High-temperature precipitation as a purification method for thermostable proteins is a well established chromatographic technique that is usually employed in the initial stages of purification. The method relies on the differences in temperature stability between proteins and its use is typically limited to heat-stable proteins. A different use of protein heating was recently described in which heat treatment was used as a protein-purification polishing step to increase the diffraction quality of protein crystals (Pusey et al., 2005 ▶). The method is based on the assumption that improperly folded proteins are less heat-stable and have a higher propensity for aggregation than the correctly folded protein. Heating will thus precipitate misfolded forms of the protein of interest, thereby increasing the protein-solution homogeneity of the remaining protein and presumably the crystal quality.
In the present study, we have applied the method of Pusey et al. (2005 ▶) to the aggregation-prone G53S/E54D/L55S triple mutant of transthyretin. By heat treatment of the purified protein, most of the aggregation-prone forms of the protein were precipitated. For successful crystallization, however, the protein material had to be heated for at least 48 h at 328 K. When we used treatments for shorter periods of time, for example 10 min, which was the time frame typically used in the experiments described by Pusey et al. (2005 ▶), we did not obtain useful well diffracting crystals.
The transthyretin triple mutant was heated to 328 K for 96 h. During the first 48 h, 40% of the protein precipitated; however, the protein left in solution was stable and well diffracting crystals could be obtained from this material.
2. Materials and methods
2.1. Cloning and expression of TTR G53S/E54D/L55S
The 127-amino-acid TTR G53S/E54D/L55S triple mutant was generated in the pET-3a plasmid using site-directed mutagenesis as described previously (Goldsteins et al., 1997 ▶).
Soluble protein was expressed in Escherichia coli BL-21 (DE3) cells. Briefly, the competent cells were transformed with triple mutant-containing pET-3a plasmids (Hanahan, 1983 ▶) and grown overnight on LB agar plates containing 50 µg ml−1 carbenicillin. All colonies were transferred to LB media supplemented with 100 µg ml−1 carbenicillin and grown with orbital shaking at 310 K. At an OD600 of 0.4, the temperature was lowered to 293 K and the cells were grown for another 1 h (to an OD600 of 0.8) before inducing protein expression for 18 h by adding IPTG to a final concentration of 0.2 mM. The cells were then collected by centrifugation at 5900g at 277 K for 30 min, resuspended in 20 mM Tris–HCl pH 7.5 and 100 mM NaCl, frozen in liquid nitrogen and stored at 193 K.
2.2. Purification of TTR G53S/E54D/L55S
All chromatography media were purchased from GE Healthcare, Uppsala, Sweden.
The cells were thawed on ice and lysed by sonication. Insoluble debris was removed by centrifugation at 54 000g and the soluble fraction was loaded onto a Q-Sepharose Fast Flow anion-exchange column. The column was equilibrated in buffer A (10 mM Tris–HCl pH 7.5 and 100 mM NaCl) and the tetrameric protein was eluted with a linear NaCl gradient (0.1–0.3 M NaCl) in 10 mM Tris–HCl pH 7.5. Fractions confirmed by SDS–PAGE to contain TTR (monomeric molecular weight of 13.8 kDa) were pooled and dialyzed overnight against buffer A and then loaded onto a MonoQ 10/100 HR anion-exchange column equilibrated with the same buffer. Proteins were eluted with a linear NaCl gradient (0.1–0.3 M in 10 mM Tris–HCl pH 7.5) and the TTR-containing fractions were pooled and concentrated to 5.0 mg ml−1 using a Centriprep YM-10 (Millipore) centrifugal concentrator. The tetrameric protein was then isolated on a HiLoad 16/60 Superdex 200pg gel-filtration column equilibrated in 5 mM Tris–HCl pH 7.5 and 50 mM NaCl. TTR fractions eluting at approximately 80 ml (corresponding to a molecular weight of 55.1 kDa) with an estimated TTR purity of >95% were pooled, frozen in liquid nitrogen and stored at 193 K. Following this purification scheme, 20–40 mg protein was isolated per litre of culture.
2.3. Probing the time-dependency of temperature-mediated precipitation of TTR G53S/E54D/L55S
To follow the level of precipitation at 328 K over time, the mutant was concentrated to 3 mg ml−1 in 5 mM Tris–HCl pH 7.5 and 50 mM NaCl, transferred as 0.2 ml aliquots to thin-walled PCR tubes, each tube corresponding to one defined incubation time, and refrozen at 193 K. Prior to incubation, the tubes were thawed at room temperature for 15 min. The tubes were then incubated in a hybridization chamber employing slow rotation at 328 K for 0–96 h. After heat-incubation, the tubes were cooled to room temperature for 15 min and refrozen at 193 K. To analyse the amount of protein remaining in solution after heating, the tubes were thawed and the precipitated material was removed by centrifugation at 14 000g for 30 min at 277 K followed by filtration through 0.2 µm centrifugal filter devices (Amicon Ultrafree-MC). The concentration of the protein solution was then determined using the Bradford method (Bradford, 1976 ▶). The precipitated material was washed twice in deionized H2O, solubilized in SDS loading buffer containing 0.7 M β-mercaptoethanol and analysed by SDS–PAGE.
2.4. Crystallization
Crystallization was carried out at 291 K using the hanging-drop vapour-diffusion method. The crystallization drops were prepared by mixing 8.0 µl protein solution (4 mg ml−1 in 5 mM Tris–HCl buffer pH 7.5 containing 50 mM NaCl) with 1.0 µl reservoir solution on Hybri-slips (Sigma–Aldrich). The drops were equilibrated against 500 µl reservoir solution. Crystallization screening of the nonheated protein yielded nondiffracting crystals after 5 d using condition No. 22 from Hampton Research Crystal Screen 1 (30% PEG 4000, 0.1 M Tris–HCl pH 8.5, 0.2 M sodium acetate trihydrate). The diffraction properties of these crystals could not be improved. Following heat treatment, the screening was repeated and crystals with maximal dimensions of 0.3 × 0.2 × 0.2 mm were obtained after 5 d using conditions C5 and C6 of the Hampton Research Grid Screen Ammonium Sulfate (2.4 M ammonium sulfate, 0.1 M Tris–HCl pH 8.0 and 2.4 M ammonium sulfate, 0.1 M Bicine pH 9.0, respectively).
2.5. Data collection, structure determination and refinement
The diffraction quality of the crystals grown from nonheated and heated protein was tested at beamline I711 at the MAX-lab synchrotron, Lund, Sweden. The crystals obtained from conditions C5 and C6 were of equal diffraction quality. One crystal grown in condition C5 was cryocooled and diffraction data were measured using X-rays of wavelength 1.115 Å at 100 K. Diffraction data were processed to 2.3 Å resolution with the program XDS (Kabsch, 1993 ▶). The crystals belong to space group P3221, with unit-cell parameters a = 58.36, b = 58.36, c = 229.16 Å, α = 90, β = 90, γ = 120°, with one tetramer in the asymmetric unit and a solvent content of 46.3%. The data-collection statistics are shown in Table 1 ▶. The structure was solved by molecular replacement using the program MOLREP available in the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶) using all protein atoms from the previously solved structure of TTR G53S/E54D/L55S as the search model (PDB code 1g1o; Eneqvist et al., 2000 ▶). Individual monomers were refined with REFMAC5 and the phenix.refine module included in the PHENIX macromolecular crystallographic package (Adams et al., 2002 ▶) to an R factor and R free of 23.1 and 28.9%, respectively, including all reflections in the resolution range 15–2.3 Å. To model large-scale thermal motions, atomic displacement parameter refinement as used in the translation, libration, screw-rotation (TLS) method was implemented during refinement (Winn et al., 2001 ▶). Figures were created using MOLSCRIPT (Kraulis, 1991 ▶) and RASTER3D (Merritt & Bacon, 1997 ▶).
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Crystal from heated protein (PDB code 2qel) | Original crystal (PDB code 1g1o; Eneqvist et al., 2000 ▶) | |
|---|---|---|
| Data collection | ||
| Crystal dimensions (mm) | 0.3 × 0.2 × 0.2 | 0.1 × 0.1 × 0.2 |
| X-ray source | Beamline I711, MAX-lab | Beamline I711, MAX-lab |
| Wavelength (Å) | 1.1150 | 0.9960 |
| Resolution range (Å) | 30–2.3 (2.43–2.30) | 20–2.3 (2.42–2.30) |
| No. of observed/unique reflections | 192555/19767 | 109159/20911 |
| Space group | P3221 | P3221 |
| Unit-cell parameters (Å, °) | a = b = 58.36, c = 229.16, α = β = 90, γ = 120 | a = b = 58.31, c = 228.95, α = β = 90, γ = 120 |
| Completeness (%) | 92.8 (94.0) | 99.2 (98.4) |
| Redundancy | 9.8 (9.0) | 5.2 (5.0) |
| 〈I/σ(I)〉 | 18.5 (3.8) | 8.8 (1.3) |
| Rsym (%) | 10.4 (53.8) | 8.2 (54.6) |
| Wilson B factor (Å2) | 43.3 | 44.0 |
| Refinement | ||
| Resolution range used in refinement | 15.0–2.3 (2.35–2.29) | 15.0–2.3 (2.38–2.30) |
| No. of reflections in working set | 18874 (1199) | 18492 (1563) |
| No. of reflections in test set | 1018 (73) | 939 (91) |
| Residues included in model | A11–A125, B11–B124, C12–C125, D10–D125 | A11–A125, B11–B124, C12–C125, D10–D125 |
| No. of protein atoms refined | 3483† | 3466 |
| No. of solvent atoms | 150 | 143 |
| R factor (%) | 23.1 (25.1) | 23.8 (33.2) |
| Rfree (%) | 28.9 (34.6) | 29.0 (37.2) |
| Average B factors (Å2) | ||
| Overall | 37.8 | 55.6 |
| Main chain | 39.4 | 56.1 |
| Side chains and waters | 36.1 | 55.0 |
| R.m.s.d. bond lengths (Å) | 0.015 | 0.007 |
| R.m.s.d. bond angles (°) | 1.567 | 1.46 |
| Ramachandran plot analysis | ||
| Most favoured regions (%) | 86.9 | 83.3 |
| Additionally allowed regions (%) | 11.6 | 15.8 |
3. Results and discussion
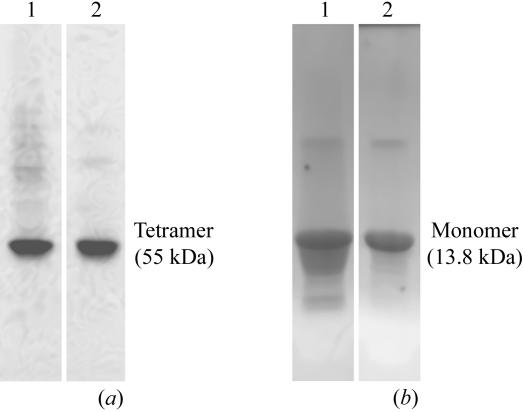
Most naturally occurring TTR mutants are soluble when overexpressed recombinantly at 310 K. However, the overexpression of TTR G53S/E54D/L55S triple mutant at this temperature leads to its precipitation in the form of inclusion bodies (Goldsteins et al., 1997 ▶). Native PAGE after refolding of the protein shows a highly heterogeneous mix of protein molecules ranging from monomers to high-molecular-weight aggregates (data not shown). Despite this protein heterogeneity, crystals could be obtained from one batch of purified protein and the structure was solved at 2.3 Å resolution (Eneqvist et al., 2000 ▶). The structure revealed structural changes that were suggested to be of relevance for the ability of the mutant to form fibrils, warranting a more thorough analysis. However, reproducing well diffracting crystals proved impossible despite extensive efforts in optimizing the purification and crystallization procedures. By expressing the triple mutant at 293 K the formation of inclusion bodies was avoided; however, the purified protein was still heterogeneous (Fig. 3 ▶ a, lane 1) and although crystals could be obtained from this material (Fig. 4 ▶) they did not diffract.
Figure 3.
TTR G53S/E54D/L55S was purified as described in the text and then analyzed by native PAGE before (lane 1) and after (lane 2) heat treatment at 328 K for 96 h (a). In (b), the dissolved precipitates (lane 1) and the soluble fraction remaining in solution (lane 2) after heat treatment were analyzed by SDS–PAGE. The figure shows that heat treatment precipitates heterogenic and oligomeric forms of TTR.
Figure 4.
Nondiffracting crystals of TTR G53S/E54D/L55S grown from soluble but nonheat-treated protein in PEG-containing conditions.
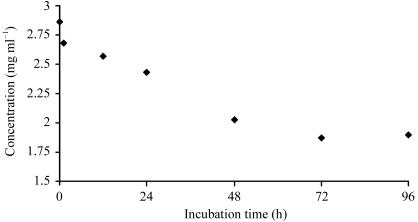
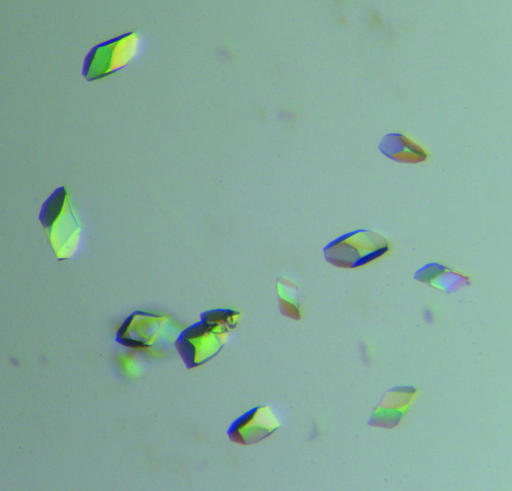
The rationale behind heating a protein solution to increase the diffraction quality of crystals has recently been described in the literature (Pusey et al., 2005 ▶). Since native tetrameric TTR is one of the most heat-stabile human plasma proteins known, with a T m of the native tetramers of approximately 373 K (Chung et al., 2001 ▶; Shnyrov et al., 2000 ▶), we postulated that it may constitute a suitable candidate to test the method of heat treatment to improve crystallization. Since irreversible tertiary-structure alterations have been shown to occur for this protein at temperatures exceeding 333 K (Chung et al., 2001 ▶), we chose to select properly folded tetramers by incubating the protein solution at 328 K. In addition, in previous studies we had shown that heat treatment at this temperature is sufficient to induce structural changes leading to amyloid formation of some highly amyloidogenic TTR mutants (Karlsson et al., 2005 ▶; Olofsson et al., 2004 ▶). In the original experiments described by Pusey and coworkers, the purified protein samples were heated for approximately 10 min at 310 K (Pusey et al., 2005 ▶) prior to the crystallization trials. In their study, the method was predominantly used to improve the diffraction quality of crystals. To elucidate the optimal incubation time for heat treatment of the TTR triple mutant, we followed the concentration of soluble protein as a function of time at 328 K (Fig. 5 ▶). The result suggested that for this particular protein ∼48 h incubation represented the optimal incubation time needed to allow all less temperature-stable substructures to precipitate out from the solution. After 48 h ∼40% of the protein material was precipitated; however, the protein molecules which remained in solution seemed perfectly stable as no further precipitation was observed during a subsequent 48 h incubation (Fig. 5 ▶). SDS–PAGE analyses verified that the precipitated protein contained transthyretin (Fig. 3 ▶ b, lane 1). As a result, the more stable and homogenous protein sample crystallized readily (Fig. 6 ▶) in comparison to the nonheated material (compare Figs. 4 ▶ and 6 ▶).
Figure 5.
Solubility of the human TTR G53S/E54D/L55S triple mutant as a function of time at 328 K.
Figure 6.
Examples of well diffracting crystals of TTR G53S/E54D/L55S grown from heat-treated protein in ammonium sulfate-containing conditions.
The heat treatment clearly led to an improvement in the behaviour of the protein in the crystallization trials. While in previous years we had been unable to reproduce the crystals from our first lucky batch, we can now grow crystals in at least two new conditions. We therefore suspect that the original batch had been unintentionally heated at some stage of the purification procedure. We wanted to investigate whether the heat treatment of the protein sample improved the diffraction quality of the subsequent crystals. The crystals of the triple mutant were therefore tested at the synchrotron beamline I711 at MAX-lab, Lund, Sweden and one full data set was collected to 2.3 Å. There was no significant improvement of resolution or data-collection statistics compared with the original data (Eneqvist et al., 2000 ▶). The structure was solved and refined and the final model includes residues A11–A125, B11–B124, C12–C125 and D10–D125 and 150 water molecules. The Ramachandran plot for the structure showed that 86.9% of nonglycine or proline residues lie in the most favoured regions of the plot, while 12.3% fall within the additionally allowed regions as defined in the program PROCHECK (Laskowski et al., 1993 ▶).
We could not detect any significant structural differences compared with the previously determined structure of this mutant (PDB code 1g1o; Eneqvist et al., 2000 ▶). The root-mean-square deviation (r.m.s.d.) between main-chain Cα atoms for residues 12–124 is 0.37, 0.28, 0.47 and 0.27 Å for monomers A, B, C and D, respectively. When comparing individual monomers with monomer A of the wild-type TTR structure (PDB code 1f41; Hörnberg et al., 2000 ▶), the r.m.s.d. of the main-chain Cα atoms is 2.5, 2.6, 2.5 and 2.6 Å for monomers A, B, C and D, respectively. In agreement with the previous mutant structure, parts of the structure have regions with high conformational flexibility and large positional errors. In particular, residues A35–A40, A50–A64, B50–B56, C33–C64 and D50–D55 are poorly described in the electron density. As a consequence of this, residues A50–A63 and C50–C64 were modelled as polyalanines, as were the same residues in the 1g1o structure. By being able to reproducibly isolate and crystallize a stable species of the transthyretin triple mutant TTR G53S/E54D/L55S, we can now proceed with further biochemical and structural characterizations of this in vitro fibril-forming mutant. Furthermore, we wish to promote the usefulness of this method for other cases in which the protein does not behave as previously in crystallization trials, for example, owing to batch differences etc., and fails to crystallize.
Supplementary Material
PDB reference: transthyretin mutant G53S/E54D/L55S, heated protein, 2qel, r2qelsf
Acknowledgments
We would like to thank the personnel of the MAX-lab biocrystallography beamlines for their help during data collection. This work was supported by the Swedish Research Council, the Gustafsson Foundation and FAMY/AMYL.
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L. W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed] [Google Scholar]
- Blake, C. C., Geisow, M. J., Oatley, S. J., Rérat, B. & Rérat, C. (1978). J. Mol. Biol.121, 339–356. [DOI] [PubMed] [Google Scholar]
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed] [Google Scholar]
- Chung, C. M., Connors, L. H., Benson, M. D. & Walsh, M. T. (2001). Amyloid, 8, 75–83. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Eneqvist, T., Andersson, K., Olofsson, A., Lundgren, E. & Sauer-Eriksson, A. E. (2000). Mol. Cell, 6, 1207–1218. [DOI] [PubMed] [Google Scholar]
- Ghiso, J. & Frangione, B. (2002). Adv. Drug Deliv. Rev.54, 1539–1551. [DOI] [PubMed] [Google Scholar]
- Goldsteins, G., Andersson, K., Olofsson, A., Dacklin, I., Edvinsson, Å., Baranov, V., Sandgren, O., Thylén, C., Hammarström, S. & Lundgren, E. (1997). Biochemistry, 36, 5346–5352. [DOI] [PubMed] [Google Scholar]
- Hamilton, J. A., Steinrauf, L. K., Braden, B. C., Liepnieks, J., Benson, M. D., Holmgren, G., Sandgren, O. & Steen, L. (1993). J. Biol. Chem.268, 2416–2424. [PubMed] [Google Scholar]
- Hanahan, D. (1983). J. Mol. Biol.166, 557–580. [DOI] [PubMed] [Google Scholar]
- Hörnberg, A., Eneqvist, T., Olofsson, A., Lundgren, E. & Sauer-Eriksson, A. E. (2000). J. Mol. Biol.302, 649–669. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800. [Google Scholar]
- Karlsson, A., Olofsson, A., Eneqvist, T. & Sauer-Eriksson, A. E. (2005). Biochemistry, 44, 13063–13070. [DOI] [PubMed] [Google Scholar]
- Kelly, J. W. (1998). Curr. Opin. Struct. Biol.8, 101–106. [Google Scholar]
- Kraulis, P. J. (1991). J. Appl. Cryst.24, 946–950. [Google Scholar]
- Laskowski, R. A., Moss, D. S. & Thornton, J. M. (1993). J. Mol. Biol.231, 1049–1067. [DOI] [PubMed] [Google Scholar]
- Merritt, E. A. & Bacon, D. J. (1997). Methods Enzymol.277, 505–524. [DOI] [PubMed]
- Olofsson, A., Ippel, J. H., Wijmenga, S. S., Lundgren, E. & Öhman, A. (2004). J. Biol. Chem.279, 5699–5707. [DOI] [PubMed] [Google Scholar]
- Pepys, M. B. (2006). Annu. Rev. Med.57, 223–241. [DOI] [PubMed] [Google Scholar]
- Pusey, M. L., Liu, Z.-J., Tempel, W., Praissman, J., Lin, D., Wang, B.-C., Gavira, J. A. & Ng, J. D. (2005). Prog. Biophys. Mol. Biol.88, 359–386. [DOI] [PubMed] [Google Scholar]
- Rochet, J. C. & Lansbury, P. T. Jr (2000). Curr. Opin. Struct. Biol.10, 60–68. [DOI] [PubMed] [Google Scholar]
- Shnyrov, V. L., Villar, E., Zhadan, G. G., Sanchez-Ruiz, J. M., Quintas, A., Saraiva, M. J. & Brito, R. M. (2000). Biophys. Chem.88, 61–67. [DOI] [PubMed] [Google Scholar]
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: transthyretin mutant G53S/E54D/L55S, heated protein, 2qel, r2qelsf