Abstract
Cells from a “knock-in” mouse expressing a NF-κB p65 mutant bearing an alanine instead of serine at position 276 (S276A) display a significant reduction of NF-κB-dependent transcription, even though the mutant p65 forms appropriate complexes that translocate normally to the nucleus and bind to DNA. Surprisingly, however, instead of the expected embryonic lethality from hepatocyte apoptosis seen in the absence of NF-κB activity, the S276A knock-in embryos die at different embryonic days due to variegated developmental abnormalities. We now demonstrate that this variegated phenotype is due to epigenetic repression resulting from the recruitment of histone deacetylases by the nonphosphorylatable form of NF-κB into the vicinity of genes positioned fortuitously near NF-κB-binding sites. Therefore, unphosphorylated nuclear NF-κB can affect expression of genes not normally regulated by NF-κB through epigenetic mechanisms.
[Keywords: Epigenetics, inflammation, NF-κB, gene expression, phosphorylation]
The transcription factor NF-κB plays a critical role in the inducible expression of genes involved in many biological processes (Baldwin 1996; Ghosh et al. 1998). In unstimulated cells, NF-κB exists in the cytosol, bound to inhibitory IκB proteins. Stimulation of cells leads to the activation of the IκB kinase (IKK) complex, which then phosphorylates IκB proteins on two N-terminal serine residues, leading to its polyubiquitination and subsequent degradation by the proteasome. The released NF-κB translocates to the nucleus, binds to cognate sites in the promoters of responsive genes, and induces their expression. This cytoplasmic-to-nuclear translocation has remained a paradigm for inducible transcription factors, and it has been assumed that, once NF-κB reached the nucleus, it would be capable of driving transcription. However, recent studies have begun to suggest that such a model might be too simplistic and may fail to account for the complexity that underlies the process of NF-κB activation (Chen and Greene 2004; Hayden and Ghosh 2004; Vermeulen et al. 2006).
A hint that the transcriptional activity of NF-κB requires additional regulatory steps first came from biochemical studies where the catalytic subunit of cAMP-dependent protein kinase (PKAc) was found to copurify with cytosolic NF-κB:IκB complexes (Zhong et al. 1997). Subsequent analysis indicated that in response to specific stimuli PKAc phosphorylates the p65 NF-κB subunit at a consensus PKA phosphorylation site located within the Rel homology domain. Overexpression experiments suggested that mutation of this serine residue at position 276 to alanine (S276A) abolishes the ability of the transfected p65 protein to drive transcription, even though the mutant protein can assemble into complexes with other Rel proteins and IκBs and bind normally to DNA (Zhong et al. 1997, 1998). Further studies showed that phosphorylation of p65 at Ser 276 allows efficient recruitment of the coactivator CREB-binding protein (CBP)/p300 and may, therefore, be a crucial event in the activation of NF-κB (Zhong et al. 1998). Remarkably, phosphorylation was not only required for stable interaction with CBP/p300, but was also required for displacement of repressive histone deacetylase (HDAC) proteins that otherwise associated with unphosphorylated, nuclear NF-κB proteins (Zhong et al. 2002). These findings suggested that the ability of NF-κB to bind to DNA and to recruit histone-modifying enzymes can be uncoupled, and that regulation of chromatin states by NF-κB may exert both positive and negative effects on transcription. Thus, phosphorylation of p65 could act as a switch regulating the association with either CBP/p300 or HDAC, ensuring that only NF-κB that enters the nucleus in response to certain stimuli is able to efficiently drive transcription.
To provide a genetic test for the role of p65 phosphorylation in NF-κB-dependent transcription, we “knocked in” a S276A mutant version of p65 into the mouse genome. Cells expressing the mutant p65 display dramatically reduced NF-κB transcriptional activity, consistent with the failure of S276A p65 to bind coactivators. Surprisingly, however, the phenotype of the embryonic lethality in the knock-in mouse is complex and variegated, and cannot be readily explained by lack of NF-κB activity alone. We hypothesized that expression of the nonphosphorylatable form of NF-κB, which is capable of DNA binding, leads to variegated embryonic phenotypes through epigenetic repression. To explore this hypothesis, we focused on delineating the mechanism(s) underlying the abnormalities exhibited in eye development, which are readily observable in the embryos and are both highly penetrant and variegated. We identified the paired homeobox transcription factor Pax6, which is known to regulate eye development, as being particularly susceptible to epigenetic repression by mutant p65. Ablating the DNA-binding capability of the nonphosphorylatable form of p65 eliminates the variegated phenotypes and results in embryonic lethality due to hepatocyte apoptosis, similar to the phenotype of p65-deficient embryos (Beg et al. 1995). Therefore, our results demonstrate that uncoupling DNA binding of nuclear NF-κB from phosphorylation-dependent coactivator recruitment can lead to variegated developmental defects through epigenetic repression of gene expression.
Results
Replacement of wild-type p65 with a S276A mutant leads to embryonic lethality with variegated phenotypes, including aberrant eye development
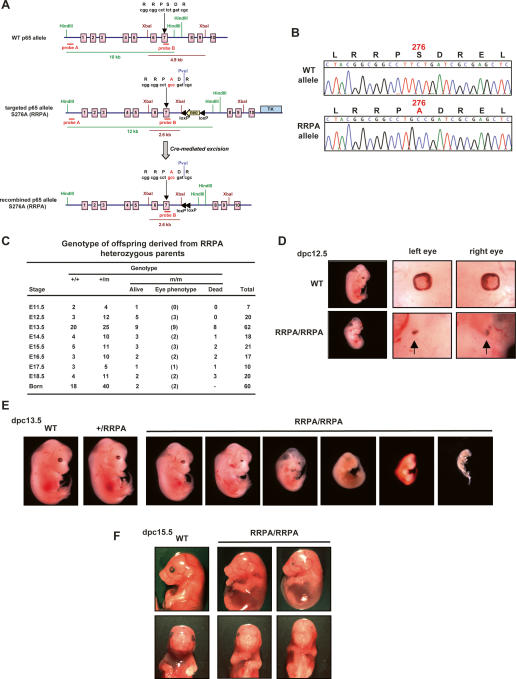
To create the knock-in mouse expressing p65 S276A (RRPA) we used a strategy presented in Figure 1A (and the Supplemental Material). The mutant allele was detected by Northern blot or PCR genotyping (Supplemental Fig. S1A) and was confirmed by sequencing the p65 cDNA (Fig. 1B). There were few homozygotes amongst the progeny derived from interbreeding between heterozygotes (two out of 60 live births) (Fig. 1C), suggesting prenatal lethality of RRPA knock-in embryos. Histological analysis and genotyping of recovered embryos revealed that the vast majority of homozygous embryos died in utero between 11 and 16 d post-coitum (dpc). The majority of embryos appeared normal until 11 dpc; however, homozygous embryos recovered on subsequent days exhibited a variety of phenotypes that ranged from normal-looking embryos to embryos that were nearly resorbed at 13.5 dpc (Fig. 1E). Even amongst the homozygous embryos that developed to between ∼14 and 16 dpc, there were multiple developmental defects, variable in severity, including deformed limbs. Most striking, however, was a frequently recurring, but variable, phenotype concerning the development of the eye. In the majority of the homozygous embryos, there were ocular abnormalities, ranging from two small eyes (Fig. 1D; Supplemental Fig. S1B), to one normal and one small eye (left or right) (data not shown), to only one normal or small eye (left or right) (Fig. 1F), to no eyes (data not shown). In embryos where there was only one eye (Fig. 1F, middle panels), the position of the eye was not altered; i.e., it was not a “cyclops” phenotype (Rebagliati et al. 1998)—rather, it appeared that eye development on one side had terminated. Histopathological analysis of sections of rudimentary eyes from the homozygous embryos revealed formation of the early eye structures, but development apparently failed to proceed to maturity (Supplemental Fig. S1C). The eye phenotypes resemble ocular malformations in humans that are recognizable at birth and are caused by abnormalities in the normal program of development, including anophthalmia (absence of the eye) and extreme microphthalmia (very small eye remnant) (Fitzpatrick and van Heyningen 2005). To minimize the possibility that this variegation might be due to some defect in the embryonic stem (ES) cell line used (this line has been used extensively for generating numerous knockouts/knock-ins and has been carefully karyotyped), we backcrossed the animals extensively with laboratory NZ-B6 strains, obtained periodically from Jackson laboratories. Following many backcross generations, we observed no change in the embryonic lethality with variable phenotypes.
Figure 1.
Generation and phenotypic characterization of p65–S276A (RRPA) knock-in mice. (A) Strategy to generate p65–S276A (RRPA) knock-in mice by homologous recombination. A PvuI restriction enzyme site was coupled to the mutation in the targeted allele for PCR analysis. Restriction enzyme sites for HindIII and XbaI and the probes used for Southern blot analysis were as indicated. (B) Sequencing of p65 cDNA obtained by RT–PCR from wild-type and RRPA homozygous mice. (C) Genotype analysis of offspring derived from RRPA heterozygous parents. The number of live embryos displaying an eye phenotype is indicated in parentheses. (D) Eye development abnormalities in RRPA homozygous embryos at 12.5 dpc. (E) Variegated developmental defects in RRPA homozygous embryos. Embryos at 13.5 dpc are shown, as well as wild-type and RRPA heterozygous control embryos. (F) Eye development abnormalities in RRPA homozygous embryos at 15.5 dpc. The embryo presented in the middle panels has a normal-sized eye on the right side and no discernible eye on the left side. The embryo in the right panels has two small eyes.
Mouse embryonic fibroblasts (MEFs) from RRPA mice display normal pattern and induction of NF-κB complexes but defective expression of many NF-κB-dependent genes
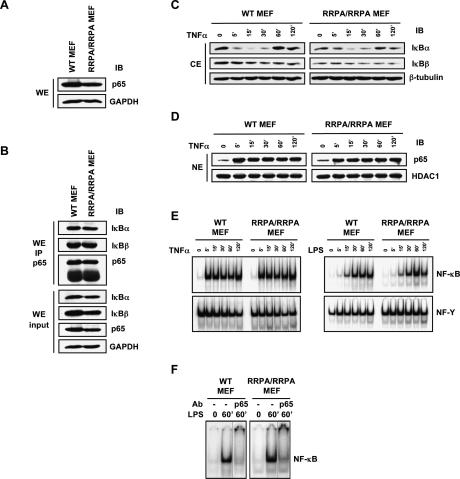
To begin addressing the role of the mutated p65 protein in regulating NF-κB-dependent transcriptional events, we derived MEFs from wild-type and homozygote littermates using 12.5-dpc embryos. The homozygous knock-in fibroblasts could be cultured, albeit with somewhat slower growth kinetics, compared with the wild-type MEFs. The level of p65 protein was nearly identical in wild-type or knock-in MEFs, indicating that the mutation did not significantly affect p65 stability (Fig. 2A). Immunoprecipitation of the p65 protein demonstrated that the mutant behaved identically to the wild-type protein in forming NF-κB:IκBα/β complexes (Fig. 2B), and stimulation with TNFα (Fig. 2C) or lipopolysaccharide (LPS) (data not shown) led to normal kinetics of IκBα and IκBβ degradation. Consistently, nuclear translocation of p65 was equivalent in wild-type and knock-in cells (Fig. 2D), as was the formation of κB-specific gel-shift complexes in response to TNFα or LPS stimulation (Fig. 2E). Supershifting with an antibody directed against p65 indicated that the DNA-bound NF-κB in both wild-type and knock-in MEFs was predominantly p65-containing heterodimers (Fig. 2F).
Figure 2.
Characterization of RRPA knock-in cells. (A) Immunoblot analysis of p65 expression levels in wild-type and RRPA homozygous MEFs generated from embryos at 12.5 dpc. (B) Interaction between p65 and IκBα or IκBβ in MEFs detected by immunoprecipitation. (C) Degradation and resynthesis of IκBα and IκBβ proteins in MEFs following TNFα stimulation (10 ng/mL). (D) p65 translocation in MEFs following TNFα stimulation. (E) EMSA using labeled κB probe and control NF-Y probe. Nuclear extracts (5 μg) from unstimulated MEFs and MEFs stimulated with TNFα or LPS (10 μg/mL) were analyzed. (F) Supershift analysis of NF-κB–DNA-binding complexes was performed using anti-p65 antibody. Nuclear extracts (5 μg) from unstimulated MEFs and MEFs stimulated with LPS for 60 min were analyzed.
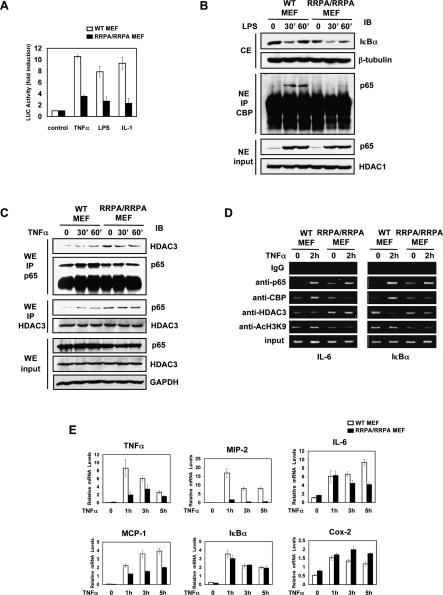
Our previous studies had suggested that phosphorylation of Ser 276 is critical for recruitment of CBP/p300 to nuclear NF-κB following stimulation (Zhong et al. 1998, 2002). CBP (and its close homolog p300) acts as a coactivator by promoting histone acetylation near target promoters, thereby facilitating chromatin unwinding and remodeling. As shown in Figure 3A, NF-κB containing the mutated p65 was dramatically less efficient in driving transcription from a transfected NF-κB luciferase reporter in response to TNFα, LPS, and IL-1. As seen in Figure 3B, the mutant p65 was unable to recruit a significant amount of CBP to nuclear NF-κB-containing complexes and instead can be immunoprecipitated with greater amounts of HDAC3, even in stimulated cells, than wild-type p65 (Fig. 3C). To verify whether the inability of the mutant p65 to recruit CBP was also maintained on promoters of responsive genes, we tested the IL-6 and IκBα promoters using chromatin immunoprecipitation (ChIP) assays. While both wild-type and mutant p65 were recruited to these two promoters, there was significantly less CBP and acetylated histone H3, along with a substantially higher amount of HDAC3, on the promoters in mutant cells relative to wild-type cells (Fig. 3D).
Figure 3.
Impaired transcriptional activity of RRPA mutant protein and the underlying mechanism. (A) Transactivation by RRPA p65 detected by reporter gene assay in MEFs. MEFs were transfected with pBIIx-luc and control construct Renilla for 24 h; stimulated with TNFα, LPS, or IL-1 (10 ng/mL) for 6 h; and analyzed. (B) The ability of p65 to interact with CBP was determined by immunoprecipitation. Immunoblotting for IκBα and nuclear p65 was performed to confirm the stimulation by LPS. (C) The ability of p65 to interact with HDAC3 was detected by immunoprecipitation. MEFs were stimulated with TNFα. (D) ChIP assay was performed with untreated or TNFα-treated (2 h) MEFs using the indicated antibodies. Precipitated IL-6 κB site DNA and IκBα κB site DNA were assayed by semiquantitative PCR. (E) Differential expression of NF-κB-regulated genes in MEFs. Total RNA isolated from unstimulated MEFs or MEFs stimulated with TNFα for 1, 3, or 5 h was quantified by real-time PCR and normalized to the level of GAPDH.
Lastly, we examined the effect of the RRPA mutation on the inducible expression of a number of well-characterized NF-κB target genes. The kinetics of expression of these genes was monitored using real-time PCR (Fig. 3E). Surprisingly, the results indicated that, whereas expression of certain genes was completely blocked (e.g., MIP-2), others were relatively unaffected (e.g., IκBα and Cox-2). Most of the NF-κB-regulated genes tested were affected partially (e.g., TNFα, MCP-1, and IL-6). Such variable responses are consistent with recent reports demonstrating that NF-κB-regulated genes can be classified into distinct groups based on promoter accessibility and their requirement for chromatin remodeling for expression (Saccani et al. 2001; Ramirez-Carrozzi et al. 2006).
RRPA embryos with eye defects and cells expressing p65–RRPA show reduced Pax6 expression
The block in ocular development in the RRPA knock-ins occurs at different stages, suggesting that eye development is interrupted, rather than being altered (Fig. 1D–F). We also observed a striking defect in the development of the ocular mesenchyme, which is reminiscent of previously characterized mutations in the regulatory gene Pax6 (Hanson 2003). The eye phenotypes are similar to those observed in various mutations of the Pax6 gene, including those that have been linked to human anophthalmia (Kleinjan et al. 2001; Hanson 2003). Pax6 encodes a transcription factor that is expressed from the earliest stages of eye morphogenesis in the optic vesicle, a structure that gives rise to the retina and the overlying ectoderm, which later forms the lens and the cornea (Pichaud and Desplan 2002; Kozmik 2005). Remarkably, in both humans and mice, heterozygous mutations in Pax6 lead to striking defects in eye development (Hanson 2003; Fitzpatrick and van Heyningen 2005). Reduction of Pax6 activity in heterozygotes leads to phenotypes such as Small eye in mice and rats (which is similar to the phenotypes observed in the RRPA knock-ins) and Aniridia in humans (Kleinjan et al. 2001). Changes in the level of Pax6 have been shown to alter expression of Pax6-regulated genes that are essential for eye development, including transcription factors (Six3, c-Maf, MafA/L-Maf, and Prox1), structural genes (crystallines and cell adhesion molecules), and signaling molecules affecting the migration of neural crest cells into the eye by inductive processes (Pichaud and Desplan 2002; Kozmik 2005).
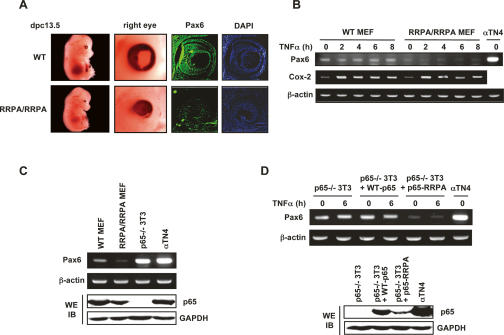
To determine whether Pax6 expression was altered in the knock-in embryos, sections of eyes from 13.5-dpc wild-type and mutant embryos were prepared and immunostained with a Pax6-specific monoclonal antibody (R&D systems). We intentionally chose mutant embryos that displayed relatively normal development until 13.5 dpc except for abnormal eye development (Fig. 4A). As apparent in these sections, eye development is severely disrupted in the mutant embryos, although at least in the embryos chosen for study, the numbers of cells detected by DAPI staining were approximately equivalent. However, there was a dramatic difference in the level of Pax6 staining in the eyes between the wild-type and mutant embryos. Notably, the levels of Pax6 are decreased but not abolished in the mutant knock-ins, similar to the situation in human patients with Pax6 mutations, or in heterozygous knockout strains of Pax6.
Figure 4.
Expression of Pax6 gene was diminished by RRPA mutant protein. (A) Pax6 expression in the eye. Embryonic eye cross-sections (13.5 dpc) were immunostained with the antibody recognizing Pax6 and then detected with secondary antibody coupled to Alexa 488. Nuclei were stained by DAPI. (B) Pax6 expression in untreated MEFs and in MEFs treated with TNFα. Gene expression levels were detected by semiquantitative RT–PCR. Cox-2 was used as a positive control for TNFα stimulation. αTN4 cells are mouse lens epithelial cells in which Pax6 is highly expressed, and were used as a positive control for Pax6 PCR analysis. (C) Pax6 was expressed at a high level in p65-deficient 3T3 cells detected by semiquantitative RT–PCR. p65 expression levels detected by immunoblotting are displayed in the bottom panels. (D) Pax6 expression was inhibited by RRPA mutant protein in stable cell lines. Wild-type p65 or p65-RRPA mutant was stably introduced into p65-deficient 3T3 cells. Pax6 levels were detected by semiquantitative RT–PCR. p65 expression levels detected by immunoblotting are displayed in the bottom panels.
The reduction in levels of Pax6 expression in mutant embryos strongly suggests that the basis of the eye phenotype in the RRPA knock-ins might be dysregulation of Pax6 expression. However, Pax6 expression has not been reported to be subject to regulation by NF-κB and it is unlikely that expression of Pax6 could be directly regulated by NF-κB, as none of the NF-κB knockouts—including the p65, c-Rel, RelB, p50, p52 (and their double and triple knockouts)—display any eye development phenotype. In addition, activation of NF-κB in wild-type MEFs leads to no detectable changes in the level of Pax6 expression (Fig. 4B, wild-type MEFs). On the other hand, the RRPA knock-in MEFs had dramatically reduced Pax6 expression, which was not changed upon TNFα stimulation, suggesting that the effect on Pax6 levels seen in the RRPA knock-in mice is probably mediated through indirect mechanisms (Fig. 4B, RRPA/RRPA MEFs).
To more definitively establish a link between Pax6 expression and the RRPA mutant p65, which was independent of the knock-in mice and MEFs derived from them, we decided to use p65 knockout 3T3 fibroblasts and stably reconstitute them with either wild-type or the RRPA mutant p65. As the p65 knockouts had been generated using a completely different ES clone (Beg et al. 1995), it also eliminated the possibility that the effect on Pax6 expression might be influenced by the genetic background of the ES cell lines. 3T3 fibroblasts derived from traditional p65 knockouts have normal levels of Pax6 expression, while cells from the RRPA knock-ins display a dramatic reduction in Pax6 levels, confirming that Pax6 is not normally regulated by p65 (Fig. 4C; data not shown). The same p65 knockouts stably reconstituted with wild-type p65 display normal levels of Pax6 (Fig. 4D, lanes 3,4), whereas reconstitution with the mutant RRPA p65 dramatically reduces the level of Pax6 expression (Fig. 4D, lanes 5,6). In agreement with the results seen in the MEFs (Fig. 4B), TNFα stimulation has no effect on Pax6 expression in these reconstituted cell lines (Fig. 4D). This result strongly suggests that while in normal cells NF-κB does not affect Pax6 expression, expression of the RRPA mutant inhibits Pax6 expression, most likely through indirect, epigenetic mechanisms. This epigenetic repression of Pax6 expression could also therefore help explain the variegated defects in eye development observed in the RRPA knock-in mice.
Inhibition of Pax6 expression by the RRPA mutant p65 can be reversed by treatment with an HDAC inhibitor
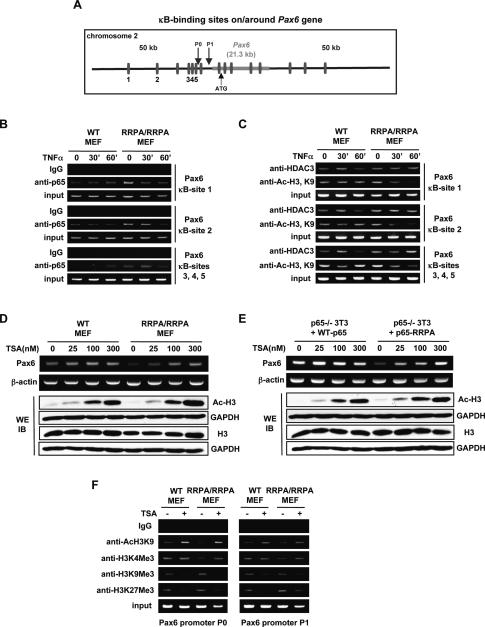
A possible explanation for how Pax6 expression might be inhibited by the RRPA p65 mutant is through binding of the mutant NF-κB to distal segments in the chromosome containing Pax6. The RRPA mutant p65-containing complexes might nucleate HDAC-containing, repressive chromatin-modifying complexes that could inhibit normal Pax6 expression by epigenetic mechanisms. We examined the DNA sequence of the chromosome around the Pax6 gene and identified a number of putative NF-κB sites upstream of, downstream from, and within the Pax6 gene (Fig. 5A). Because expression of Pax6 has been reported to be influenced by regulatory elements >150 kb away from the gene (Kleinjan et al. 2001), it is possible that binding of the RRPA mutant NF-κB to any or all of these sites could influence its expression. For experimental convenience, we focused on a few of the upstream sites (sites 1–5): Site 1 (at position −35 kb) and site 2 (at position −22.8 kb) were analyzed individually, whereas sites 3, 4, and 5 are localized within an ∼0.5-kb fragment, and hence were analyzed together (Fig. 5A; Supplemental Fig. S2A). ChIP analysis revealed that sites 1 and 2 bound to low amounts of p65 in unstimulated, wild-type MEFs, while the fragment containing sites 3–5 was bound to far less p65. In mutant cells, the amounts of p65 bound to these sites were greater (Fig. 5B), and there was a corresponding increase in the recruitment of HDAC3 (Fig. 5C). We also stimulated the MEFs with TNFα to increase the amount of NF-κB in the nucleus, and used ChIP to examine histone acetylation around these sites. While no changes were observed in the wild-type cells, acetylation at histone H3 Lys 9 (K9), which is a marker of active chromatin, was significantly reduced when the mutant cells were stimulated with TNFα (Fig. 5C). A likely explanation is that some of the newly imported RRPA p65 binds to the NF-κB sites around Pax6 and, by bringing in additional HDACs, begins to trigger deacetylation and compaction of the surrounding chromatin. Out of practical necessity, we had to limit our analysis to a few sites that may or may not be the main sites affected by the mutant p65, and additional studies are necessary to better analyze the state of chromatin around the Pax6 gene.
Figure 5.
RRPA mutant p65 affects Pax6 gene expression through epigenetic mechanisms. (A) κB-binding sites on/around the Pax6 gene on chromosome 2 in the mouse genome. (B) Binding of p65 to κB sites upstream of the Pax6 gene. ChIP assay was performed in MEFs using antibody against p65. (C) HDAC3 and acetylated H3K9 levels at the κB sites upstream of the Pax6 gene were determined by ChIP assay. (D) Impaired Pax6 expression in RRPA homozygous MEFs was rescued by TSA. MEFs were treated with TSA for 18 h and Pax6 expression was determined by semiquantitative RT–PCR. (E) Impaired Pax6 expression in p65-deficient 3T3 cells stably transfected with RRPA mutant was rescued by TSA. Cells were treated with TSA for 18 h and Pax6 expression was determined by semiquantitative RT–PCR. (F) Analysis of histone modifications on Pax6 promoters P0 and P1 using ChIP assays.
These ChIP analyses implicate recruitment of HDACs by the mutant p65, and we therefore reasoned that treatment with trichostatin A (TSA), a well-characterized HDAC inhibitor, should reverse the observed inhibition of Pax6 expression. Wild-type or knock-in MEFs (Fig. 5D) and p65 knockout 3T3 cells reconstituted with wild-type or RRPA mutant p65 (Fig. 5E) were treated with increasing amounts of TSA, and Pax6 expression was analyzed using RT–PCR. While TSA did not affect Pax6 expression in wild-type MEFs or rela−/− 3T3 cells reconstituted with wild-type p65, treatment with TSA led to a striking increase of Pax6 expression in the knock-in MEFs (Fig. 5D; Supplemental Fig. S2B) and rela−/− 3T3 cells reconstituted with the RRPA mutant p65 (Fig. 5E; Supplemental Fig. S2C). These results are therefore fully in agreement with our proposal that recruitment of HDACs by the RRPA mutant p65 is responsible for epigenetic repression of Pax6 expression.
Repression induced by recruitment of HDACs triggers a series of modifications on histones, including methylation of specific lysine residues. These modifications can mark either actively transcribed genes or repressed/inactive genes. We used ChIP assays to examine two activation marks and two repression marks on the two well-characterized promoters of the mouse Pax6 gene (Xu et al. 1999). The activation marks were acetylation at Lys 9 and trimethylation of Lys 4 on histone H3. The repression marks analyzed were trimethylation of Lys 9 and Lys 27 on histone H3. Consistent with the data in Figure 3D, the activation marks on H3K4 and H3K9 are significantly reduced in the RRPA knock-ins (Fig. 5F). More dramatic, however, were the changes upon TSA treatment, which led to increased acetylation on H3K9, increased methylation on H3K4, and more significant loss of methylation on H3K9 and H3K27. These results, therefore, strongly suggest that the RRPA mutant induces changes around the Pax6 promoters that are consistent with the mechanism of increased HDAC recruitment.
Genome-wide expression analysis identifies genes that are epigenetically affected by aberrant recruitment of HDACs in the RRPA knock-ins
Although the defects in eye development are the most pronounced, and hence easiest to trace back to aberrant expression of a specific effector gene, it is clear from the overall variegation of the phenotype that a large number of additional genes must also be affected. To identify such genes we performed a genome-wide Affymetrix microarray expression analysis using RNA isolated from wild-type and RRPA knock-in cells, either untreated or treated with TSA. Our expectation was that expression of genes affected epigenetically would be reduced in the knock-in cells and, if this reduction was due to aberrant recruitment of HDACs, then expression of many of these genes should be rescued with TSA treatment. As shown in Figure 6, we identified 133 genes that fell into this category (Group I and Group II) with differing degrees of TSA-induced recovery. Genes listed in Group I (75 genes) had at least twofold increase upon treatment with TSA in the knock-in cells, while expression of genes in Group II (58 genes) was not affected by TSA. Therefore, Group I genes resemble Pax6 most closely in that their RRPA-induced repression can be reversed by TSA; however, genes in Group II are also epigenetically affected by the mutant p65, as none of these genes are known to be regulated by NF-κB. The vast majority of genes that are expressed at detectable levels in unstimulated wild-type cells (∼60% of the genes on the microarray) are unaffected by the knock-in (Group III). Interestingly, there are a number of homeobox genes (Hoxd8, Hoxa9, Hoxd9, Hoxd10, and Hoxd13) in Group II whose expression is dramatically repressed in the RRPA knock-in and fails to be rescued by TSA. The wide variety of developmental defects seen in the knock-in embryos therefore could result from repression of these Hox genes. Hence, these results suggest that the epigenetic, variegated effect is manifested over a subset of genes located on different chromosomes, which are probably united in possessing certain features that lead to their repression by HDAC recruited by the mutant p65 protein.
Figure 6.
Comparison of gene expression in wild-type versus RRPA knock-in cells using Affymetrix microarrays. Gene expression levels in untreated MEFs or in MEFs treated with TSA (300 nM, 18 h) were analyzed. Representative genes in each of three groups were hierarchically clustered and displayed, including 75 genes in Group I that were significantly repressed in RRPA/RRPA MEFs and could be rescued by TSA, 58 genes in Group II that were significantly repressed in RRPA/RRPA MEFs but not rescued by TSA, and 20 representative genes shown for Group III that were not significantly affected in RRPA MEFs. Each row represents a single gene, and each column represents an experimental sample. The ratio of the abundance of transcripts of each gene to the median abundance of the gene’s transcript among all of the samples is represented by the color in the matrix as indicated. Relative expression levels are shown at the bottom. Homeobox genes are marked with a pink box.
Knock-in of a nonphosphorylatable form of p65 that cannot bind DNA abolishes the variegated embryonic phenotype
If the variegated phenotype of the RRPA knock-in mice is due to the recruitment of transcriptional corepressor complexes, rather than simply a failure of the mutant NF-κB to drive transcription, then abolishing the ability of the nonphosphorylatable p65 mutant to bind to DNA should eliminate the variegation of the phenotype observed. Testing of this hypothesis was facilitated by the serendipitous discovery that alteration of arginine at position 274 of p65 to alanine (in addition to changing the serine at position 276 to alanine; i.e., changing RRPS to RAPA) dramatically decreases the ability of this mutant p65 to bind DNA as a heterodimer with p50 (Fig. 7A). This RAPA mutation, however, does not affect the stability of p65 or its ability to dimerize with p50 or interact with IκBs (data not shown).
Figure 7.
Characterization of p65–R274A–S276A (RAPA) knock-in mice. (A) p50 (lane 1), wild-type p65 (lane 2), p50 and wild-type p65 (lane 3), or p50 and RAPA p65 mutant (lane 4) were transiently transfected into rela−/− 3T3 cells. EMSA using labeled κB probe was performed with nuclear extracts of transfected cells. (B) Sequencing of p65 cDNA obtained by RT–PCR from wild-type and RAPA homozygous mice. (C) Genotype analysis of offspring derived from RAPA heterozygous parents. (D) Wild-type and RAPA/RAPA embryos from 15.5 dpc are shown. The RAPA embryos develop normally and are in general of similar size as the wild-type embryos. (E) Haematoxylin and eosin staining for sections of liver from 15.5-dpc wild-type and RAPA/RAPA embryos is shown. The massive apoptosis in the RAPA/RAPA knock-in livers is apparent. (F) p65 translocation in MEFs upon LPS stimulation detected by immunoblotting of nuclear extracts. (G) EMSA using labeled κB probe and control NF-Y probe. (Top panels) Nuclear extracts (5 μg) from unstimulated MEFs and MEFs stimulated with TNFα or LPS were analyzed. (Bottom panels) Degradation and resynthesis of IκBα protein in MEFs following TNFα or LPS stimulation. (H) Supershift analysis of NF-κB–DNA-binding complexes was performed using indicated antibodies. Nuclear extracts (5 μg) from unstimulated MEFs and MEFs stimulated with TNFα were analyzed. In RAPA MEFs, p50:p65–RAPA complex failed to bind to DNA; instead, the major NF-κB complex on DNA is p50:c-Rel. (I) Transactivation by RAPA p65 detected by reporter gene assay in MEFs. MEFs were transfected with pBIIx-luc and control Renilla construct for 24 h and were stimulated with TNFα, IL-1, or LPS for 6 h, and luciferase activity was analyzed. (+/+) Wild-type; (+/m) +/RAPA; (m/m) RAPA/RAPA.
To address the hypothesis that the variegated phenotype could be abolished by the non-DNA-binding RAPA mutant p65, we created an additional knock-in strain of mice expressing this form of p65 using a strategy similar to that used to generate the RRPA knock-in mice (Supplemental Fig. S3A). The mutant allele was genotyped using an introduced BanI digestion site (Supplemental Fig. S3B), and was further confirmed by RT–PCR and DNA sequencing (Fig. 7B). As might be predicted for a mutant form of p65 that fails to bind to DNA, nearly all of the RAPA knock-in mice die during embryogenesis (Fig. 7C) at ∼15 dpc due to massive hepatocyte apoptosis (Fig. 7D,E), similar to the phenotype of p65-deficient mice (Beg et al. 1995). No variegation of the phenotype was observed in these mice. MEFs were derived from these embryos, and immunoblotting analysis indicated that the amount of RAPA mutant protein was somewhat reduced compared with p65 in wild-type MEFs (Supplemental Fig. S3C). Immunoprecipitation analysis showed that the RAPA p65 forms cytosolic complexes with IκBα and IκBβ (Supplemental Fig. S3C) and stimulation of these MEFs leads to degradation of IκBα (Fig. 7G, bottom panels). Consistent with the lower amount of p65 in these cells, the amount of p65 that translocates into the nucleus is also reduced (Fig. 7F; Supplemental Fig. S3D); however, the nuclear complexes detected by EMSA were dramatically reduced (Fig. 7G, top panels). Supershifting of these weak nuclear complexes from the RAPA MEFs indicated that they were composed predominantly of p50 and c-Rel, and did not contain p65, confirming that p65–RAPA complexes do not bind DNA (Fig. 7H, right half). This resulted in an inability to drive NF-κB-reporter gene expression (Fig. 7I), and a failure to resynthesize IκBα following stimulation with TNFα or LPS (Fig. 7G, bottom panels). The cells were also highly susceptible to TNFα-induced apoptosis (Supplemental Fig. S3E), which explains the observed embryonic hepatocyte apoptosis. Therefore, the absence of variegated phenotype in the RAPA knock-in mice is consistent with the model that epigenetic repression of genes such as Pax6 in the RRPA knock-in mice is dependent on the mutant protein binding to DNA, thus allowing recruitment of repressive complexes to the chromosome.
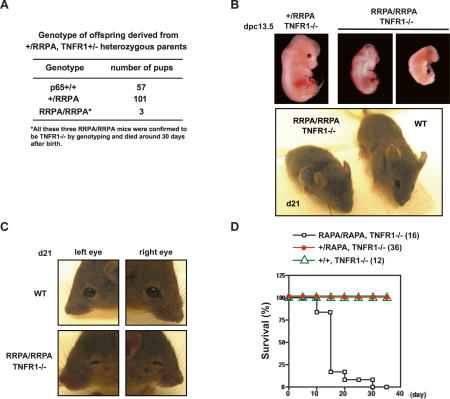
TNFR1 knockouts rescue the embryonic lethality of RAPA mutants, but not the majority of RRPA mutants
Although the variegated developmental defect is the most striking feature of the RRPA knock-in mice, it is important to note that almost all of the embryos die in utero. Some of the embryos that develop normally up to ∼16 dpc begin to exhibit liver hemorrhage, probably as a result of hepatocyte apoptosis, consistent with the finding that the RRPA p65 is quite crippled in transcriptional activity (Fig. 3A). However, other embryos that do not display such overt signs of liver apoptosis also fail to develop and die during embryogenesis. Hepatocyte apoptosis has been observed in p65 (Beg et al. 1995), IKKβ (Q. Li et al. 1999; Z.W. Li et al. 1999), and NEMO knockouts (Makris et al. 2000; Rudolph et al. 2000; Schmidt-Supprian et al. 2000), and is a result of TNFα-mediated apoptosis in the developing liver. Hence, crossing the p65 knockouts with TNF receptor 1 knockouts prevents hepatocyte apoptosis, allowing the mice to survive for a few months in clean, barrier facilities (Alcamo et al. 2001). To determine whether sensitivity to TNFα-mediated apoptosis contributes to the embryonic death observed in the RRPA knock-in mice, we crossed a TNFR1 knockout mouse strain with the RRPA knock-in mice. As predicted by our model, this cross failed to rescue the variegated embryonic lethality (Fig. 8A,B), and out of 26 independent litters only three mice with the genotype RRPA/RRPA, TNFR1−/− were obtained (these mice died ∼1 mo after birth). Remarkably, the rescued mice exhibited a strong eye phenotype and were blind (Fig. 8B,C). Therefore, while crossing with the TNFR1 knockout rescues the few RRPA knock-in embryos that would otherwise die due to hepatocyte apoptosis, these are still susceptible to the epigenetic repression of developmental genes affecting eye development. Further, the epigenetic repression mediated by RRPA p65 affects multiple crucial developmental pathways because only a small proportion of the RRPA/RRPA TNFR1−/− mice survive to birth. In contrast, crossing the RAPA knock-in mice with the TNFR1−/− mice leads to complete rescue of the embryonic lethality, supporting the idea that the variegated embryonic lethality is a consequence of DNA binding by the phosphorylation site mutant form of p65 (Fig. 8D). Therefore, it appears that unphosphorylated, nuclear p65 may be capable of broadly exerting epigenetic repression, as suggested by the microarray results (Fig. 6).
Figure 8.
Rescue of p65 knock-in mice by TNFR1 knockout. (A) Genotype analysis of offspring derived from +/RRPA, TNFR1+/− heterozygous parents. Among 161 pups from 26 litters, only three RRPA/RRPA homozygous mice were rescued by introducing the TNFR1 knockout. The three rescued mice had eye developmental defects, small body size, and lethargic behavior, and died ∼30 d after birth. (B) Knocking out TNFR1 could not rescue the phenotype induced by RRPA mutant protein in the majority of RRPA homozygous mice. (C) Eye developmental defect in RRPA/RRPA, TNFR1−/− mice. (D) Survival curve of TNFR1 knockout rescued RAPA homozygous mice.
Discussion
Inducible nuclear translocation of NF-κB has served as a model for explaining the regulation of many inducible transcription factors (Ziegler and Ghosh 2005). It has, however, become apparent that post-translational modifications of nuclear NF-κB also play an important role in determining the ability of nuclear NF-κB to drive transcription (Chen and Greene 2004; Hayden and Ghosh 2004; Vermeulen et al. 2006). Our previous studies have established the importance of phosphorylation at Ser 276 of NF-κB p65 in regulating the recruitment of the coactivator/histone acetylase, CBP/p300 (Zhong et al. 1997, 1998, 2002). Unphosphorylated nuclear NF-κB, in contrast, binds to HDACs, and therefore phosphorylation of NF-κB p65 acts as a switch to determine whether nuclear NF-κB promotes or inhibits transcription. We previously proposed that this additional layer of regulation helps ensure that only NF-κB that translocates to the nucleus in response to external signals, but not NF-κB that is present in the nucleus of unstimulated cells, is able to drive transcription.
Despite a significant amount of previous work using biochemical and molecular approaches that has supported the hypothesis that phosphorylation regulates NF-κB activity, genetic studies definitively establishing the biological importance of such a regulatory process in NF-κB function have been lacking. The major impediment to genetic analyses of this problem has been the multiplicity of protein kinases implicated in phosphorylation of NF-κB p65. Therefore, instead of targeting the protein kinases, we decided to mutate Ser 276 to alanine and “knock in” the mutated p65 protein into the germline. As our prior studies had indicated that the major role for Ser 276 was to promote recruitment of CBP/p300, we anticipated that mice expressing the S276A mutant would fail to properly express the subset of NF-κB p65-regulated genes that are dependent on CBP/p300. Consistent with our expectations, we observed that following stimulation of the knock-in RRPA cells, some NF-κB-regulated genes (e.g., MIP-2) were completely inhibited in their expression, while others (e.g., IκBα) were unaffected. Most other NF-κB-regulated genes lay within these extremes, with interesting variations in their pattern of expression. Thus, some genes appeared to be induced rapidly in the knock-in cells, but then their expression was gradually reduced compared with that in wild-type cells (e.g., IL-6). Interestingly, these results do not correlate with the accessibility status of the genes examined, as reported in previous studies (Saccani et al. 2001), and hence it appears that accessibility of NF-κB-dependent promoters is not solely determined by recruitment of histone acetylases such as CBP. This knock-in therefore represents a unique model for analyzing the differential requirement for NF-κB p65 coactivator binding for the expression of specific target genes. We are currently making efforts to understand the basis for the differential effects on specific genes described here to determine more precisely how NF-κB interactions with the chromatin-modifying machinery affect gene expression.
A surprising finding in these studies was the variegation of embryonic phenotypes that resulted in nearly complete embryonic lethality in the RRPA knock-in mice. We believe that these phenotypic effects are due to an epigenetic phenomenon similar to that invoked to explain “position-effect variegation” (PEV) in Drosophila (Wakimoto 1998). In the classical description of PEV, a translocation event places the white eye color gene adjacent to heterochromatin, and a popular model invokes linear spreading of repression from the heterochromatin to affect the expression of the translocated gene. The degree of repression varies between individual cells, resulting in a mosaic effect seen in the color of individual eye cells. While details of the mechanism by which PEV occurs remains an area of active research, linear spread of repression is a generally accepted model for the generation of variable phenotypes (Wakimoto 1998; Schotta et al. 2003).
The phenotype of the S276A embryos displays features resembling PEV, although the effects are not limited to a single gene. The knock-in embryos display a large number of variable phenotypes, including lack of limbs, abnormal head development, or more serious developmental defects, leading to early resorption of the embryos (Fig. 1). We therefore hypothesize that the DNA-bound RRPA NF-κB:HDAC complex exerts epigenetic repression on neighboring genes (Fig. 9). We suspect that the sites that exert the most epigenetic repression share certain common characteristics, including an open chromatin conformation and a cluster of NF-κB sites, which would thereby allow recruitment of a large amount of HDAC proteins. The spread of repression from such cis-elements is variable, however, thereby leading to differing degrees of severity in individual embryos. The microarray results support this hypothesis and indicate that basal expression of a significant number of genes that are not normally regulated by NF-κB is dramatically repressed in the RRPA knock-in cells. The inclusion of a number of Hox genes in the list of repressed genes probably helps explain the wide variety of embryonic developmental defects observed. However, the frequent occurrence of defects in eye morphology might be both because this defect is compatible with life, and because a region with multiple NF-κB-binding sites that exists in an accessible conformation ∼11 dpc is located relatively near Pax6, a gene that is known to play a critical role in eye development (Kozmik 2005). Consequently, the expression of Pax6 is affected in the knock-in; however, the degree to which it is affected determines the severity of the eye phenotype. Pax6 expression is regulated by two promoters that specify tissue and developmental stage-specific expression, each of which is located upstream of the gene (Anderson et al. 2002). While neither of these promoters has been shown to be dependent on NF-κB, Pax6 gene expression has been shown to be regulated by an element located ∼150 kb downstream from the gene. This sensitivity of Pax6 expression to long-range elements may explain its susceptibility to mechanisms of epigenetic repression. As haplotype insufficiency of Pax6 has been shown to be responsible for eye development defects in humans (Hanson 2003), even a partial reduction in Pax6 levels is likely to give rise to the observed phenotypes.
Figure 9.
A model of the mechanism through which p65 epigenetically affects gene expression. Upon stimulation, p50:p65 heterodimers containing phosphorylated p65 at Ser 276 enter the nucleus and recruit the transcription coactivator CBP/p300 to κB-binding sites on DNA, leading to normal gene transcription. In contrast, p50:p65–RRPA heterodimers lacking phosphorylation of Ser 276 cannot bind to CBP/p300 efficiently and instead recruit HDAC3 corepressor complexes to DNA, resulting in direct repression of a subset of NF-κB target genes and repression of non-NF-κB-regulated genes through epigenetic mechanisms. p50:p65–RAPA heterodimers cannot bind to DNA and thus cannot mediate the epigenetic effects induced by RRPA mutant p65.
One of the interesting questions raised by our analysis is the source of the inducing signals that lead to accumulation of nuclear complexes that bind to κB sites during embryonic development. There are, in fact, many stimuli of NF-κB that are active during development, although these have not been thoroughly characterized in most settings. This was suggested from studies using a NF-κB-driven lacZ reporter mouse that showed NF-κB activation in early embryos, including the brain, beginning embryonic day 12.5 (Schmidt-Ullrich et al. 1996). Although it is nearly impossible to know which NF-κB-inducing signals might be at play in the current model, nevertheless, it is the aberrant response to such stimuli that manifests as the embryonic lethality exhibited by several NF-κB-deficient mouse models. Therefore, NF-κB stimuli are present and of great importance during embryonic development. It is also possible that MEFs, both in the course of embryonic development as well as during derivation ex vivo, might have been exposed to both specific NF-κB-inducing stimuli as well as less specific cell stressors that could have led to low levels of NF-κB activation. We believe that it is the persistent nuclear localization of the unphosphorylated p65 complexes that is responsible for epigenetic repression of gene expression. In addition, cells that are currently unstimulated but may have been exposed previously to diverse stimuli may maintain low levels of nuclear complexes, which, if of the S276A variety, would begin to exert epigenetic repression.
The ability to recapitulate the inhibition of Pax6 seen in the knock-ins, by stably reconstituting p65 knockout 3T3 cells with vectors expressing the RRPA mutant p65, opens up the possibility of better elucidating the mechanism responsible for epigenetic regulation of Pax6 expression. As Pax6 is clearly not directly regulated by NF-κB, since its expression is unaffected by either removal of NF-κB activity or activation of NF-κB, the inhibitory effect of the RRPA mutant must therefore be through indirect mechanisms. Since HDAC inhibitors can reverse the observed inhibition, it is very likely that the observed recruitment of HDACs by the mutant p65 is responsible for the effect on Pax6, which therefore must be epigenetic in nature. While numerous transcription factors have been implicated in PEV, these effects have been generally dependent on transcription factor dose; i.e., genetic ablation or transgenic overexpression. Therefore, we anticipate that examining Pax6 regulation in the S276A knock-in system will provide a novel system for studying the principles of epigenetic silencing of mammalian gene expression (Lewin 1998).
Materials and methods
Immunoblotting, immunoprecipitation, and ChIP
For immunoblotting, whole-cell lysates or cytosolic and nuclear extracts were resolved in SDS-PAGE gels, transferred to PVDF membranes, and incubated with the following antibodies: anti-p65 (Biomol); anti-IκBα, anti-IκBβ, and anti-HDAC3 (Santa Cruz Biotechnology); anti-HDAC1 and anti-β-tubulin (Sigma); anti-GAPDH (Fitzgerald); anti-H3 (Abcam); and anti-Ac-H3 (Upstate Biotechnology). Immunoprecipitation was performed as described previously (Marienfeld et al. 2003). Anti-CBP used for immunoprecipitation was from Santa Cruz Biotechnology. ChIP assays were performed essentially according to manufacturer’s instructions (Upstate Biotechnology). Anti-p65, anti-CBP, and anti-HDAC3 antibodies were as described above. Anti-AcH3K9, anti-H3K9Me3, and anti-H3K27Me3 antibodies and mouse IgG were from Upstate Biotechnology. Anti-H3K4Me3 antibody was from Abcam. The extracted DNA was used for semiquantitative PCR to amplify the DNA fragments of interest using HotStarTaq DNA polymerase (Qiagen). Primer sequences used are available upon request.
Luciferase reporter assays
MEFs generated from wild-type, RRPA knock-in, or RAPA knock-in embryos were transfected with FuGene6 (Roche). Luciferase activities of total cell lysates were measured using the dual-luciferase reporter assay system (Promega) following the manufacturer’s instructions.
Real-time PCR and semiquantitative PCR analyses
Total RNA was isolated from cultured cells using RNeasy mini kit (Qiagen) and reverse-transcribed with QuantiTect reverse transcription system (Qiagen) to produce cDNA. cDNA was used as template for PCR analysis. Real-time PCR was performed using QuantiTect SYBR green PCR mix (Qiagen) with Stratagene Mx3000P QPCR system following instructions. Semiquantitative PCR was carried out with HotStarTaq DNA polymerase. Primer sequences are available upon request.
Histology and immunostaining
Tissues were fixed in Streck tissue fixative (STRECK), and paraffin-embedded tissue sections (5 μm) were stained with haematoxylin and eosin using standard methods. Cryostat sections (5 μm) containing the eye were prepared, fixed with acetone, and immunostained with a Pax6-specific monoclonal antibody (R&D systems) and secondary antibody coupled to Alexa 488 (Molecular Probes). Sections were counterstained with DAPI to visualize nuclei.
Acknowledgments
We thank Dr. Tian Chi, Mr. Phillip West, and Dr. Brian Poligone for carefully reading the manuscript, and Ms. Crystal Bussey for managing the mouse colony. We also thank Dr. Paul Russell, NEI, NIH for providing the αTN4 cell line. Analysis of the Affymetrix microarrays was carried out at the Yale W.M. Keck Biostatistics Resource by Dr. Aiping Lin. The research in this paper was supported by grants from the National Institutes of Health (R37-AI33443 and RO1-AI066109).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1657408.
References
- Alcamo E., Mizgerd J.P., Horwitz B.H., Bronson R., Beg A.A., Scott M., Doerschuk C.M., Hynes R.O., Baltimore D. Targeted mutation of TNF receptor I rescues the RelA-deficient mouse and reveals a critical role for NF-κB in leukocyte recruitment. J. Immunol. 2001;167:1592–1600. doi: 10.4049/jimmunol.167.3.1592. [DOI] [PubMed] [Google Scholar]
- Anderson T.R., Hedlund E., Carpenter E.M. Differential Pax6 promoter activity and transcript expression during forebrain development. Mech. Dev. 2002;114:171–175. doi: 10.1016/s0925-4773(02)00051-5. [DOI] [PubMed] [Google Scholar]
- Baldwin A.S. The NF-κB and I κB proteins: New discoveries and insights. Annu. Rev. Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- Beg A.A., Sha W.C., Bronson R.T., Ghosh S., Baltimore D. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κB. Nature. 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- Chen L.F., Greene W.C. Shaping the nuclear action of NF-κB. Nat. Rev. Mol. Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D.R., van Heyningen V. Developmental eye disorders. Curr. Opin. Genet. Dev. 2005;15:348–353. doi: 10.1016/j.gde.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Ghosh S., May M.J., Kopp E.B. NF-κB and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Hanson I.M. PAX6 and congenital eye malformations. Pediatr. Res. 2003;54:791–796. doi: 10.1203/01.PDR.0000096455.00657.98. [DOI] [PubMed] [Google Scholar]
- Hayden M.S., Ghosh S. Signaling to NF-κB. Genes & Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Kleinjan D.A., Seawright A., Schedl A., Quinlan R.A., Danes S., van Heyningen V. Aniridia-associated translocations, DNase hypersensitivity, sequence comparison and transgenic analysis redefine the functional domain of PAX6. Hum. Mol. Genet. 2001;10:2049–2059. doi: 10.1093/hmg/10.19.2049. [DOI] [PubMed] [Google Scholar]
- Kozmik Z. Pax genes in eye development and evolution. Curr. Opin. Genet. Dev. 2005;15:430–438. doi: 10.1016/j.gde.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Lewin B. The mystique of epigenetics. Cell. 1998;93:301–303. doi: 10.1016/s0092-8674(00)81154-x. [DOI] [PubMed] [Google Scholar]
- Li Q., Van Antwerp D., Mercurio F., Lee K.F., Verma I.M. Severe liver degeneration in mice lacking the IκB kinase 2 gene. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- Li Z.W., Chu W., Hu Y., Delhase M., Deerinck T., Ellisman M., Johnson R., Karin M. The IKKβ subunit of IκB kinase (IKK) is essential for nuclear factor κB activation and prevention of apoptosis. J. Exp. Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makris C., Godfrey V.L., Krahn-Senftleben G., Takahashi T., Roberts J.L., Schwarz T., Feng L., Johnson R.S., Karin M. Female mice heterozygous for IKKγ/NEMO deficiencies develop a dermatopathy similar to the human X-linked disorder incontinentia pigmenti. Mol. Cell. 2000;5:969–979. doi: 10.1016/s1097-2765(00)80262-2. [DOI] [PubMed] [Google Scholar]
- Marienfeld R., May M.J., Berberich I., Serfling E., Ghosh S., Neumann M. RelB forms transcriptionally inactive complexes with RelA/p65. J. Biol. Chem. 2003;278:19852–19860. doi: 10.1074/jbc.M301945200. [DOI] [PubMed] [Google Scholar]
- Pichaud F., Desplan C. Pax genes and eye organogenesis. Curr. Opin. Genet. Dev. 2002;12:430–434. doi: 10.1016/s0959-437x(02)00321-0. [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi V.R., Nazarian A.A., Li C.C., Gore S.L., Sridharan R., Imbalzano A.N., Smale S.T. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes & Dev. 2006;20:282–296. doi: 10.1101/gad.1383206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebagliati M.R., Toyama R., Haffter P., Dawid I.B. cyclops encodes a nodal-related factor involved in midline signaling. Proc. Natl. Acad. Sci. 1998;95:9932–9937. doi: 10.1073/pnas.95.17.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph D., Yeh W.C., Wakeham A., Rudolph B., Nallainathan D., Potter J., Elia A.J., Mak T.W. Severe liver degeneration and lack of NF-κB activation in NEMO/IKKγ-deficient mice. Genes & Dev. 2000;14:854–862. [PMC free article] [PubMed] [Google Scholar]
- Saccani S., Pantano S., Natoli G. Two waves of nuclear factor κB recruitment to target promoters. J. Exp. Med. 2001;193:1351–1359. doi: 10.1084/jem.193.12.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Supprian M., Bloch W., Courtois G., Addicks K., Israel A., Rajewsky K., Pasparakis M. NEMO/IKKγ-deficient mice model incontinentia pigmenti. Mol. Cell. 2000;5:981–992. doi: 10.1016/s1097-2765(00)80263-4. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ullrich R., Mémet S., Lilienbaum A., Feuillard J., Raphaël M., Israel A. NF-κB activity in transgenic mice: Developmental regulation and tissue specificity. Development. 1996;122:2117–2128. doi: 10.1242/dev.122.7.2117. [DOI] [PubMed] [Google Scholar]
- Schotta G., Ebert A., Dorn R., Reuter G. Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila. Semin. Cell Dev. Biol. 2003;14:67–75. doi: 10.1016/s1084-9521(02)00138-6. [DOI] [PubMed] [Google Scholar]
- Vermeulen L., Vanden Berghe W., Haegeman G. Regulation of NF-κB transcriptional activity. Cancer Treat. Res. 2006;130:89–102. [PubMed] [Google Scholar]
- Wakimoto B.T. Beyond the nucleosome: Epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- Xu P.X., Zhang X., Heaney S., Yoon A., Michelson A.M., Maas R.L. Regulation of Pax6 expression is conserved between mice and flies. Development. 1999;126:383–395. doi: 10.1242/dev.126.2.383. [DOI] [PubMed] [Google Scholar]
- Zhong H., SuYang H., Erdjument-Bromage H., Tempst P., Ghosh S. The transcriptional activity of NF-κB is regulated by the IκB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell. 1997;89:413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]
- Zhong H., Voll R.E., Ghosh S. Phosphorylation of NF-κB p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- Zhong H., May M.J., Jimi E., Ghosh S. The phosphorylation status of nuclear NF-κB determines its association with CBP/p300 or HDAC-1. Mol. Cell. 2002;9:625–636. doi: 10.1016/s1097-2765(02)00477-x. [DOI] [PubMed] [Google Scholar]
- Ziegler E.C., Ghosh S. Regulating inducible transcription through controlled localization. Sci. STKE. 2005;2005:re6. doi: 10.1126/stke.2842005re6. [DOI] [PubMed] [Google Scholar]