Abstract
Synthesis of ribosomal RNAs (rRNAs) is the major transcriptional event in proliferating cells. In eukaryotes, ribosomal DNA (rDNA) is transcribed by RNA polymerase I from a multicopy locus coexisting in at least two different chromatin states. This heterogeneity of rDNA chromatin has been an obstacle to defining its molecular composition. We developed an approach to analyze differential protein association with each of the two rDNA chromatin states in vivo in the yeast Saccharomyces cerevisiae. We demonstrate that actively transcribed rRNA genes are largely devoid of histone molecules, but instead associate with the high-mobility group protein Hmo1.
[Keywords: Chromatin, transcription, ribosomal DNA]
In the yeast Saccharomyces cerevisiae (hereafter called yeast), >60% of total cellular transcripts are produced from the ribosomal DNA (rDNA) locus by a specific multiprotein enzyme, RNA Polymerase I (Pol I), and an assorted set of additional transcription factors (Reeder 1999; Moss and Stefanovsky 2002; Russell and Zomerdijk 2006). The basal Pol I transcription machinery has been largely defined and consists of two multiprotein complexes, the upstream activating factor (UAF) (Keys et al. 1996), and the core factor (CF) (Keys et al. 1994), as well as the TATA-box-binding protein (TBP) (Steffan et al. 1996).
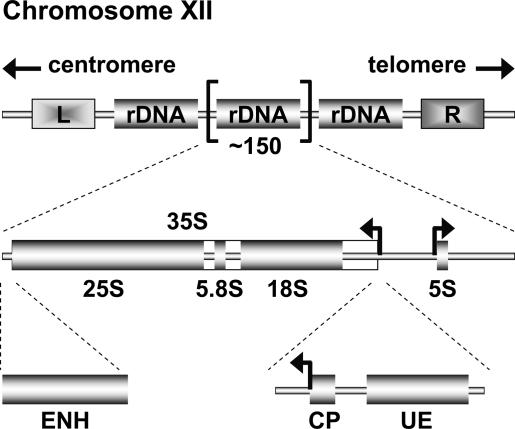
The template of RNA Pol I transcription, the yeast ribosomal RNA (rRNA) genes are located on the right arm of chromosome XII in a tandem array of 150–200 copies (Fig. 1), representing almost 10% of the yeast genome. Each of the repeats contains the Pol I-transcribed 35S rRNA gene (precusor of the 5.8S, 18S, and 25S rRNAs) and the RNA Polymerase III (Pol III)-transcribed 5S rRNA gene (Fig. 1). Three different regulatory DNA elements have been identified within the 35S rRNA gene sequence, two of which are located at the 5′ end of the 35S rDNA, spanning only ∼170 base pairs (bp) (Musters et al. 1989; Kulkens et al. 1991): the upstream element (UE), which is the binding site for the UAF; and the core promoter (CP), where CF can be recruited (Fig. 1). The third element, called the enhancer (ENH), is located at the 3′ end of the 35S transcription unit (Fig. 1) and was originally identified as exhibiting a stimulatory effect on Pol I transcription in experiments using episomal reporter systems (Elion and Warner 1984, 1986). However, this sequence has been shown subsequently to be dispensable for RNA Pol I transcription in the chromosomal context (Wai et al. 2001).
Figure 1.
Schematic representation of the yeast rDNA locus. The position of the rDNA repeat cluster on chromosome XII including the left (L) and the right (R) flanking regions with respect to the centromere and telomere is shown. Each rDNA repeat consists of the Pol I-transcribed 35S rRNA gene (precursor for the 18S, 5.8S, and 25S rRNAs) and the RNA Pol III-transcribed 5S rRNA gene. Arrows mark the transcription start sites and direction. The positions of three regulatory DNA elements—ENH, CP, and UE—are indicated.
In each cell, Pol I-transcribed rDNA coexists in two different chromatin states (Conconi et al. 1989). In these studies, cells were treated with the DNA cross-linking drug psoralen, and it was shown that actively transcribed and transcriptionally inactive rRNA genes differed substantially in their degree of psoralen incorporation. The different migration behavior of the corresponding restriction fragments in agarose gel electrophoresis led to the terms s-bands and f-bands (slow- and fast-migrating bands), originating from actively transcribed and nontranscribed rDNA, respectively (Conconi et al. 1989). The characteristic cross-linking pattern of the f-band visualized by electron microscopy suggested that the DNA had been assembled into nucleosomes in the living cell (Dammann et al. 1993). Furthermore, it was demonstrated that nascent rRNA is selectively associated with the heavily psoralen-cross-linked s-band (Conconi et al. 1989; Dammann et al. 1993), leading to the conclusion that this (presumably) nucleosome-free fraction of rDNA is actively transcribed in vivo.
In agreement with the above data, electron microscopy of rDNA chromatin spread from yeast cells shows repetitive structures of 35S rDNA transcription units densely packed with polymerases from which extending transcripts form a Christmas-tree-like structure (Rattner et al. 1982). Clusters of these actively transcribed rRNA genes are sometimes separated by longer linear DNA stretches, which are believed to represent the transcriptionally inactive rDNA population (French et al. 2003). Thus, different approaches independently monitor the two states of rDNA chromatin.
Direct physical association of proteins with rDNA was investigated by chromatin immunoprecipitation (ChIP). To date, a large number of factors have been mapped to the rRNA gene sequence, among them abundant structural chromatin components like histone proteins and the high-mobility group (HMG) protein Hmo1. A more recent ChIP study demonstrated that actively transcribed rRNA genes are assembled into a dynamic chromatin structure of unphased nucleosomes (Jones et al. 2007), challenging the interpretation of the psoralen cross-linking analyses. Hmo1 was found to bind throughout the entire 35S rDNA sequence, suggesting that it may be implicated in the formation of rDNA-specific chromatin (Hall et al. 2006; Berger et al. 2007; Kasahara et al. 2007). Although cells can survive in the absence of Hmo1, they are compromised in their growth. There is genetic evidence that Hmo1 assists Pol I in transcribing rDNA, but the molecular mechanism underlying this phenomenon is not understood (Gadal et al. 2002).
So far, ChIP has been the only technique used to analyze rDNA chromatin composition in vivo. The coexistence of different rDNA chromatin states is problematic for the interpretation of such analyses, since it cannot be determined with which population of rRNA genes an identified protein factor is associated. Thus, additional approaches are needed to obtain a molecular description of rDNA chromatin.
Here, we investigated 35S rDNA chromatin by Chromatin Endogenous Cleavage (ChEC). This method allows the precise localization of chromatin-associated factors on the genomic DNA sequence (Schmid et al. 2004). We combined this approach with psoralen cross-linking to assess the differential association of proteins with the two rDNA chromatin structures. We show the physical association of Pol I with s-band but not f-band chromatin, demonstrating that the s-band rDNA state is actively transcribed in the cell. Vice versa, histone proteins are components of f-band but not s-band chromatin, arguing that the presence of nucleosomes plays only a negligible role on Pol I-transcribed rDNA. Instead, the HMG-box protein Hmo1 colocalizes with Pol I on actively transcribed rRNA genes, and remains associated with the DNA even when Pol I leaves the template.
Results
To analyze yeast rDNA chromatin composition in vivo, we employed ChEC (Schmid et al. 2004). In brief, a factor of interest is expressed as a fusion protein with a C-terminal Micrococcal nuclease (MNase). After formaldehyde cross-linking of yeast cells, nuclei are prepared and incubated in a calcium-containing buffer. The MNase is activated and cleaves the genomic DNA nearest the binding site of the tagged factor. After DNA isolation, specific cleavage events can be mapped precisely to the DNA sequence in indirect end-labeling Southern blot analysis.
We created a library of yeast strains in which components of the Pol I transcription machinery are expressed as MNase fusion proteins from their endogenous loci, replacing the wild-type alleles (Table 1). Expression of the fusion proteins was verified by Western blot analysis detecting a triple-HA epitope at the C terminus of each fusion protein (data not shown). Relative protein levels determined by comparative Western blot analysis agreed with published data (Ghaemmaghami et al. 2003; Bier et al. 2004). For most of the Pol I transcription factors, nucleolar localization was confirmed by immunofluorescence microscopy (data not shown). Except for one slow-growing strain expressing an RNA Polymerase II (Pol II) subunit (Rpb4) and a strain with a reduced growth rate at 37°C expressing yeast histone H3 (Hht1), all strains used in this study behaved similarly to the wild type. Thus, the MNase fusion proteins were substituting the function of their wild-type counterparts in vivo.
Table 1.
Yeast strains used in this study
rRNA genes are present in multiple copies in the cell and rDNA copy number is dynamic, partly depending on the physiological state of the cell (Gaubatz et al. 1976; Rustchenko et al. 1993). Since rDNA copy number may have an impact on the ratio of actively transcribed and transcriptionally inactive rRNA genes, we determined the average number of rRNA genes in each of the above strains by quantitative PCR (qPCR) and/or Southern blotting (see Fig. 6C, below, as an example). We did not observe significant differences (i.e., >10%–20%, which is in the range of the measurement error) between the copy number in our MNase fusion protein-expressing strains and the copy number in the respective wild-type strains. However, already slight changes in rDNA repeat number may influence the proper interpretation of experimental results (see below).
Figure 6.
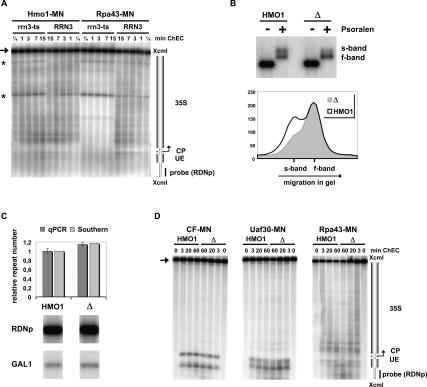
Hmo1 is a structural constituent of actively transcribed 35S rDNA chromatin. (A) Hmo1 remains associated with rDNA in the absence of Pol I. ChEC was performed with nuclei isolated from yeast strains cultured for 2 h at 37°C, carrying either a wild-type (RRN3) or a temperature-sensitive (rrn3-ts) allele of the essential, Pol I-specific initiation factor Rrn3, and expressing the MNase fusion protein indicated at the top of the panel. Isolated nuclei were incubated in calcium-containing buffer at 30°C for the minutes depicted above each lane. DNA was isolated, digested with XcmI, and analyzed in a Southern blot as described in the legend for Figure 2. A cartoon of the genomic region is depicted on the right as described in the legend for Figure 2. An arrow points to the uncut XcmI fragment of the genomic DNA region. Two fragments of unknown origin are labeled by asterisks. (B) S-band chromatin can be established in the absence of Hmo1. Psoralen cross-link analyses were performed with nuclei isolated from yeast strains carrying either a wild-type (HMO1, y881) or a complete deletion (Δ, y1159) of the HMO1 gene. DNA was isolated, digested with EcoRI, and analyzed in a Southern blot as described in the legend for Figure 5B. A profile analysis after quantification of the signal intensity by the FLA-5000 imaging system and MultiGauge software (FujiFilm) is shown. The signal intensity in arbitrary units was plotted against the distance of migration in the gel. (C) rDNA copy number is slightly increased in the HMO1 deletion strain. rDNA copy number was determined in genomic DNA isolated from yeast strains carrying either a wild-type (HMO1, y881) or a complete deletion (Δ, y1159) of the HMO1 gene. The DNA was analyzed by quantitative Southern blot analysis of XcmI restriction fragments detecting either a 4.9-kb fragment of the promoter and coding region of the 35S rRNA gene (RDNp) or a 6.8-kb fragment containing the GAL gene locus (GAL1). Signal intensities of the respective restriction fragments were determined using the FLA-5000 imaging system and MultiGauge software (FujiFilm). The same DNA was analyzed by qPCR amplifying either a region of the 18S rDNA or a region of the single-gene PHO5 locus. The primary data of the Southern blot analysis is shown, with the name of the probes used for hybridization indicated on the left. The diagram is a graphical representation of the results derived by the two kinds of analyses. The relative rDNA copy number was determined by normalizing the ratio of the amount of rDNA to the ratio of the amount of the single-copy genes. (D) Deletion of Hmo1 does not affect transcription factor binding at rDNA. ChEC was performed with nuclei isolated from yeast strains carrying either a wild-type (HMO1) or a complete deletion (Δ) of the HMO1 gene and expressing the MNase fusion protein indicated at the top of the panel. Incubation in calcium-containing buffer was performed for the minutes depicted above each lane. DNA was analyzed as described in the legend for Figure 2. A cartoon of the genomic region is depicted on the right as described in the legend for Figure 2. An arrow points to the uncut XcmI fragment of the genomic DNA region.
ChEC patterns suggest a loop structure at the rDNA promoter
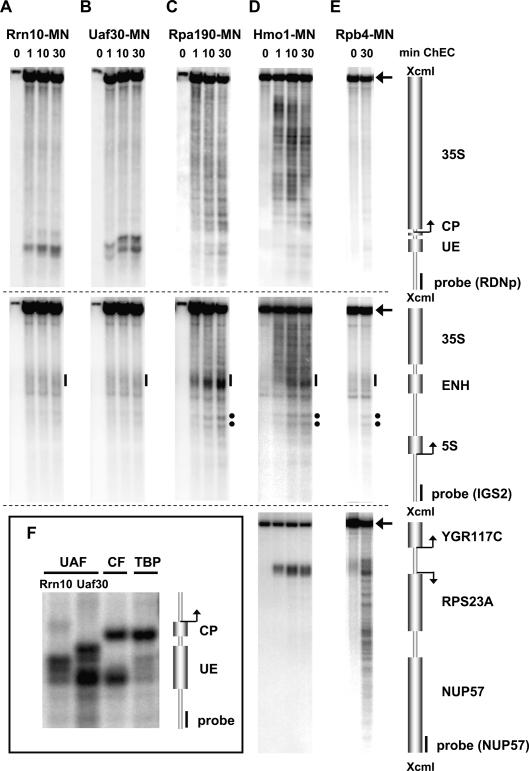
To validate the ChEC technique as a suitable tool for investigating rDNA chromatin, we first localized MNase fusion proteins of various RNA Pol I transcription factors on the 35S rDNA sequence. These analyses revealed two classes of binding: (1) single-site binding, producing distinct cleavage events at specific locations; and (2) spread binding, with multiple cleavage events along the entire transcribed rDNA sequence. Proteins belonging to the first class were Rrn10 and Uaf30, components of the UAF (Fig. 2A,B [top panels], F); Rrn7, a CF component (Fig. 2F); and Spt15, the yeast TBP (Fig. 2F).
Figure 2.
ChEC analyses suggest DNA looping at the yeast rDNA promoter. ChEC was performed with nuclei isolated from yeast strains expressing the MNase fusion protein indicated at the top of each panel. Isolated nuclei were incubated in calcium-containing buffer at 30°C (24°C for Rpb4-MN) for the times depicted above each lane (A–E), or for 30 min (F). (A–F) DNA was isolated, digested with XcmI, separated in a 1% agarose gel, and analyzed in a Southern blot by indirect end-labeling with probes visualizing either a 4.9-kb fragment of the promoter and coding region of the 35S rRNA gene (top panels), a 4.3-kb fragment of IGS1 and IGS2 flanking the 5S rRNA gene, and the 3′ end of the 35S rRNA gene (middle panels), or a 4.2-kb fragment encompassing the RPS23A gene locus (bottom panels). Cartoons of the genomic regions including positions and names of the respective genes with their corresponding transcription start sites, and of probes used for indirect end-labeling, are depicted on the right. Arrowheads point to the uncut XcmI fragment of the corresponding genomic DNA region. (Top panels) (CP) Core promoter; (UE) upstream element; (ENH) enhancer of the 35S rDNA. (Middle panels) Black bars indicate cleavage by promoter-bound factors within the ENH region, and black dots show two hypersensitive sites (C–E). (F) An enlarged view of cleavage events within the rDNA promoter region is presented. In the input lanes of A–C (0) only one-tenth of the DNA was applied to the gel.
Cleavage by UAF components fused to MNase occurred at their known binding site, the UE (Fig. 2F). Cuts mediated by Uaf30-MNase flank Rrn10-MNase cleavage sites (Fig. 2F). A MNase fusion protein of the CF component Rrn7 cleaves at the 5′ end of the CP (Fig. 2F), but also produces a well-defined cut at the 5′ end of the UE (Fig. 2F). The same was observed for another CF component, Rrn11 (data not shown). This distinct cleavage pattern accorded well with a suggested model that rDNA promoter DNA loops around UAF (Keener et al. 1997), thus making the DNA sequence at the 5′ end of the UE available for cutting CF-MNase fusion proteins bound to CP (see Discussion).
TBP-MNase produced a clear cut in the spacer between the UE and CP (Fig. 2F), indicating that the protein is in direct contact with DNA. This location fit a model derived from protein–protein interaction studies in vitro, with TBP bridging between the UAF and CF (Steffan et al. 1996).
All of the above factors showed weak and diffuse cleavage in the rDNA ENH region, as exemplified for Rrn10 and Uaf30 in the middle panel of Figure 2, A and B (black bar). None of the UAF-MNase or CF-MNase fusion proteins cut within the rDNA-unrelated RPS23A or RPS22B gene loci (data not shown).
Proteins spreading along the transcribed rDNA region were components of the Pol I enzymes Rpa190 and Rpa43 (Figs. 2C [top and middle panels], 6A,D [below]), and the HMG protein Hmo1 (Figs. 2D, [top and middle panels], 6A [below]). The cleavage pattern produced by these factors was very similar. Pol I and Hmo1-MNase cutting was restricted to the transcribed region, ending at the ENH element (Fig. 2C,D, middle panels).
Hmo1 association with the 35S rDNA locus has been observed in ChIP experiments (Hall et al. 2006; Berger et al. 2007; Kasahara et al. 2007). In these studies, Hmo1 interaction with the promoters of many ribosomal protein genes (RPs), transcribed by RNA Pol II, was also reported. We detected strong cleavage within the promoter region of one Hmo1 target gene, RPS23A, in ChEC experiments (Fig. 2D, bottom panel). In contrast to the spreading observed along rDNA, Hmo1-mediated cleavage was restricted to the RPS23A promoter. No significant cleavage was observed at another ribosomal gene locus, RPS22B (data not shown), in agreement with the published data (Hall et al. 2006; Berger et al. 2007; Kasahara et al. 2007).
As a control, the Pol II subunit Rpb4 fused to MNase did not cut within the transcribed rDNA sequence (Fig. 2E, top panel). However, cuts were observed in the intergenic spacer 1 (IGS1) (Fig. 2E, middle panel, black dots) and the ENH region (Fig. 2E, middle panel, black bar). The latter cleavage is presumably unspecific since it occurs even in the absence of MNase activity (Fig. 2E, middle panel, cf. lanes “0” and “30”). The two distinct cuts in IGS1 correspond to hypersensitive sites, detected in ChEC experiments with most of the MNase fusion proteins (Fig. 2, cf. middle panels) and after digestion of yeast chromatin with exogenously added MNase (Fig. 3B, lane E, black dots). These sites match sequences identified previously as a bidirectional Pol II promoter (Ganley et al. 2005; Kobayashi and Ganley 2005). As expected, Rpb4-MNase cleaved at the promoter and within the coding sequence of the highly transcribed RPS23A gene (Fig. 2E, bottom panel).
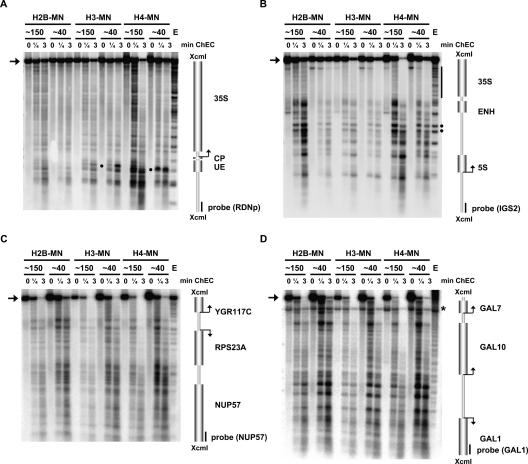
Figure 3.
Histone protein association with the 35S rDNA region depends on the transcriptional state of the locus. ChEC was performed with nuclei isolated from yeast strains carrying either a wild-type rDNA locus (∼150) or a locus with a reduced rDNA repeat copy number (∼40) and expressing the MNase fusion protein indicated at the top of the panel. Isolated nuclei were incubated in calcium-containing buffer at 30°C for the minutes depicted above each lane. (Lane E) As a control, nuclei of a wild-type strain were treated with exogeneously added MNase. DNA was analyzed as described in the legend for Figure 2 with probes visualizing either a 4.9-kb fragment of the promoter and coding region of the 35S rRNA gene (A), a 4.3-kb fragment of IGS1 and IGS2 flanking the 5S rRNA gene and the 3′ end of the 35S rRNA gene (B), a 4.2-kb fragment encompassing the RPS23A gene locus (C), or a 6.8-kb fragment containing the GAL gene locus (D). Cartoons of the genomic regions are depicted on the right as described in the legend for Figure 2. Arrowheads on the left point to the uncut XcmI fragment of the corresponding genomic DNA region. (A) Specific cleavage events within the UE occurring selectively in the H3-MNase- and H4-MNase-expressing strains are indicated by black dots on the right. (B) A black bar on the right indicates cleavage within the 3′ region of the 35S rRNA gene, and black dots on the right show two hypersensitive sites at a bidirectional Pol II promoter element (Ganley et al. 2005). (D) An asterisk on the right indicates a background signal from the hybridization with the probe used in B, still detectable after probe removal and rehybridization.
Thus, ChEC analysis is an appropriate method to characterize rDNA chromatin. Besides the accurate localization of factors on the rDNA sequence, the occurrence of cleavage events mediated by MNase fusion proteins far from their known DNA-binding site provides some structural information.
Actively transcribed rDNA is largely devoid of histone molecules
As outlined above, the nucleosomal nature of rDNA chromatin is discussed controversially in the literature. Whereas early analyses based on psoralen accessibilities in rDNA led to the conclusion that actively transcribed rRNA genes are devoid of nucleosomes (Dammann et al. 1993), more recent data suggest that active rDNA has nucleosomal structure (Jones et al. 2007). We reasoned that ChEC could give more insights into the association of histone molecules with Pol I-transcribed loci.
ChEC experiments were carried out in yeast strains expressing MNase fusion proteins of either one of the yeast histones H2B (Htb2), H3 (Hht1), or H4 (Hhf2) in two different genetic backgrounds: NOY505 (Nogi et al. 1993), a yeast strain carrying a wild-type copy number of rDNA repeats (∼150 in Fig. 3); and NOY886, a yeast strain carrying only ∼40 rRNA genes (hereafter called “40-copy strain”) (∼40 in Fig. 3). In this strain, most of the 35S transcription units are actively transcribed (French et al. 2003). As a control for nonspecific cleavage at hypersensitive sites, we analyzed DNA isolated from nuclei of a wild-type yeast strain treated with exogenously added MNase (Fig. 3, lanes E).
In all strains carrying a wild-type rDNA copy number, cleavage was observed in the rDNA promoter region and throughout the entire coding region (Fig. 3A, lanes ∼150). The overall cleavage patterns were similar, but not identical, to that obtained in DNA isolated from nuclei after incubation with exogenously added MNase (Fig. 3A, lane E). In the 40-copy strains, cleavage events within the 35S rDNA coding sequence were reduced (Fig. 3A, cf. lanes ∼150 and ∼40). In addition, full-length 35S rDNA XcmI fragments were degraded more slowly in the 40-copy strains than in the corresponding strains carrying a wild-type copy number of rDNA repeats (Fig. 3A, cf. signal intensities of the band labeled by an arrow in ∼40 and ∼150 strains). The observed differences between wild-type and 40-copy strains were strongest in the strains expressing yeast histone H2B-MNase (Fig. 3A, lanes H2B-MN). We conclude that histone molecules can associate with rDNA in general, but that the histone density on actively transcribed rDNA regions is low.
Closer inspection of the specific cleavage events within the rDNA promoter region revealed cuts within the UE sequence, which were observed only in strains expressing either histone H3-MNase or H4-MNase (Fig. 3A, dots in lanes H3-MN and H4-MN). This specific cleavage was stronger in the 40-copy strains, and did not correlate with hypersensitive sites detected when chromatin was digested with exogenously added MNase (Fig. 3A, lane E). The result is in accordance with histones H3 and H4 being part of the UAF, as suggested previously by biochemical fractionation (Keener et al. 1997).
The same membrane used for analysis in Figure 3A was hybridized with a probe detecting the 3′ end of the 35S rDNA, the 5S rDNA, and parts of the noncoding intergenic spacer regions (Fig. 3B). We again observed that cleavage events within the 3′ region of the 35S rDNA transcription unit (Fig. 3B, cf. lanes ∼40 and ∼150, black bar) and degradation of the full-length XcmI fragment (Fig. 3B, cf. signal intensities of the band labeled by an arrow in ∼40 and ∼150 strains) were reduced in the 40-copy strains. We observed a very similar cleavage pattern within the intergenic spacer regions in all strains analyzed (Fig. 3B). These distinct cuts could be mediated by positioned nucleosomes, whose presence has been suggested by psoralen analyses and MNase mapping experiments (Dammann et al. 1993; Vogelauer et al. 1998). We note that there was an apparent reduction in the relative amounts of fragments produced by the histone-MNases (reflected in the different signal intensities in the Southern blot) when a 40-copy strain was compared with the respective wild-type strain. Again, this became most obvious in the strains expressing a MNase fusion protein of H2B (Fig. 3B, lanes H2B-MN).
XcmI fragments bearing rDNA sequences were still detected at all time points of ChEC (Fig. 3A,B, bands labeled by an arrow). This was in sharp contrast to XcmI fragments generated from Pol II-dependent loci, encompassing either the actively transcribed RPS23A gene or the GAL1, GAL10, and GAL7 genes being repressed and, presumably, fully assembled into nucleosomes in the growth conditions chosen: At these loci, most of the full-length DNA was rapidly degraded (Fig. 3C,D, bands labeled by an arrow). This indicates that association of histone molecules with rDNA is reduced compared with histone association with DNA from Pol II-dependent loci.
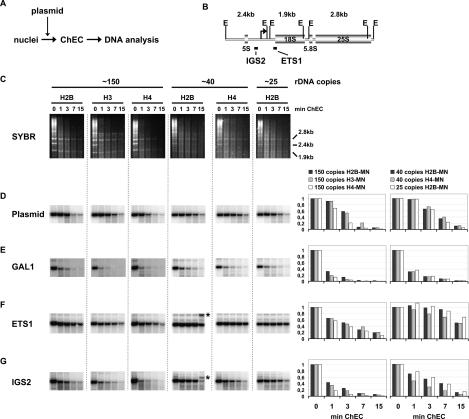
We wanted to measure histone density more quantitatively at the rDNA locus. Extended ChEC time-course experiments using nuclei from strains carrying a wild-type rDNA copy number and expressing either histone H2B-MNase, H3-MNase, or H4-MNase were performed. We also included two 40-copy strains expressing either H2B-MNase or H4-MNase, and a strain expressing H2B-MNase but carrying only 25 rDNA repeats on average (25-copy strain) (∼25 in Fig. 4).
Figure 4.
Actively transcribed rRNA genes are largely devoid of histone molecules. (A) Schematic representation of the experimental layout. (B) A cartoon of the yeast rDNA locus is presented. The coding sequences for the ribosomal RNAs (5S, 18S, 5.8S, 25S) and the positions of the two rDNA probes used in the Southern blot analyses (IGS2, ETS1) are indicated. EcoRI restriction sites (E) and positions of the 2.4-kb, 1.9-kb, and 2.8-kb rDNA fragments are depicted. (C–G) ChEC was performed with nuclei isolated from yeast strains carrying either a wild-type rDNA locus (∼150), or a locus with a reduced rDNA repeat copy number (∼40 or ∼25) and expressing the MNase fusion protein indicated at the top of the panel. Before ChEC, purified, linearized plasmid DNA was added to the reaction. Isolated nuclei were incubated in calcium-containing buffer at 30°C for the minutes depicted above each lane. DNA was isolated, digested with EcoRI, and separated in a 1% agarose gel. (C) Analysis by staining with SYBR Safe. Positions and size of three EcoRI fragments originating from the rDNA locus are indicated on the right. (D–G) The DNA was transferred to a nylon membrane and analyzed in a Southern blot with probes indicated on the left, visualizing either a 3.4-kb fragment of plasmid DNA (D), a 1.9-kb fragment containing the GAL gene locus (E), the 1.9-kb fragment containing the 18S rDNA sequence (F), or the 2.4-kb fragment containing the intergenic spacer rDNA region (G). (E,G) Asterisks on the right in lane “∼40 H2B” mark DNA fragments presumably resulting from incomplete EcoRI digestion. Signal intensities of the respective uncut restriction fragments were determined using the FLA-5000 imaging system and MultiGauge software (FujiFilm). The ratios relative to the input value were calculated and plotted against the time of incubation in calcium-containing buffer. A graphical representation of the results is depicted on the right of each row.
Histones belong to the most abundant proteins in the nucleus. Thus, histone-MNases released from the chromosomes in the course of ChEC will digest DNA randomly to some extent. For an accurate quantitative analysis, it is therefore important to assess the contribution of random digestion to the overall degradation. To determine the extent of random digestion of DNA not associated with histone molecules, nuclei were mixed with a purified plasmid containing yeast-unrelated DNA sequences before commencing ChEC (Fig. 4A).
After ChEC, the DNA was isolated and digested with EcoRI. EcoRI digestion of rDNA yielded three characteristic fragments: a 1.9-kb fragment, containing most of the 18S rDNA sequence; a 2.8-kb fragment, including 5.8S rDNA and large parts of 25S rDNA; and finally a 2.4-kb segment bearing the intergenic spacer regions and the 5S rDNA (Fig. 4B). Since rDNA represents ∼10% of the total genomic DNA in wild-type yeasts (and ∼2.5% in the strains with a low copy number), the fragments could be detected in an agarose gel stained with SYBR Safe (Fig. 4C). Degradation of these three regions of rDNA by histone MNase fusion proteins followed different kinetics. Whereas the 2.4-kb fragment encompassing the intergenic spacers was rapidly digested, the 18S and 25S rDNA-containing fragments (1.9 kb; 2.8 kb) were more resistant to degradation (Fig. 4C). This resistance is a special feature of the transcribed rDNA regions because we observed almost complete degradation of the bulk genomic DNA (visible as a high-molecular-weight smear in the untreated samples in Fig. 4C) at the earliest time point of ChEC by histone-MNases.
For quantitative Southern blot analysis, the DNA in the gel was transferred to a membrane and subsequently hybridized with various probes. We first measured the extent of nonspecific, random DNA degradation of naked plasmid DNA by MNase fusion proteins liberated from the chromosome in the course of ChEC. At the earliest time point, only weak degradation was observed (Fig. 4D, 1 min). After 15 min of incubation, most of the full-length plasmid DNA was digested (Fig. 4D, 15 min).
In contrast, digestion of an EcoRI fragment of a Pol II-dependent gene locus was very fast in all strains analyzed, being almost complete already after 3 min of ChEC (Fig. 4E, 3 min). This was in accordance with our above observations (Fig. 3C,D). Taken together, these results demonstrate that the kinetics of nonspecific, random degradation of DNA by histone-MNase fusion proteins liberated in the course of ChEC and the specific degradation by histone-MNase fusion proteins covalently linked to the DNA differ significantly, and allow us to differentiate between these two processes.
We next analyzed the kinetics of degradation of the 1.9-kb DNA fragment, spanning the 18S rRNA coding sequence (Fig. 4F). For all strains encompassing a wild-type number of rDNA repeats, we observe biphasic kinetics with an initial rapid digestion followed by slow degradation of the remainder of the rDNA fragments (Fig. 4F, lanes ∼150). The kinetics of the latter reaction were slower than the kinetics of degradation of the exogenously added plasmid (Fig. 4D, lanes ∼150). Presumably, the naked plasmid was a better substrate for the free MNase than the cross-linked chromosomal DNA. After 3 min of ChEC—a time point when the majority of DNA fragments from the RNA Pol II-dependent reference locus were digested (Fig. 4E, lanes ∼150)—38% (H4-MNase) to 51% (H3-MNase) of the original 18S rDNA input was still detected (Fig. 4F). Importantly, in the 40-copy and 25-copy strains expressing histone H2B-MNase, >90% of the original 18S rDNA input could be recovered after 3 min of ChEC (Fig. 4F, lanes ∼40 H2B and ∼25 H2B). Degradation of rDNA by H4-MNase expressed in a 40-copy strain followed slightly faster kinetics (Fig. 4F, lanes ∼40 H4). Still, close to 80% of the input rDNA was recovered from the 40-copy strain after 3 min of ChEC (Fig. 4F, lanes ∼40 H4). Taken together, the numbers derived for “ChEC-resistant” DNA fragments correlated well with the numbers obtained for the psoralen-accessible fraction of 35S rRNA genes (typically 35%–50% in wild-type strains and >80% in 40-copy strains) (Fig. 6B, below; Supplemental Fig. 1). These quantitative analyses agreed also with the qualitative statement of a low histone density correlating with active transcription of rRNA genes, and led us to the conclusion that these regions are largely devoid of histone molecules.
As mentioned above, the 2.4-kb rDNA fragment carrying the intergenic spacers was more susceptible to early degradation (Fig. 4C). In fact, the degradation kinetics of the intergenic spacers in strains carrying a wild-type rDNA locus were similar to those seen for the Pol II-dependent gene locus (Fig. 4, cf. rows G and E). Interestingly, in the 25- and 40-copy strains expressing H2B-MNase, >50% of the input intergenic DNA was resistant after 3 min of ChEC (Fig. 4G). Again, H4-MNase digested this DNA sequence more efficiently, but to a considerably lower extent than the DNA at the chromosomal GAL1 locus (Fig. 4, cf. rows G and E). Thus, in yeast strains with a wild-type rDNA locus, the majority of intergenic regions was associated with histone molecules. However, in the genetic background with a lowered rDNA copy number, we observed a reduced histone density on the same DNA sequence.
Kinetic analyses of the type presented in Figure 4 have been repeated up to five times, but at least twice for each of the strains expressing the respective MNase fusion proteins (data not shown). The outcome of these experiments was very similar (with 5%–15% deviation in cleavage efficiencies). Most of the variability probably resulted from differences in the extent of formaldehyde cross-linking: Prolonged formaldehyde fixation will interfere with efficient cutting of the MNase fusion proteins, whereas low cross-linking efficiency will dramatically increase the nonspecific random degradation of chromosomal DNA.
ChEC–psoralen analysis determines protein association with different rDNA chromatin states
To demonstrate the specific association of histones with only one of the two different rDNA chromatin states, we combined a ChEC experiment with psoralen cross-linking analysis. If a MNase fusion protein is associated with either s-band or f-band chromatin, the corresponding fragment will be selectively degraded and undetectable in the subsequent Southern blot analysis (Fig. 5A).
Figure 5.
Hmo1 and Histone H3 associate with different states of yeast rDNA chromatin. (A) Schematic representation of a combined ChEC–psoralen cross-linking experiment. An example of specific association of a MNase fusion protein (square) with s-band chromatin is shown. Nucleosomes as components of f-band chromatin are depicted as ovals. Psoralen molecules are represented by black dots, and specific restriction sites used are represented as a pair of scissors. (B) ChEC was performed with nuclei isolated from yeast strains expressing the MNase fusion protein indicated at the top of each panel. Isolated nuclei were incubated in calcium-containing buffer at 30°C for the minutes depicted above each lane. After ChEC, nuclei were treated with psoralen and DNA was isolated. The genomic DNA was either separated in a 1% agarose gel and analyzed by ethidium bromide staining (bottom panel), or digested with EcoRI, separated in a 0.8% agarose gel, and analyzed in a Southern blot with a probe detecting an EcoRI fragment within the coding region of the 35S rRNA gene (top panel). EcoRI restriction sites (E) and the positions of the fragment and probe are indicated in the cartoon below the panels.
When ChEC–psoralen analysis was carried out with a strain expressing a histone H3-MNase fusion protein, we observed selective degradation of the f-band, originating from transcriptionally inactive rRNA genes. This is consistent with our conclusion that actively transcribed rDNA—represented by the nondegraded, highly psoralen-cross-linked s-band—is largely devoid of histone molecules (Fig. 5B, lanes H3-MN, top panel). As mentioned above, the resistance to cleavage by H3-MNase is specific for actively transcribed rRNA genes, since bulk genomic DNA is degraded instantly in such an experiment (Fig. 5B, lanes H3-MN, bottom panel).
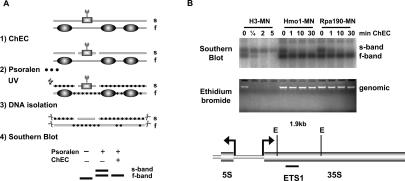
In addition, we examined two different strains expressing either Rpa190 or Hmo1 in fusion with MNase by ChEC–psoralen analysis. Rpa190-MNase specifically degraded the s-band (Fig. 5B, top panel). Interestingly, the same was observed in a Hmo1-MNase-expressing strain (Fig. 5B, top panel). Thus, our data demonstrate for the first time the selective physical association of Pol I with s-band chromatin. We further define Hmo1 as a specific constituent of s-band chromatin.
Hmo1 is a structural component of active rRNA genes
Association of Hmo1 with rDNA has been described (Hall et al. 2006; Berger et al. 2007; Kasahara et al. 2007), but apart from the observation that it genetically interacts with the Pol I subunit Rpa49 (Gadal et al. 2002), little is known about its function.
In the study by Kasahara et al. (2007) it was shown that Hmo1 associated with 35S rDNA in a Pol I-dependent manner. To address the question of whether Hmo1 travels with the polymerase or if it is a rDNA-associated factor, Hmo1-MNase and Rpa43-MNase fusion proteins were expressed in two different genetic backgrounds: a strain carrying a temperature-sensitive allele of the Pol I-specific transcription initiation factor Rrn3 (Ycc95-rrn3-8) (rrn3-ts in Fig. 6A) and the isogenic RRN3 wild-type strain (CG379) (RRN3 in Fig. 6A; Cadwell et al. 1997). The rrn3-ts strain supports rDNA transcription at 24°C, while incubation at 37°C leads to transcriptional shutdown, presumably because the initiation-competent Rrn3–Pol I complex can no longer be formed. At the permissive temperature, Hmo1-MNase and Rpa43-MNase fusion proteins cleave rDNA to a similar extent in both rrn3-ts and RRN3 strains (A. Philippi, R. Steinbauer, S. Fath, K. Merz, P. Milkereit, J. Griesenbeck, and H. Tschochner, in prep.). At the nonpermissive temperature, no cleavage by Rpa43-MNase could be detected, while digestion was still observed in the RRN3 strain (Fig. 6A). Cleavage by Hmo1-MNase in both rrn3-ts and RRN3 strains was not affected at the elevated temperature (Fig. 6A). Thus, Hmo1 is a structural component of rDNA chromatin, remaining bound to rDNA even in the absence of Pol I.
S-band chromatin is established in the absence of Hmo1
We next asked if the presence of Hmo1 is required to establish s-band chromatin. HMO1 wild-type and HMO1 deletion cells were subjected to psoralen cross-linking analysis (Fig. 6B). S-band chromatin was still established in the absence of the HMO1 gene. However, psoralen incorporation into rDNA was compromised in the HMO1 knockout strain tested, as revealed by a reduced s-band formation (Fig. 6B). We note that the HMO1 deletion strain used in this experiment had a 15% increase in rDNA copy number when compared with its wild-type counterpart (Fig. 6C). It thus cannot be excluded that the observed changes in psoralen accessibility are a consequence of rDNA locus expansion, as opposed to a structural change in rDNA chromatin in the absence of HMO1.
We then examined if the absence of Hmo1 affected recruitment of Pol I transcription factors to rDNA by performing ChEC experiments in strains expressing MNase fusion proteins of components of the CF (Rrn7) and UAF (Uaf30) in HMO1 wild-type and HMO1 deletion strains (Fig. 6D, first two panels). Neither cleavage pattern nor cleavage efficiencies were affected significantly in the deletion strains, indicating that chromatin structure at the rDNA promoter does not change dramatically in the absence of Hmo1. Furthermore, the ChEC pattern observed in HMO1 wild-type and deletion strains expressing MNase fusion proteins of a Pol I subunit were almost identical (Fig. 6D, third panel).
Taken together, we demonstrate that Hmo1 is a constituent of transcriptionally active rRNA genes. Its function within the specialized, histone-reduced rDNA chromatin awaits further analysis.
Discussion
Previous data have suggested that the coding regions of 35S rRNA genes exist in two different chromatin states—one being nucleosomal and transcriptionally inactive, and the other one devoid of nucleosomes and associated with the transcribing Pol I (Dammann et al. 1993). However, these results were based solely on the interpretation of psoralen cross-linking patterns of rDNA fragments—a rather indirect proof. In this study, we show for the first time the physical association of Pol I and histones with the different rDNA chromatin states (Fig. 5B). Our findings thus confirm the conclusions drawn by Dammann et al. (1993).
Our results that actively transcribed rRNA genes are virtually histone-free are in apparent conflict with the interpretation of recent ChIP experiments and MNase digestion analyses, suggesting that Pol I-transcribed rDNA repeats have nucleosomal structure (Jones et al. 2007). The conclusions drawn in the latter study rely on the observation that in a strain with a reduced rDNA repeat copy number, all of the rRNA genes are transcriptionally active (French et al. 2003). We used the same genetic background in some of our analyses. Psoralen cross-linking analyses in some of the 40-copy strains revealed a minor f-band fraction (∼10%–20% in the experiment shown in Supplemental Fig. 1) corresponding to nucleosomal nontranscribed rRNA genes (Fig. 5B). This was corroborated by a low degree of rDNA degradation at intermediate time points of ChEC in low-copy strains expressing histone-MNase fusion proteins (e.g., ∼20% after 3 min of ChEC for H4-MNase) (Fig. 4F). Since electron microscopy provides no evidence for the presence of nontranscribed rRNA genes within the rDNA cluster (French et al. 2003), it might be that within the 40-copy strain cell population a fraction of cells does not transcribe rDNA at all. Such cells would be dismissed in the electron microscopy analyses, because the rDNA locus could not be distinguished from the rest of the chromatin. The presence of a subpopulation of transcriptionally inactive rDNA repeats in the 40-copy strain offers a simple explanation for both the detection of histone molecules on 35S rDNA by ChIP and the occurrence of a protected 150-bp rDNA fragment after degradation of chromatin by exogenous MNase (Jones et al. 2007).
The histone density on the intergenic spacer DNA in strains carrying a wild-type rDNA copy number was very similar to the histone density observed on fully nucleosomal RNA Pol II-dependent loci (Fig. 4, cf. G and E). Instead, we observed a reduced histone H2B and (to a lesser extent) H4 density on the intergenic spacer region in yeast strains with a lower rDNA copy number (Fig. 4G). In order to maintain a low copy number of rDNA, yeast strains need to be deleted in the FOB1 gene (Kobayashi et al. 1998). FOB1 codes for a protein that binds to DNA elements within the rDNA enhancer region defining the so-called replication fork barrier (Kobayashi and Horiuchi 1996). It has been shown that FOB1 is required for regional Pol II silencing in the intergenic spacer region and thus seems to influence local chromatin structure at this locus (Huang and Moazed 2003). Furthermore, it has been reported recently that Pol II can transcribe intergenic spacer DNA from a bidirectional promoter (E-pro) located within IGS1 (Ganley et al. 2005; Kobayashi and Ganley 2005). Transcription from this site drives repeat expansion to obtain a wild-type rDNA copy number by displacing cohesin from this region, thereby promoting unequal sister chromatid exchange (Kobayashi and Ganley 2005). Accordingly, E-pro-dependent transcription is elevated in the low-copy strains (Kobayashi and Ganley 2005), which could lead to nucleosome displacement from intergenic spacer sequences, resulting in reduced histone density. Additionally, the RNA Pol III-dependent 5S rRNA gene is located within the intergenic spacer region analyzed in Figure 4, and enhanced transcription from this locus might influence histone density as well. Further analyses are required to explore these possibilities.
Recently, it has been reported that Pol II can transcribe intergenic spacer DNA from a bidirectional promoter (E-pro) located within IGS1 (Ganley et al. 2005; Kobayashi and Ganley 2005). Transcription from this site presumably drives repeat expansion to obtain a wild-type rDNA copy number by displacing cohesin from this region, thereby promoting unequal sister chromatid exchange (Kobayashi and Ganley 2005). Accordingly, E-pro-dependent transcription is elevated in the low-copy strains (Kobayashi and Ganley 2005), which could lead to nucleosome displacement from intergenic spacer sequences resulting in reduced histone density. Additionally the RNA Pol III-dependent 5S rRNA gene is located within the intergenic spacer region analyzed, and enhanced transcription from this locus might influence histone density as well. Further analyses are required to explore these possibilities.
The HMG protein Hmo1 binds to 35S rDNA in the promoter region and throughout the entire coding region (Fig. 2D). Thus, our analyses confirm previous ChIP data (Hall et al. 2006; Kasahara et al. 2007), but add the important information that Hmo1 specifically associates with actively transcribed 35S rRNA genes (Fig. 5B). We define Hmo1 as a structural component of psoralen-accessible rDNA chromatin, which remains bound to this chromatin state even in the absence of Pol I (Fig. 6A). Taken together with the observation that Hmo1 associates with 35S rDNA in a Pol I-dependent manner (Kasahara et al. 2007), one can speculate that Pol I recruits Hmo1 to this locus. Alternatively, Hmo1 may preferentially recognize DNA structures transcribed by Pol I. Our data further suggest that there may be a cellular mechanism by which Hmo1 is removed from rDNA when Pol I transcription ceases. One possibility is that Hmo1 displacement occurs when the replication machinery enters the gene followed by re-establishment of a nucleosomal structure on the newly replicated rDNA coding regions (Lucchini and Sogo 1995).
A MNase fusion protein of a CF component (Rrn7) showed cleavage within sequences at the 5′ end of the UE (Fig. 1F). This observation could be explained by the existence of a local loop structure of rDNA promoter DNA around UAF in vivo, as proposed previously (Keener et al. 1997). It was suggested that the presence of histones H3 and H4 as part of the multiprotein complex UAF could support such a structure (Keener et al. 1997). Accordingly, we localize these proteins at the UE of actively transcribed 35S rRNA genes in vivo (Fig. 3A). An alternative explanation for the ChEC pattern observed would be the presence of two binding sites for this complex at the rDNA promoter. The interaction of a CF component with 5′ regulatory sequences provides a strong argument for the functional equivalence of yeast CF and the selectivity factor (SL1) in higher eukaryotes, for which such an interaction has been shown in in vitro footprinting experiments (Bell et al. 1988, 1990).
In conclusion, we demonstrate that the combination of ChEC and psoralen analyses is a powerful tool to study rDNA chromatin composition and structure in vivo. We provide insights into the organization of rDNA promoter chromatin, suggesting that the process of transcriptional initiation in yeast and higher eukaryotes, with very little homology at the protein sequence level, is structurally conserved. We find that actively transcribed rRNA genes are largely devoid of histones and insofar unique compared with other genomic loci investigated. Since the psoralen accessibility of actively Pol I-transcribed rDNA chromatin is also conserved from yeast to humans (Conconi et al. 1989; Dammann et al. 1993), it is reasonable to speculate that our observations hold true in higher eukaryotes. Our future research aims to further define the molecular nature of yeast rDNA chromatin states and the mechanisms involved in their establishment and maintenance.
Materials and methods
Plasmids and yeast strains
A complete list of oligonucleotides and yeast strains used in this study is presented in Tables 1 and 2.
Table 2.
Oligonucleotides used in this study
Plasmid K643 was generated by inserting the DNA sequence for a 3xHA epitope amplified by PCR from pYM1 (Knop et al. 1999) into BbsI- and BssHII-restricted pFA6a-MN-KanMX6 (Schmid et al. 2004). K643 served as a template in PCRs using the primers listed in Table 2 to amplify MN-3HA∷KanMX6 cassettes. The amplified fragments were framed by 35–50 bp of DNA sequence homologous with the 3′ end excluding the stop codon (in-frame with the MNase coding sequence at the 3′ end), and 35–50 bp of sequence homologous with the DNA downstream from the stop codon of the respective target gene. The DNA in the PCR reactions was precipitated with ethanol and used for transformation of yeast cells, as described (Knop et al. 1999). After selection, proper integration was confirmed by PCR and/or Southern blot analysis. Expression of the MNase fusion proteins was verified by Western blot analysis using the 3F10 antibody, recognizing the HA epitope (Roche).
To establish HMO1 deletion strains, the URA3 gene from Kluyveromyces lactis (kl URA3) was amplified with primers 1354 and 1355 from plasmid pBS1539 (Puig et al. 2001). The amplified fragment was framed by 50 bp of DNA sequence homologous with the 5′ sequence upstream of the ATG, and 50 bp of sequence homologous with the DNA downstream from the stop codon of HMO1. The amplicon was directly used for transformation as described above. HMO1 deletion was confirmed by PCR.
Plasmid pM49.2 (Griesenbeck et al. 2004) was used to amplify a 200-bp sequence by PCR containing a binding site for R recombinase (RS) and binding cluster for the bacterial Lex A protein (LEX A). The amplicon served as a template for Southern probe preparation to detect a 3.4-kb NotI fragment from plasmid pM53.1, encompassing this sequence, which was added to the ChEC reaction in the experiment presented in Figure 4.
Yeast cultures, fixation, and isolation of nuclei
Yeast cells were grown in YPD at 30°C, unless otherwise noted. When the cultures reached an OD600 of 0.5, formaldehyde was added to a final concentration of 1% and incubation was continued for 15 min at growth temperature. Excess formaldehyde was removed by the addition of glycine to a final concentration of 0.125 M. Cells were harvested and nuclei were isolated as described (Schmid et al. 2004), except that a VXR basic IKA Vibrax orbital shaker was used for cell lysis, and that a Protease inhibitor cocktail (PI) containing 330 μg/mL Benzamidin, 170 μg/mL PMSF, 1.37 μg/mL Pepstatin A, 0.284 μg/mL Leupeptin, and 2 μg/mL Chymostatin was added to all buffers.
MNase digestion of yeast nuclei
Nuclei isolated from formaldehyde-cross-linked yeast cells (strain NOY505) were digested as described previously (Boeger et al. 2003). For the experiment presented in Figure 4, nuclei from ∼85 mg of cells (wet weight) were incubated in 200 μL of reaction buffer containing 5 U/mL MNase (Sigma) for 20 min at 37°C.
ChEC
ChEC was carried out using nuclei from formaldehyde-cross-linked yeast cells as described (Schmid et al. 2004) with slight modifications. Where indicated in the figure legends, purified, NotI-restricted pM53.1 plasmid DNA was added to reach a final concentration of 30 pM in the ChEC reaction. The reactions were stopped by addition of an equal volume of IRN buffer (50 mM Tris-HCl at pH 8, 20 mM EDTA, 0.5 M NaCl). RNase A was added to a final concentration of 100 μg/mL, and the reactions were incubated for 1 h at 37°C. After addition of 1/40 vol of 20% SDS and Proteinase K to a final concentration of 100 μg/mL, incubation was continued for 1 h at 56°C. Formaldehyde cross-links were removed overnight (or at least 6 h) at 65°C. DNA was extracted with phenol–chloroform–isoamyl alcohol (25:24:1), precipitated with ethanol, and suspended in TE. One-third of the DNA was digested with the appropriate restriction enzyme and separated in a 1% agarose gel in Tris-borate-EDTA with 4 V/cm.
ChEC–psoralen cross-linking analyses
Nuclei suspensions prepared as described above were either subjected to ChEC or directly mixed with IRN buffer. Two-hundred microliters of the suspension were transferred to a 24-multiwell plate, and 4,5′,8-trimethylpsoralen (Sigma) in ethanol was added to a final concentration of 10 μg/mL. After incubation for 5 min on ice in the dark, samples were irradiated with UV light (288 nm) at a distance of 2–3 cm for 15 min on ice. Addition of psoralen and UV irradiation were repeated twice. DNA was isolated as described above. One-tenth of the DNA was digested with the appropriate restriction enzyme and separated in a 0.8% agarose gel in Tris-borate-EDTA with 3 V/cm. To reverse the psoralen cross-link, the gel was irradiated with UV light (254 nm) for 2 min on each side before transfer of the DNA to a membrane for Southern blot analysis.
Blot analysis and quantitation
A list of probes used for blot hybridization is presented in Table 3.
Table 3.
Description of Southern blot probes and targets
Nucleic acids from agarose gels were blotted onto nylon membranes (Positive Membrane, Qbiogen) by capillary transfer in 1 M ammonium acetate as described previously (Allefs et al. 1990). DNA probes for hybridization were generated using the RadPrime DNA labeling system (Invitrogen) with incorporation of [α-32P]dATP (GE Healthcare) according to the instructions of the manufacturer. All probes used in this study were in the size range of 200–300 bp. Imaging and quantitation were performed using the FLA-5000 imaging system and MultiGauge software (FujiFilm).
For some blots (Figs. 2A–C [top and middle panel], D [top panel], 3A), probes were prepared using the PCR DIG Labeling Mix (Roche). Hybridization and detection were performed according to the DIG Application Manual (Roche). Images were collected using the LAS-3000 imaging system (FujiFilm).
Before rehybridization of a membrane, probe removal was achieved by adding boiling 0.1× SSPE buffer (18 mM sodium chloride, 1 mM sodium phosphate at pH 7.7, 0.1 mM EDTA) containing 0.5% SDS to the membrane and incubation for 15 min at 80°C. The procedure was repeated twice.
qPCR analysis
Yeast cells were grown in YPD to an OD600 of 0.5, and genomic DNA was prepared according to a standard protocol (Philippsen et al. 1991). The DNA was diluted in water to a final concentration of 5 fM, and 4 μL were used to inoculate a PCR reaction resulting in a final volume of 20 μL. All reactions were performed in triplicate using either primers 688 and 689 specific for the PHO5 single-gene locus, or primers 712 and 713 to amplify a region within the 18S rDNA. Detection was performed via fluorescence measurement of the DNA-intercalating dye SYBR Green (Roche) using a Rotor-Gene 3000 system (Corbett Life Science). Data were evaluated using the comparative quantitation analysis tool included in the Rotor-Gene software.
Acknowledgments
We are grateful to Drs. Masayasu Nomura and Manfred Schmid for providing us with yeast strains and the plasmid pFA6a-MN-KanMX6, respectively. We thank Axel Berger, Andreas Ehrensberger, and Drs. Philipp Milkereit, Anne Rascle, and Herbert Tschochner for carefully reading the manuscript and for constructive discussions. This work was supported by the 6th framework program of the EU, IRG 016489 (J.G.). M.H. and H.G. received fellowships of the Studienstiftung des Deutschen Volkes.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.466908.
References
- Allefs J.J., Salentijn E.M., Krens F.A., Rouwendal G.J. Optimization of non-radioactive Southern blot hybridization: Single copy detection and reuse of blots. Nucleic Acids Res. 1990;18:3099–3100. doi: 10.1093/nar/18.10.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P., Learned R.M., Jantzen H.M., Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- Bell S.P., Jantzen H.M., Tjian R. Assembly of alternative multiprotein complexes directs rRNA promoter selectivity. Genes & Dev. 1990;4:943–954. doi: 10.1101/gad.4.6.943. [DOI] [PubMed] [Google Scholar]
- Berger A.B., Decourty L., Badis G., Nehrbass U., Jacquier A., Gadal O. Hmo1 is required for TOR-dependent regulation of ribosomal protein transcription. Mol. Cell. Biol. 2007;27:8015–8026. doi: 10.1128/MCB.01102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier M., Fath S., Tschochner H. The composition of the RNA polymerase I transcription machinery switches from initiation to elongation mode. FEBS Lett. 2004;564:41–46. doi: 10.1016/S0014-5793(04)00311-4. [DOI] [PubMed] [Google Scholar]
- Boeger H., Griesenbeck J., Strattan J.S., Kornberg R.D. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- Cadwell C., Yoon H.J., Zebarjadian Y., Carbon J. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol. Cell. Biol. 1997;17:6175–6183. doi: 10.1128/mcb.17.10.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioci F., Vu L., Eliason K., Oakes M., Siddiqi I.N., Nomura M. Silencing in yeast rDNA chromatin: Reciprocal relationship in gene expression between RNA polymerase I and II. Mol. Cell. 2003;12:135–145. doi: 10.1016/s1097-2765(03)00262-4. [DOI] [PubMed] [Google Scholar]
- Conconi A., Widmer R.M., Koller T., Sogo J.M. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- Dammann R., Lucchini R., Koller T., Sogo J.M. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E.A., Warner J.R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984;39:663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Elion E.A., Warner J.R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol. Cell. Biol. 1986;6:2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S.L., Osheim Y.N., Cioci F., Nomura M., Beyer A.L. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol. Cell. Biol. 2003;23:1558–1568. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadal O., Labarre S., Boschiero C., Thuriaux P. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 2002;21:5498–5507. doi: 10.1093/emboj/cdf539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganley A.R., Hayashi K., Horiuchi T., Kobayashi T. Identifying gene-independent noncoding functional elements in the yeast ribosomal DNA by phylogenetic footprinting. Proc. Natl. Acad. Sci. 2005;102:11787–11792. doi: 10.1073/pnas.0504905102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaubatz J., Prashad N., Cutler R.G. Ribosomal RNA gene dosage as a function of tissue and age for mouse and human. Biochim. Biophys. Acta. 1976;418:358–375. doi: 10.1016/0005-2787(76)90297-5. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., Weissman J.S. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Griesenbeck J., Boeger H., Strattan J.S., Kornberg R.D. Purification of defined chromosomal domains. Methods Enzymol. 2004;375:170–178. doi: 10.1016/s0076-6879(03)75011-3. [DOI] [PubMed] [Google Scholar]
- Hall D.B., Wade J.T., Struhl K. An HMG protein, Hmo1, associates with promoters of many ribosomal protein genes and throughout the rRNA gene locus in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:3672–3679. doi: 10.1128/MCB.26.9.3672-3679.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Moazed D. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes & Dev. 2003;17:2162–2176. doi: 10.1101/gad.1108403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H.S., Kawauchi J., Braglia P., Alen C.M., Kent N.A., Proudfoot N.J. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct. Mol. Biol. 2007;14:123–130. doi: 10.1038/nsmb1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara K., Ohtsuki K., Ki S., Aoyama K., Takahashi H., Kobayashi T., Shirahige K., Kokubo T. Assembly of regulatory factors on rRNA and ribosomal protein genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:6686–6705. doi: 10.1128/MCB.00876-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J., Dodd J.A., Lalo D., Nomura M. Histones H3 and H4 are components of upstream activation factor required for the high-level transcription of yeast rDNA by RNA polymerase I. Proc. Natl. Acad. Sci. 1997;94:13458–13462. doi: 10.1073/pnas.94.25.13458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys D.A., Vu L., Steffan J.S., Dodd J.A., Yamamoto R.T., Nogi Y., Nomura M. RRN6 and RRN7 encode subunits of a multiprotein complex essential for the initiation of rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae. Genes & Dev. 1994;8:2349–2362. doi: 10.1101/gad.8.19.2349. [DOI] [PubMed] [Google Scholar]
- Keys D.A., Lee B.S., Dodd J.A., Nguyen T.T., Vu L., Fantino E., Burson L.M., Nogi Y., Nomura M. Multiprotein transcription factor UAF interacts with the upstream element of the yeast RNA polymerase I promoter and forms a stable preinitiation complex. Genes & Dev. 1996;10:887–903. doi: 10.1101/gad.10.7.887. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., Nasmyth K., Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Ganley A.R. Recombination regulation by transcription-induced cohesin dissociation in rDNA repeats. Science. 2005;309:1581–1584. doi: 10.1126/science.1116102. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Horiuchi T. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells. 1996;1:465–474. doi: 10.1046/j.1365-2443.1996.d01-256.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Heck D.J., Nomura M., Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes & Dev. 1998;12:3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkens T., Riggs D.L., Heck J.D., Planta R.J., Nomura M. The yeast RNA polymerase I promoter: Ribosomal DNA sequences involved in transcription initiation and complex formation in vitro. Nucleic Acids Res. 1991;19:5363–5370. doi: 10.1093/nar/19.19.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchini R., Sogo J.M. Replication of transcriptionally active chromatin. Nature. 1995;374:276–280. doi: 10.1038/374276a0. [DOI] [PubMed] [Google Scholar]
- Moss T., Stefanovsky V.Y. At the center of eukaryotic life. Cell. 2002;109:545–548. doi: 10.1016/s0092-8674(02)00761-4. [DOI] [PubMed] [Google Scholar]
- Musters W., Knol J., Maas P., Dekker A.F., van Heerikhuizen H., Planta R.J. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1989;17:9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogi Y., Yano R., Dodd J., Carles C., Nomura M. Gene RRN4 in Saccharomyces cerevisiae encodes the A12.2 subunit of RNA polymerase I and is essential only at high temperatures. Mol. Cell. Biol. 1993;13:114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P., Stotz A., Scherf C. DNA of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:169–182. doi: 10.1016/0076-6879(91)94014-4. [DOI] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Seraphin B. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- Rattner J.B., Saunders C., Davie J.R., Hamkalo B.A. Ultrastructural organization of yeast chromatin. J. Cell Biol. 1982;93:217–222. doi: 10.1083/jcb.93.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R.H. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog. Nucleic Acid Res. Mol. Biol. 1999;62:293–327. doi: 10.1016/s0079-6603(08)60511-5. [DOI] [PubMed] [Google Scholar]
- Russell J., Zomerdijk J.C. The RNA polymerase I transcription machinery. Biochem. Soc. Symp. 2006;73:203–216. doi: 10.1042/bss0730203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustchenko E.P., Curran T.M., Sherman F. Variations in the number of ribosomal DNA units in morphological mutants and normal strains of Candida albicans and in normal strains of Saccharomyces cerevisiae. J. Bacteriol. 1993;175:7189–7199. doi: 10.1128/jb.175.22.7189-7199.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M., Durussel T., Laemmli U.K. ChIC and ChEC; Genomic mapping of chromatin proteins. Mol. Cell. 2004;16:147–157. doi: 10.1016/j.molcel.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Steffan J.S., Keys D.A., Dodd J.A., Nomura M. The role of TBP in rDNA transcription by RNA polymerase I in Saccharomyces cerevisiae: TBP is required for upstream activation factor-dependent recruitment of core factor. Genes & Dev. 1996;10:2551–2563. doi: 10.1101/gad.10.20.2551. [DOI] [PubMed] [Google Scholar]
- Vogelauer M., Cioci F., Camilloni G. DNA protein-interactions at the Saccharomyces cerevisiae 35 S rRNA promoter and in its surrounding region. J. Mol. Biol. 1998;275:197–209. doi: 10.1006/jmbi.1997.1451. [DOI] [PubMed] [Google Scholar]
- Wai H., Johzuka K., Vu L., Eliason K., Kobayashi T., Horiuchi T., Nomura M. Yeast RNA polymerase I enhancer is dispensable for transcription of the chromosomal rRNA gene and cell growth, and its apparent transcription enhancement from ectopic promoters requires Fob1 protein. Mol. Cell. Biol. 2001;21:5541–5553. doi: 10.1128/MCB.21.16.5541-5553.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]