Abstract
Mutations in ATM (Ataxia telangiectasia mutated) result in Ataxia telangiectasia (A-T), a disorder characterized by progressive neurodegeneration. Despite advances in understanding how ATM signals cell cycle arrest, DNA repair, and apoptosis in response to DNA damage, it remains unclear why loss of ATM causes degeneration of post-mitotic neurons and why the neurological phenotype of ATM-null individuals varies in severity. To address these issues, we generated a Drosophila model of A-T. RNAi knockdown of ATM in the eye caused progressive degeneration of adult neurons in the absence of exogenously induced DNA damage. Heterozygous mutations in select genes modified the neurodegeneration phenotype, suggesting that genetic background underlies variable neurodegeneration in A-T. The neuroprotective activity of ATM may be negatively regulated by deacetylation since mutations in a protein deacetylase gene, RPD3, suppressed neurodegeneration, and a human homolog of RPD3, histone deacetylase 2, bound ATM and abrogated ATM activation in cell culture. Moreover, knockdown of ATM in post-mitotic neurons caused cell cycle re-entry, and heterozygous mutations in the cell cycle activator gene String/CDC25 inhibited cell cycle re-entry and neurodegeneration. Thus, we hypothesize that ATM performs a cell cycle checkpoint function to protect post-mitotic neurons from degeneration and that cell cycle re-entry causes neurodegeneration in A-T.
[Keywords: Drosophila, neurodegeneration, Ataxia telangiectasia, ATM, cell cycle, HDAC]
In 1995, Savitsky et al. (1995) linked the ataxia telangiectasia mutated (ATM) gene to the human autosomal recessive disorder Ataxia telangiectasia (A-T). Since then, cell-based and animal models have been used to understand how loss-of-function mutations in ATM give rise to A-T phenotypes, including immunodeficiency, radiation sensitivity, cancer predisposition, and neurodegeneration (Lavin and Shiloh 1997; Crawford 1998; McKinnon 2004). These studies have shown that the ATM serine/threonine protein kinase plays a central role in maintaining genome stability.
ATM monitors the genome for DNA double-strand breaks (DSBs) and responds to this signal by phosphorylating hundreds of proteins, including DNA repair factors, cell cycle regulators, and apoptosis regulators (Abraham 2001; Shiloh 2003; Matsuoka et al. 2007). ATM is recruited to DSBs by the trimeric MRE11–RAD50–NBS1 (MRN) DNA repair complex, which possesses ATP- dependent nuclease (MRE11) and DNA-tethering (RAD50) activities (Jackson 2002; Carson et al. 2003; Lee and Paull 2004, 2005). Recruitment of ATM to DSBs is mediated by the NBS1 subunit and is associated with autophosphorylation of ATM on Ser 1981 (pS1981) and conversion of inactive ATM dimers to active ATM monomers (Bakkenist and Kastan 2003; van den Bosch et al. 2003; Abraham and Tibbetts 2005; Falck et al. 2005; Lee and Paull 2005; You et al. 2005; Dupré et al. 2006). Although each of the above processes (ATM recruitment, autophosphorylation, and dimer dissociation) is important for ATM activation, the precise order of these events remains controversial. In addition, other post-translational modifications of ATM contribute to its regulation. Of particular relevance to this study, ATM undergoes acetylation by the Tip60 acetyltransferase in response to DNA damage, and ATM acetylation is required for activation of ATM kinase activity (Sun et al. 2005, 2007).
The most well-understood functions of ATM relate to its control of the cell cycle (Abraham 2001). ATM-deficient cells are defective in ionizing radiation (IR)-induced G1/S-, intra-S-, and G2/M-phase cell cycle checkpoints. Defective checkpoint function in ATM-deficient cells can be attributed, in part, to defective regulation of the CDC25 family of protein phosphatases. In mammals, Checkpoint kinase 1 (CHK1) and CHK2 are major effectors of ATM-dependent checkpoint activation. ATM phosphorylates and activates both kinases, which in turn directly phosphorylate and inactivate one of three CDC25 family members (CDC25A, CDC25B, and CDC25C) that mediate dephosphorylation and activation of cyclin-dependent kinases during cell cycle phase transitions (Boutros et al. 2006).
To date, it is unclear why mutation of ATM causes progressive degeneration of cerebellar Purkinje and granule neurons in A-T patients (Lavin and Shiloh 1997; Crawford 1998; McKinnon 2004). Other A-T phenotypes, such as radiation sensitivity and cancer predisposition, are attributed to the failure of cell cycle checkpoint activation, normally mediated by ATM in response to DSBs. However, chromosome breaks are presumed to occur rarely in neurons since they do not undergo DNA replication (Vilenchik and Knudson 2003). Studies of neurodegeneration in A-T have also been hampered by the lack of an experimental animal model. In particular, ATM knockout (ATM−/−) and knockin (ATMΔSRI/ΔSRI) mice do not display pronounced cerebellar degeneration (Barlow et al. 1996; Elson et al. 1996; Xu et al. 1996; Spring et al. 2001). However, recent studies hinted that Drosophila might be a useful model to investigate ATM function in neurons. The mammalian ATM signaling pathway is largely conserved in Drosophila, and analysis of ATM mutant flies has shown that ATM is involved in early activation of the G2/M-phase cell cycle checkpoint elicited by IR-induced DNA damage and in maintaining chromosome integrity (Bi et al. 2004, 2005; Brodsky et al. 2004; Ciapponi et al. 2004; Oikemus et al. 2004; Silva et al. 2004; Song et al. 2004; Song 2005).
To understand the mechanisms underlying neurodegeneration in A-T, we generated a Drosophila model in which RNAi was used to knock down ATM expression in neurons. ATM knockdown resulted in progressive degeneration of neurons through a programmed cell death pathway. Neurons in ATM knockdown flies re-entered the cell cycle, as determined by assays for DNA content, DNA replication, or mitosis. Heterozygous mutation of the cell cycle activator gene String/CDC25 suppressed both cell cycle re-entry and degeneration of ATM knockdown neurons, suggesting that cell cycle re-entry is causative for neurodegeneration. Additionally, the RPD3 deacetylase was implicated as a negative regulator of ATM function in neurons. A human homolog of RPD3, histone deacetylase 2 (HDAC2), physically interacted with ATM and negatively regulated ATM activation in the absence of exogenously induced DNA damage. Thus, mechanistic insights provided by the Drosophila A-T model may lead to the development of therapies for the most debilitating clinical manifestation of A-T, neuromotor dysfunction.
Results
Generation of ATM knockdown flies
To investigate the molecular mechanisms that underlie neurodegeneration in A-T, we generated transgenic flies (pATM flies) in which the yeast GAL4-UAS system was used to induce conditional or tissue-specific knockdown of ATM by RNAi (Brand and Perrimon 1993). The pWIZ vector was used to construct the pATM transgene, which contained inverted copies of exon 6 from the Drosophila ATM gene (Lee and Carthew 2003). When transcribed, pATM produced dsRNA that targeted ATM mRNA for degradation by the RNAi machinery (Fig. 1A). Knockdown of ATM in pATM transgenic lines was verified by RT–PCR of adult flies and Western blot analysis of embryos carrying the heat-shock-inducible hsp70-GAL4 driver (hsp70-GAL4/+; pATM/+ flies) (Fig. 1B,C). In addition, consistent with the essential nature of ATM in flies, ubiquitous knockdown of ATM by the actin-GAL4 driver or neuron-specific knockdown of ATM by the Elav-GAL4 driver (at 25°C, but not 18°C) resulted in lethality (Oikemus et al. 2004; Silva et al. 2004; Song et al. 2004). These data indicate that dsRNA transcribed from pATM reduces ATM expression in flies.
Figure 1.
ATM RNAi in pATM flies reduced the level of ATM mRNA and protein. (A) A schematic diagram of the pWIZ-ATM (pATM) transformation vector (Lee and Carthew 2003). (B) Quantitative PCR analysis of ATM mRNA in heat-shocked hsp70-GAL4/+; pATMT4/+ (hs-ATMi) and hsp70-GAL4/+ (hs-GAL4) flies. Graphed is the level of ATM mRNA relative to actin 5C mRNA in hs-ATMi flies normalized to hs-GAL4 flies. Error bars represent the standard error of the mean of experiments performed in triplicate. (C, lanes 3,4) Western blot analysis of ATM in hsp70-GAL4/pATMT4 and hsp70-GAL4/+ embryos. Extracts were probed with an α-ATM antibody and an α-SIN3 antibody as a loading control (Pile and Wassarman 2000). Note that lane 4 was overloaded relative to lane 3. (Lanes 1,2) RNAi-mediated knockdown of ATM in S2 cells by long dsRNA served as a marker for ATM.
Based on the algorithm described by Dietzl et al. (2007), the ATM dsRNA was unlikely to have off-target RNAi effects. A specificity score of 1.0 reflects no off-target effects, and the ATM dsRNA had specificity scores of S19 = 0.9987 (on all on-target 19-mer matches) and S18 = 0.9975. Furthermore, the ATM dsRNA did not contain CAN repeats, a common cause of off-target RNAi effects (Ma et al. 2006). Finally, as described below, phenotypes of ATM knockdown flies were similar to ATM mutant flies, and genetic modifiers of ATM knockdown phenotypes were enriched for recognized components of the ATM signaling pathway, such as RAD50 and CDC25. These data suggest that ATM expression is exclusively affected in ATM knockdown flies.
ATM knockdown causes degeneration of photoreceptor neurons
To determine whether ATM is required for neuron survival in flies, we examined photoreceptor neurons in the eye. The adult fly eye is made up of ∼800 unit eyes called ommatidia, each of which contains eight post-mitotic photoreceptor neurons (Fig. 2A; Wolff and Ready 1993). ATM knockdown in all cells or in neurons of the eye was achieved using the GMR-GAL4 or Elav-GAL4 driver, respectively (Hay et al. 1994; Yao and White 1994). Morphological analysis of neurons was carried out by transmission electron microscopy (TEM) of thin sections of 3-d-old adult flies. ATM knockdown induced by either driver resulted in neurodegeneration, as marked by swelling, fragmentation, and loss of rhabdomeres (the light-sensitive plasma membranes of photoreceptor neurons) throughout the eye (Fig. 2B,D; Supplemental Fig. 1). We consider these phenotypes signs of degeneration as they are observed in flies with light-induced or age-related photoreceptor degeneration due to mutations in phototransduction pathway genes (Davidson and Steller 1998; Hsu et al. 2004). In contrast, these phenotypes were not observed in GMR-GAL4, Elav-GAL4, or pATM flies, although GMR-GAL4 flies had a low rate of ommatidial defects (Figs. 2A,C, 3; data not shown). Thus, ATM protects post-mitotic photoreceptor neurons from degeneration.
Figure 2.
RNAi-mediated knockdown of ATM causes degeneration of photoreceptor neurons. Shown are representative TEM micrographs of single ommatidia (A–H) and high-magnification micrographs of single photoreceptor neurons from flies of the indicated genotypes (A′–H′). (A) Three-day-old Elav-GAL4/+ (Elav-GAL4). (B) Three-day-old Elav-GAL4/pATMT4 (Elav-ATMi). (C) Three-day-old GMR-GAL4/+ (GMR-GAL4). (D) Three-day-old GMR-GAL4/+; pATMT4/+ (GMR-ATMi). (E) Three-day-old hs-ATMi not heat-shocked. (F) Three-day-old hs-ATMi heat-shocked throughout development. (G) Fifteen-day-old hs-ATMi not heat-shocked. (H) Fifteen-day-old hs-ATMi heat-shocked days 3–15 of adulthood. (A′–H′) Red circles indicate the photoreceptor neuron that is shown in high magnification. Red arrows indicate defects in rhabdomere structure.
Figure 3.
ATM knockdown causes progressive degeneration of adult neurons. Zero-day-old to 4-d-old GMR-GAL4, GMR-ATMi, and GMR-GAL4/+; pATMT4/StgEY12388 (GMR-ATMi, Stg) adult flies were collected and aged for 9, 27, and 48 d. The number of morphologically wild-type photoreceptor neurons per ommatidium was determined by analyzing TEM images of sections at the R7 level in the center of each fly eye. Error bars represent standard error of the mean. P-values were calculated using two-way ANOVA and Bonferonni post-tests. (A) Flies of the same genotype were compared at each time point. (B) Flies of different genotypes were compared at each time point.
ATM knockdown in post-mitotic, adult neurons is sufficient to cause degeneration
To determine when ATM is required during the lifetime of a neuron for neuron survival, we used the hsp70-GAL4 driver to knock down ATM either during eye development or in the adult eye. Specifically, hsp70-GAL4/+; pATMT4/+ (hs-ATMi) flies were either heat-shocked twice daily from the time of egg laying to day 3 of adulthood, or from day 3 to day 15 of adulthood. TEM analysis revealed that, in both cases, ATM knockdown resulted in degeneration of adult photoreceptor neurons (Fig. 2F,H). In contrast, no degeneration was observed in non-heat-shocked hs-ATMi flies or control heat-shocked hsp70-GAL4/+ (hs-GAL4) flies (Fig. 2E,G; Supplemental Fig. 2). Since ATM knockdown in post-mitotic, adult neurons is sufficient to cause degeneration, neurodegeneration can occur independently of developmental abnormalities.
ATM knockdown causes progressive degeneration of adult neurons
In humans, mutation of ATM causes progressive neurodegeneration. Typically, A-T patients are asymptomatic at birth, present with an ataxic gait around the age of two, and are wheelchair-bound by the age of 10 (Crawford 1998). Analysis of GMR-GAL4/+; pATMT4/+ (GMR-ATMi) flies revealed that the extent of neurodegeneration became more severe as adult flies aged. GMR-ATMi flies, but not control GMR-GAL4 flies, had significantly higher levels of neurodegeneration at 27–31 d versus 48–52 d (Fig. 3A). In addition, GMR-ATMi flies had significantly higher levels of neurodegeneration than GMR-GAL4 flies at all time points examined (Fig. 3B). No significant difference was observed in the level of neurodegeneration between GMR-ATMi flies at any given time point, indicating that the progressive neurodegeneration phenotype is robust. Thus, in flies, as in humans, loss of ATM causes age-dependent, progressive degeneration of post-mitotic neurons.
ATM knockdown causes neurodegeneration by programmed cell death
Necrosis and programmed cell death are the primary mechanisms by which a neuron can die (Yuan et al. 2003). To determine whether a programmed cell death pathway mediates degeneration of ATM knockdown neurons, we examined the requirement for caspase activity in GMR-ATMi flies. In Drosophila, both apoptotic and autophagic programmed cell death can be blocked by expression of P35, a broad-specificity caspase inhibitor encoded by the baculovirus Autographa californica (Clem and Miller 1994; Hay et al. 1994; Chen et al. 1996; White et al. 1996; Martin and Baehrecke 2004). Expression of P35 or dIAP1 (Drosophila inhibitor of apoptosis 1), a Drosophila homolog of P35, suppressed the external rough eye phenotype of GMR-ATMi flies (discussed below) (Fig. 4C; Supplemental Table 2; Hay et al. 1995). In contrast, heterozygous mutation of dIAP1 enhanced the rough eye phenotype (Supplemental Table 2). Moreover, while expression of GMR-P35 alone did not affect photoreceptor morphology or number, expression of GMR-P35 significantly suppressed the neurodegeneration phenotype of GMR-ATMi flies (Fig. 6 [below]; Supplemental Fig. 3). Thus, ATM knockdown causes caspase-dependent programmed cell death of post-mitotic photoreceptor neurons.
Figure 4.
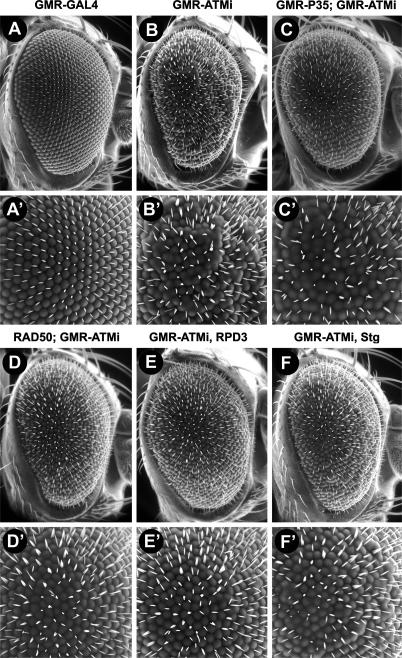
Suppressors of the GMR-ATMi rough eye phenotype. Shown are representative SEM micrographs of 0- to 4-d-old control flies (A,B) and suppressors of GMR-ATMi (C–F). (A′–F′) High-magnification micrographs of the center of the eye are shown below. (A) GMR-GAL4. (B) GMR-ATMi. (C) GMR-P35/+; GMR-GAL4/+; pATMT4/+ (GMR-P35; GMR-ATMi). (D) GMR-GAL4/RAD50EP1; pATMT4/+ (RAD50; GMR-ATMi). (E) GMR-GAL4/+; pATMT4/RPD304556 (GMR-ATMi, RPD3). (F) GMR-GAL4/+; pATMT4/StgEY12388 (GMR-ATMi, Stg).
Figure 6.
Genetic background modified the severity of the GMR-ATMi neurodegeneration phenotype. The number of morphologically wild-type photoreceptor neurons per ommatidium was determined by analyzing TEM images of sections at the R7 level in the center of eyes from 0- to 4-d-old flies of the indicated genotypes. More than 260 ommatidia were scored from at least four eyes of each genotype. RAD50EP1, RPD304556, StgEY12388, or GMR-P35 alleles were used for this analysis. (A) Graphed is the average number of normal photoreceptors per ommatidium for each genotype. Error bars represent the standard error of the mean. P-values were calculated using one-way ANOVA and Dunnett’s multiple comparison post-tests where all genotypes were compared with GMR-ATMi flies. The statistical difference between genotypes is not attributable to the variability seen in flies of a given genotype. (B) Graphed is the percentage of total ommatidia with zero to seven normal photoreceptor neurons for each genotype.
Identification of genetic modifiers of neurodegeneration in ATM knockdown flies
Having established a Drosophila model that recapitulates aspects of neurodegeneration that occur in A-T, we next sought to identify genetic modifiers of this condition. The existence of genes that modify ATM loss-of-function phenotypes is hinted at by the fact that some A-T patients have little or no ATM protein yet have mild neurological presentation (Hassin-Baer et al. 1999; Lavin et al. 2006; Alterman et al. 2007). Also, cell fusion studies showed that ATM−/− cells derived from different A-T patients fall into four distinct genetic complementation groups, suggesting that A-T phenotypes are caused by at least four different mutant genes (Murnane and Painter 1982; Buchwald 1995).
Several lines of evidence indicated that GMR-ATMi flies could be used to identify genes that modulate neurodegeneration. GMR-ATMi flies had an external rough eye phenotype that could be detected by scanning electron microscopy (SEM) as well as light microscopy (Fig. 4B; data not shown). Among the pATM lines, the severity of the rough eye phenotype positively correlated with the severity of neurodegeneration. To illustrate, when pATM lines were driven by GMR-GAL4, the severity of rough eye and neurodegeneration phenotypes fit the allelic series pATMT7 < pATMT4 < pATMT4/pATMT4 (pATMT4 and pATMT7 are alleles of pATM in which the transgene is inserted at different positions on the third chromosome) (data not shown). In addition, expression of GMR-P35 suppressed both the rough eye and neurodegeneration phenotypes of GMR-ATMi flies, indicating that the rough eye phenotype could be used as readout for the neurodegeneration phenotype (Figs. 4C, 6 [below]). Thus, using the scheme outlined in Supplemental Figure 4, we performed an unbiased screen of a random collection of 650 P-element mutants for modifiers of the GMR-ATMi rough eye phenotype.
The genetic screen identified alleles of String (Stg), RAD50, and RPD3 as suppressors of the GMR-ATMi rough eye phenotype and MEKK4 (mitogen-activated protein [MAP] kinase kinase kinase 4), PP2A-B′ (protein phosphatase 2A-B′), and Delta as enhancers of the GMR-ATMi rough eye phenotype (Figs. 4D–F, 5). Supplemental Table 1 provides a summary of the P-element alleles identified in the screen and the phenotypes of allelic mutants used to confirm the modifying genes. In all cases, the severity of the rough eye phenotype reflected the severity of neurodegeneration. Suppressors of the rough eye phenotype caused a significant increase in the average number of morphologically normal photoreceptors per ommatidium relative to GMR-ATMi flies (Fig. 6). In contrast, enhancers of the rough eye phenotype reduced the number of morphologically normal photoreceptors to such an extent that it was not possible to distinguish individual ommatidia (data not shown). Control flies heterozygous mutant for the identified genes had wild-type eye phenotypes (data not shown). Finally, heterozygous mutations in Stg significantly reduced the severity of progressive neurodegeneration in GMR-ATMi flies (Fig. 3). Thus, the extent of degeneration of ATM knockdown neurons is affected by the dose of second site genes, which presumably function in cellular processes that regulate neuron survival.
Figure 5.
Enhancers of the GMR-ATMi rough eye phenotype. Shown are representative SEM micrographs of 0- to 4-d-old control flies (A,B) and enhancers of GMR-ATMi (C–E). (A′–E′) High-magnification micrographs of the center of the eye are shown below. Note the increased incidence of ommatidium fusions in the presence of enhancer mutations. (A) GMR-GAL4. (B) GMR-ATMi. (C) GMR-GAL4/+; pATMT4/ MEKK4EY02276 (GMR-ATMi, MEKK4). (D) GMR-GAL4/+; pATMT4/PP2A-B′A131 (GMR-ATMi, PP2A-B′). (E) GMR-GAL4/+; pATMT4/Delta05151 (GMR-ATMi, Delta).
Cell cycle regulatory genes modify the ATM knockdown phenotype
Several lines of evidence suggest that cell cycle re-entry plays a role in the mechanism that underlies neurodegeneration in human A-T (Allen et al. 2001; Copani et al. 2001; Yang and Herrup 2005). In particular, aberrant expression of cell cycle regulatory genes in post-mitotic Purkinje and granule cells of A-T patients supports a role for cell cycle re-entry in neurodegeneration (Yang and Herrup 2005). In addition, the importance of cell cycle regulation in the process of neurodegeneration was suggested by our finding that Stg mutations inhibited degeneration of ATM knockdown neurons. Stg encodes the Drosophila homolog of mammalian CDC25 proteins that mediate dephosphorylation and activation of cyclin-dependent kinases during cell cycle phase transitions (Edgar and O’Farrell 1990; Edgar et al. 2001; Park et al. 2003). In a small-scale screen of other cell cycle regulatory genes, we found that heterozygous mutation of Cdk2 (cyclin-dependent kinase 2, also known as cdc2c) and dE2F2 suppressed the GMR-ATMi rough eye phenotype, whereas heterozygous mutation of dE2F1 enhanced the GMR-ATMi rough eye phenotype (Supplemental Table 2). Cdk2, together with its partner Cyclin E, is a critical regulator of entry into S phase, and dE2F1 and dE2F2 are transcription factors that regulate cell proliferation, differentiation, and apoptosis (Moon et al. 2005; Baker 2007). Consistent with the identification of dE2F1 mutants as enhancers of GMR-ATMi, E2F1 functions as a cell cycle suppressor in differentiated post-mitotic mouse neurons (Wang et al. 2007). These data suggest that activities of cell cycle regulatory genes modulate the extent of degeneration of ATM knockdown post-mitotic photoreceptor neurons.
ATM knockdown causes neurons to express S-phase and M-phase cell cycle markers
To explore the connection between cell cycle re-entry and neurodegeneration, we examined photoreceptor neurons in larval eye imaginal discs. In the eye disc, the morphogenetic furrow (MF) marks key cell cycle events important for cell proliferation and differentiation (Wolff and Ready 1993). The MF is initiated at the posterior of the eye disc and then traverses the disc. Cells anterior to the MF divide asynchronously but upon entry into the MF go into synchronous cell cycle arrest. Upon emerging from the posterior edge of the MF, a subset of photoreceptor neurons remain arrested in G1 phase and presumably remain post-mitotic for the lifetime of the fly. Surrounding cells undergo synchronous division and the resulting cells, including photoreceptor neurons, remain post-mitotic.
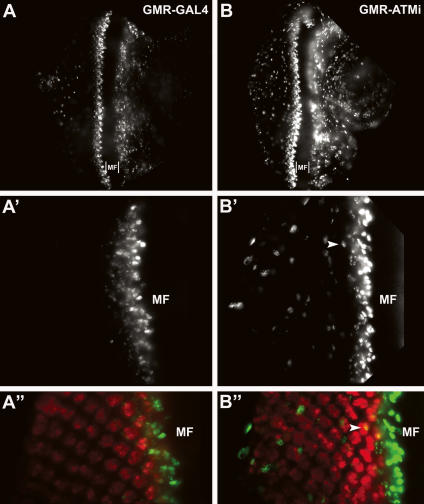
To determine the extent to which DNA replication or mitosis occurs in post-mitotic neurons, eye discs were costained with antibodies to Elav and 5-bromodeoxyuridine (α-BrdU) or Elav and phosphorylated Ser 10 on histone H3 (α-PH3), respectively. Elav is an early marker of neuronal differentiation that in the eye is exclusively expressed in all post-mitotic neurons (Soller and White 2004). BrdU incorporation occurs in cells actively undergoing DNA replication, and phosphorylation of Ser 10 on histone H3 occurs in cells undergoing mitosis (Yao and White 1994; Wei et al. 1998). As shown in Figure 7, A and B, a substantial increase in BrdU incorporation was observed posterior to the MF in GMR-ATMi eye discs compared with control GMR-GAL4 eye discs. Furthermore, α-Elav and α-BrdU costained cells were sporadically observed in GMR-ATMi but not GMR-GAL4 eye discs (Fig. 7A′, A″, B′, B″). The α-PH3 antibody detected an equivalent frequency and pattern of cells undergoing mitosis juxtaposed to the MF in GMR-ATMi and GMR-GAL4 discs (Supplemental Fig. 5). However, many more cells further posterior to the MF were α-PH3-positive in GMR-ATMi than control GMR-GAL4 eye discs. Additionally, cells costained with α-Elav and α-PH3 were observed in GMR-ATMi but not control GMR-GAL4 eye discs. These data indicate that ATM knockdown disrupts normal cell cycle control and drives neurons that are normally post-mitotic into the cell cycle.
Figure 7.
DNA replication occurred in neurons of GMR-ATMi flies. Eye discs from control GMR-GAL4 (A,A′,A″) or GMR-ATMi (B,B′,B″) third instar larvae were costained with α-Elav (red) and α-BrdU (green) antibodies. A′ and A″ and B′ and B″ are higher-magnification images of A and B, respectively. A″ and B″ are merged images of α-Elav and α-BrdU. Arrows indicate a neuron in which DNA replication has occurred. MF indicates the position of the MF.
ATM knockdown causes post-mitotic neurons to re-enter the cell cycle
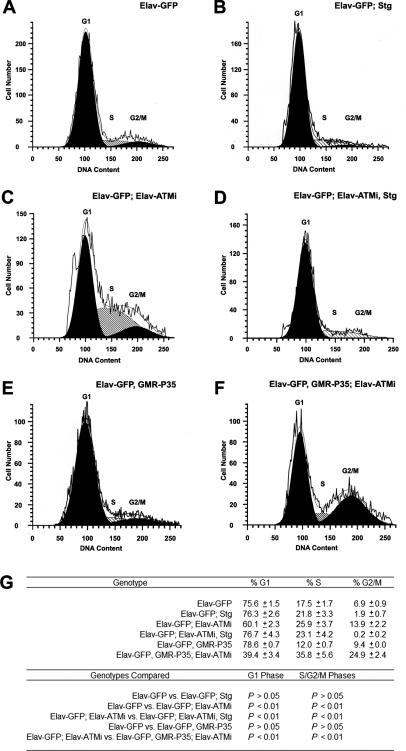
As an alternative approach to examine the cell cycle in ATM knockdown neurons, we analyzed the DNA content of neurons using a fluorescence-activated cell sorter (FACS). For this analysis, Elav-GAL4 was used to drive expression of pATMT4 and UAS-GFP (UAS-Green Fluorescent Protein) (Elav-GAL4, UAS-GFP; pATMT4/+: Elav-GFP; Elav-ATMi flies) in neurons. Larval eye discs were dissociated into single cells, and FACS was used to quantitate Hoechst staining of DNA in GFP-positive neurons. This analysis revealed that Elav-GFP; Elav-ATMi eye discs had a significantly lower fraction of neurons in G1 phase and higher fraction of cells in S/G2/M phases than control Elav-GAL4, UAS-GFP (Elav-GFP) eye discs (Fig. 8A,C,G). Thus, the FACS studies provide independent support for the conclusion that ATM knockdown causes re-entry of post-mitotic photoreceptor neurons into the mitotic cell cycle.
Figure 8.
Cell cycle re-entry in Elav-ATMi flies is inhibited by mutation of Stg but not expression of P35. FACS analysis was carried out on eye imaginal discs dissected from control Elav-GAL4,UAS-GFP/+ (Elav-GFP, n = 9) (A); Elav-GAL4, UAS-GFP/+; StgEY12388/+ (Elav-GFP; Stg, n = 4) (B); Elav-GAL4, UAS-GFP/+; pATMT4/+ (Elav-GFP; Elav-ATMi, n = 8) (C); Elav-GAL4, UAS-GFP/+; pATMT4/StgEY12388 (Elav-GFP; Elav-ATMi, Stg, n = 6) (D); Elav-GAL4, UAS-GFP/GMR-P35 (Elav-GFP, GMR-P35, n = 2) (E); and Elav-GAL4, UAS-GFP/GMR-P35; pATMT4/+ (Elav-GFP, GMR-P35; Elav-ATMi, n = 4) (F) third instar larvae. (G) Quantitation of cell cycle phases from multiple, independent samples. For each sample, 5000 live, single, GFP-positive events were analyzed. P-values were calculated using one-way ANOVA and Tukey’s multiple comparison post-tests.
Mutation of Stg blocks cell cycle re-entry of ATM knockdown neurons
We hypothesized that reducing the dose of Stg in GMR-ATMi flies suppresses neurodegeneration by suppressing cell cycle re-entry. In response to DSBs, ATM activates cell cycle checkpoints by phosphorylating CHK1 and CHK2 kinases, which in turn phosphorylate and inactivate CDC25 proteins (Abraham 2001; Shiloh 2003; McKinnon 2004). FACS was used to examine whether mutation of Stg affected the cell cycle re-entry phenotype of ATM knockdown neurons. As shown in Figure 8, heterozygous mutation of Stg had no significant effect on the cell cycle profile of neurons in control Elav-GFP eye discs; however, it significantly increased the fraction of neurons in G1 phase and reduced the fraction of cells in S/G2/M phases in Elav-GFP; Elav-ATMi eye discs (Fig. 8B,D,G). Given that Stg mutations suppressed degeneration of ATM knockdown neurons, these data indicate that cell cycle re-entry is causative for degeneration of ATM knockdown neurons.
Cell cycle re-entry precedes neurodegeneration of ATM knockdown neurons
To examine the extent to which ATM knockdown causes apoptosis of photoreceptor neurons in eye discs, control GMR-GAL4 and GMR-ATMi eye discs were analyzed by TUNEL (terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling) and acridine orange staining. Both analyses revealed a reduction in apoptotic cells directly posterior to the MF and an increase in apoptotic cells more posterior to the MF in GMR-ATMi eye discs relative to control GMR-GAL4 eye discs (Supplemental Fig. 6). The TUNEL staining pattern of GMR-ATMi eye discs was similar to that of ATM mutant eye discs (Song et al. 2004). Additionally, occasional TUNEL-positive signals in GMR-ATMi eye discs colocalized with neurons, as marked by α-Elav fluorescence. These data indicate that ATM knockdown blocks apoptosis of mitotic cells directly posterior to the MF and promotes apoptosis of older post-mitotic neurons more posterior to the MF.
To examine the epistatic relationship between cell cycle re-entry and apoptosis of neurons, we used FACS to analyze the cell cycle profile of neurons from Elav-GFP; Elav-ATMi eye discs that expressed the apoptosis inhibitor P35. We hypothesized that if cell cycle re-entry caused apoptosis, then expression of P35 would prevent cycling neurons from dying and increase the percentage of cycling neurons. In contrast, if apoptosis caused cell cycle re-entry, then expression of P35 would inhibit cell cycle re-entry and reduce the percentage of cycling neurons. This analysis revealed that in Elav-GFP; Elav-ATMi flies, GMR-P35 caused a significant reduction in G1-phase neurons and increase in S/G2/M-phase neurons, but in control Elav-GFP flies, GMR-P35 had no effect on the cell cycle profile (Fig. 8E–G). Thus, inhibiting apoptosis did not inhibit cell cycle re-entry of ATM knockdown neurons but instead caused them to accumulate in S/G2/M phases, suggesting that cell cycle re-entry precedes apoptosis in ATM knockdown neurons. In other words, these data provide evidence that ATM knockdown neurons that re-entered the cell cycle are fated to die.
HDAC2 physically and functionally interacts with ATM in human cells
Another suppressor identified in the screen, RPD3, encodes the Drosophila homolog of human class 1 HDAC1, HDAC2, HDAC3, and HDAC8 (Pandey et al. 2002). RPD3 has several potential links to ATM. Inhibition of HDACs by trichostatin A (TSA) induces ATM autophosphorylation in the absence of DNA damage and hyperphosphorylation upon IR-induced DNA damage (Bakkenist and Kastan 2003). HDAC1 physically associates with ATM in vitro and in vivo, and the extent of association is increased by exposure of cells to IR (Kim et al. 1999). Acetylation of ATM by the Tip60 acetyltransferase activates ATM kinase activity in response to DNA damage (Sun et al. 2005). Finally, the lysine residue acetylated by Tip60 in human ATM is conserved in Drosophila ATM (Sun et al. 2007). These observations suggest that RPD3 negatively regulates ATM activation, perhaps by directly antagonizing Tip60-mediated ATM acetylation.
In support of this model, we found that HDAC2 physically associated with ATM in human HEK 293T cells. ATM coprecipitated with overexpressed, Flag-epitope-tagged HDAC2 and endogenous HDAC2 (Fig. 9A,B). In addition, the interaction between ATM and HDAC2 was observed under basal and DNA damage conditions (Fig. 9A). To examine the functional consequence of the ATM–HDAC2 interaction, we assayed ATM activation, as measured by autophosphorylation of Ser 1981 (pS1981) and trans-phosphorylation of 53BP1 (an ATM kinase substrate), both of which are detected by an antibody generated against pS1981 (Bakkenist and Kastan 2003; Jowsey et al. 2007; R.S. Tibbetts, unpubl.). In the absence of IR, ATM activation, as indicated by ATM S1981 and 53BP1 phosphorylation, was enhanced by siRNA knockdown of HDAC2 in HEK 293T cells (Fig. 9C). In contrast, while HDAC1 was found to associate with ATM, HDAC1 knockdown did not cause ATM activation (data not shown). These data suggest that ATM is a direct substrate for deacetylation and repression of kinase activity by HDAC2. However, our inability to consistently observe acetylated ATM by Western blot with α-acetyl-lysine antibodies precluded a direct test of this hypothesis. Thus, it is formally possible that mutation of RPD3/HDAC suppresses degeneration of ATM knockdown neurons through increased acetylation of proteins other than ATM.
Figure 9.
HDAC2 interacts with ATM and represses ATM activation. (A) Western blot analysis of lysates and immunoprecipitates from HEK 293T cells transfected with empty plasmid (vector) or plasmids that expressed Flag-epitope-tagged human HDAC1 (FLAG-H1) or HDAC2 (FLAG-H2). Cells were either exposed to 20 Gy of IR (+) or not exposed (−), and cell lysates were immunoprecipitated with an α-Flag antibody. Immunoprecipitates (IP) or total lysates (Input) were immunoblotted (WB) for ATM or the Flag tag. (B) Lysates prepared from HEK 293T cells were immunoprecipitated with a rabbit IgG antibody (C), an α-ATM antibody from Santa Cruz Biotechnology (A1), and α-ATM antibody from Genetex (A2), or an α-HDAC2 antibody (H2). Immunoprecipitates (IP) were immunoblotted (WB) for ATM or HDAC2. (C) HEK 293T cells were transfected with control scrambled (S) or HDAC2 (H2)-specific siRNAs. Cell lysates were immunoblotted (WB) for pS1981 on ATM (pS1981-ATM), total ATM (ATM) as a loading control, or HDAC2. The triangle indicates increasing amounts of siRNAs.
Discussion
Human A-T can be modeled in Drosophila
Our data indicate that ATM knockdown by RNAi causes degeneration of Drosophila post-mitotic photoreceptor neurons. The neurodegeneration phenotype of ATM knockdown flies is similar to that observed in A-T patients. Neurodegeneration in the fly model occurred in the absence of exogenously induced DNA damage, it occurred independently of developmental defects, and it was progressive, increasing in severity as flies aged (Figs. 2, 3). Thus, ATM knockdown flies appear to be an appropriate model to study the cellular mechanisms underlying neurodegeneration in A-T.
Analysis of ATM knockdown flies identified genetic modifiers of neuron survival
A-T is a monogenic disease resulting from mutation of the ATM gene; however, our genetic screen identified second site genes that affect an A-T phenotype, neurodegeneration (Figs. 4, 5; Supplemental Tables 1, 2). Remarkably, independent lines of evidence, from the literature and from our studies, support the relevance of each of the modifier genes to the mechanism underlying neurodegeneration in A-T.
Three of the six identified genes—Stg, RAD50, and PP2A–B′—are known components of the ATM signaling pathway that responds to DNA damage in mammals (Abraham 2001; Shiloh 2003). The relevance of Stg to ATM signaling and neurodegeneration is described in a subsequent section of the Discussion. RAD50 encodes a component of the MRN complex that mediates ATM activation at DSBs (Abraham and Tibbetts 2005). A role for RAD50 in promoting neurodegeneration is not specific to the eye, as mutation of RAD50 suppressed the lethality of Elav-ATMi flies (data not shown). In addition, mutation of the gene encoding the NBS1 subunit of MRN suppressed the GMR-ATMi rough eye phenotype, suggesting that suppression by RAD50 and NBS1 mutants is due to reduced activity of the MRN complex (Supplemental Table 2). Nevertheless, the mechanism underlying suppression of neurodegeneration is unclear since reduced levels of the MRN complex would intuitively be expected to enhance GMR-ATMi phenotypes. One possibility is that the MRN complex is deregulated in the absence of ATM and carries out activities that are lethal to neurons. Finally, PP2A has been shown to dephosphorylate several ATM signaling pathways substrates, including ATM (Guo et al. 2002; Goodarzi et al. 2004; Chowdhury et al. 2005; Petersen et al. 2006). Mutation of PP2A–B′, which encodes a regulatory subunit of the PP2A complex, may enhance neurodegeneration in ATM knockdown flies by affecting the phosphorylation state of ATM substrates (Liu et al. 2007).
Two of the identified genes, MEKK4 and Delta, have potential links to ATM. Mutation of MEKK4 may enhance neurodegeneration in GMR-ATMi flies by allowing cell cycle progression. Published studies suggest a model whereby ATM and MEKK4 pathways collaborate to prevent cell cycle re-entry of post-mitotic neurons by maintaining the latency of CDC25 proteins (Ben-Levy et al. 1998; Abraham 2005; Lopez-Aviles et al. 2005; Manke et al. 2005; Boutros et al. 2006). Delta encodes a ligand for the Notch receptor, which regulates cell cycle progression and differentiation in many tissues, including the eye (Baonza and Freeman 2005; Yang and Baker 2006). Thus, there may be cross-talk between the ATM and Notch signaling pathways in neurons.
Finally, studies in cultured cells revealed a direct link between HDAC2, the human homolog of Drosophila RPD3, and ATM. HDAC2 was found to directly associate with ATM and regulate its kinase activity in the absence of exogenously induced DNA damage (Fig. 9). Thus, HDAC2 is likely the TSA-sensitive deacetylase that negatively regulates ATM kinase activity (Bakkenist and Kastan 2003). HDAC2 may function by counteracting acetylation of ATM or downstream components of the ATM signaling pathway (Sun et al. 2005, 2007). It is important to note that although deacetylation of ATM by HDAC2 may regulate ATM activity, HDAC2 is not necessarily an important factor in A-T since the majority of mutations in A-T patients are nonsense or frameshift mutations that result in complete loss or truncation of ATM protein. Nevertheless, the demonstrated physical and functional interactions between HDAC2 and ATM indicate that HDAC2 is an important component of the ATM signaling paradigm and that information garnered from studies of ATM knockdown flies can advance our understanding of ATM function in humans.
Results from the genetic screen predict that A-T patients with mild neurodegeneration will carry heterozygous mutations in suppressor genes, such as CDC25 family members, whereas A-T patients with severe neurodegeneration will carry heterozygous mutations in enhancer genes, such as MEKK4. It will be interesting to see if genes that enhance neurodegeneration in ATM knockdown flies also enhance neurodegeneration in mice. For example, do ATM−/− MEKK4+/− mice exhibit progressive degeneration of cerebellar neurons? If so, this would make mice a practical model for studying the neurodegenerative aspects of A-T.
Cell cycle re-entry may cause neurodegeneration in A-T
Our data indicate a causal relationship between cell cycle re-entry and neurodegeneration in the Drosophila model of A-T presented here. First, ATM knockdown in photoreceptor neurons resulted in cell cycle re-entry and neurodegeneration, implicating ATM in both processes (Figs. 2, 6–8). Second, heterozygous mutation of cell cycle regulatory genes Stg/CDC25, Cdk2, dE2F1, and dE2F2 modified the neurodegeneration phenotype of ATM knockdown flies, highlighting the importance of cell cycle regulation in neurodegeneration (Supplemental Table 2). Third, inhibition of cell cycle re-entry by mutation of Stg/CDC25 also inhibited degeneration of ATM knockdown neurons. In contrast, inhibition of neurodegeneration by expression of P35 did not inhibit cell cycle re-entry (Figs. 6, 8). Finally, inhibition of neurodegeneration by expression of P35 caused the accumulation of neurons in S/G2/M phases of the cell cycle, indicating that the neurons that re-entered the cell cycle are the ones that degenerated (Fig. 8E–G).
These findings add to a growing literature linking cell cycle re-entry and neurodegeneration (Herrup and Yang 2007; Khurana and Feany 2007). The observations that terminally differentiated neurons are resistant to oncogenic transformation and that brain tumors of neuronal origin rarely occur suggest that cell cycle re-entry of post-mitotic neurons results in death rather than proliferation (Heintz 1993). In fact, it has been shown in a variety of systems, including flies and humans, that when neurons re-enter the cell cycle, the result is degeneration rather than proliferation and that ectopic cell cycle activation in neurons is sufficient to trigger degeneration (al-Ubaidi et al. 1992; Feddersen et al. 1992; Herrup and Busser 1995; Du et al. 1996; Kranenburg et al. 1996; Park et al. 1996; Athanasiou et al. 1998; Kruman et al. 2004). Furthermore, up-regulation of cell cycle genes, such as proliferating cell nuclear antigen, cyclin A, and cyclin B, has been shown to occur in post-mitotic Purkinje and granule cells of A-T patients; and neurons of ATM−/− mice have been found to undergo DNA replication (Yang and Herrup, 2005). Similarly, studies in both fly and mammalian models of Alzheimer’s disease support a causative link between cell cycle re-entry and neurodegeneration (Khurana and Feany 2007; Yang and Herrup 2007). Thus, failure of cell cycle regulation may be a common cause of neurodegenerative disorders, including A-T.
It is important to keep in mind that equally plausible and nonexclusive models have been put forth for why neurodegeneration occurs in A-T. The oxidative stress model proposes that neurodegeneration occurs as a consequence of increased oxidative stress, and the DNA damage model proposes that neurodegeneration occurs as a consequence of the accumulation of DNA damage (Rolig and McKinnon 2000; Barzilai et al. 2002; Shiloh 2003; Watters 2003). While we describe these models as functioning independently, it is unlikely that this is the case. For example, oxidative stress could lead to cell cycle re-entry through several different pathways, and in response to DNA damage, neurons may re-enter the cell cycle before undergoing cell death.
Concluding remarks
Here we have described a powerful experimental model in Drosophila to study the molecular mechanisms that underlie neurodegeneration in the human disease A-T. ATM knockdown in flies caused post-mitotic neurons to re-enter the cell cycle and die by programmed cell death. This finding suggests that ATM performs a cell cycle checkpoint function in post-mitotic neurons, as it does in response to DNA damage in proliferating nonneuronal cells and neuroblasts. Heterozygous mutation of Stg/CDC25 suppressed neurodegeneration in ATM knockdown flies and inhibited cell cycle re-entry, suggesting that cell cycle re-entry is causative for neurodegeneration in A-T. In the future, further genetic, cell biological, and molecular analysis of the Drosophila A-T model will allow us to address unresolved issues, such as the extent to which oxidative stress and DNA damage contribute to neurodegeneration in the absence of ATM, what factors trigger cell cycle re-entry in the absence of ATM, and what factors link cell cycle re-entry to programmed cell death as opposed to cell division in the absence of ATM.
Materials and methods
Drosophila genetics
Flies were maintained and crosses were carried out at 25°C unless otherwise noted. pATM was generated by cloning a 642-base-pair (bp) fragment corresponding to exon 6 (nucleotides 1185–1827 of CG6535) of the Drosophila ATM gene into the pWIZ transformation vector (Lee and Carthew 2003). pATM was injected into a y1w1118 strain by standard methods (Rubin and Spradling 1982). GMR-P35 flies were provided by B. Hay, RAD50 flies were provided by M. Gatti, Elav-GFP flies were provided by G. Boekhoff-Falk, and GAL4-driver lines and P-element strains used in the screen were obtained from the Bloomington Drosophila Stock Center.
The genetic screen was performed as described in Supplemental Figure 4. So as not to bias the analysis, stock numbers, not genotypes, were used as identifiers during the screen. To assess the developmental requirement for ATM, pATMT4 flies were crossed to hsp70-GAL4/CyO flies and hsp70-Gal4/+; pATMT4/+ (hs-ATMi) progeny were heat-shocked for 1 h at 37°C, twice a day until 3 d post-eclosion, when the flies were processed for TEM. To control for effects of GAL4 expression on neuron morphology, hsp70-GAL4/CyO flies were crossed to w1118 flies, and hsp70-GAL4/+ (hs-GAL4) progeny were subjected to the same heat-shock regime. To assess the requirement for ATM in adult flies, 3-d-old hs-ATMi and hs-GAL4 flies were heat-shocked for 1 h at 37°C, twice a day for 12 d, at which time the flies were processed for TEM.
To examine whether neurodegeneration was progressive, 20–30 0- to 4-d-old female GMR-GAL4/+ (GMR-GAL4), GMR-GAL4/+; pATMT4/+ (GMR-ATMi), and GMR-GAL4/+; pATMT4/StringEY12388 (GMR-ATMi, Stg) flies were maintained in food-containing vials and turned into new vials every 7 d. Flies were processed for TEM 9, 27, or 48 d later.
For FACS analysis, Elav-GAL4, UAS-GFP (Elav-GFP) flies were crossed to w1118 flies; Elav-GAL4, UAS-GFP; pATMT4/TM6b, Tb (Elav-GFP; Elav-ATMi) flies were crossed to w1118 flies; Elav-GFP; Elav-ATMi flies were crossed to StringEY12388/TM6b, Tb flies; Elav-GFP flies were crossed to StringEY12388/TM6b, Tb flies; Elav-GFP; Elav-ATMi flies were crossed to GMR-P35 flies; and Elav-GFP flies were crossed to GMR-P35 flies. Crosses were maintained for 8–9 d at 18°C and then placed for 3 d at 25°C before eye discs were dissected from non-Tb wandering third instar larvae.
Quantitation of ATM
To assess the efficacy of ATM mRNA knockdown in pATM flies, 3-d-old hs-ATMi or hs-GAL4 flies were heat-shocked for 1 h at 37°C, and total RNA was isolated using an RNeasy Mini Kit (Qiagen), according to the manufacturer’s protocol, at 2, 5, and 8 h post-heat-shock. Quantitative real-time RT–PCR was performed and quantitated as described (Katzenberger et al. 2006).
To assess the efficacy of ATM protein knockdown in pATM flies, hsp70-GAL4/hsp70-GAL4 flies were crossed to pATMT4/pATMT4 flies, and hsp70-GAL4/pATMT4 embryos were heat-shocked for 2 h at 37°C and allowed to recover for 1 h at 25°C. Embryos were homogenized in Laemmli protein loading buffer (Bio-Rad) containing β-mercaptoethanol. Embryo extracts were fractionated by SDS-PAGE and subjected to Western blot analysis. The SIN3 antibody was described previously, and the ATM polyclonal antibody was raised in rabbits against a recombinant protein that included the kinase domain (encoded by nucleotides 6202–7290 of CG6535) (Pile and Wassarman 2000).
Electron microscopy
For SEM images, anesthetized 0- to 4-d-old female flies were imaged on a Quanta 200 Environmental Electron Scanning Electron Microscope (FEI Company) using a gaseous secondary electron detector. The cooling stage was set to 0°C to preserve the head while imaging.
For TEM images, adult fly heads were fixed for 4 h rotating at 4°C in 2% paraformaldehyde and 2% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.3). The initial fixative was then replaced with 2% paraformaldehyde, 2% glutaraldehyde, and 1% tannic acid in 0.1 M cacodylate buffer (pH 7.3), and the samples were rotated overnight. Heads were washed (five times for 5–10 min) in 0.1 M cacodylate buffer (pH 7.3) and were post-fixed with 2% osmium tetroxide in 0.1 M cacodylate buffer for 2 h at room temperature. Heads were washed (five times for 15–30 min) in 0.1 M cacodylate buffer (pH 7.3), dehydrated in an ethanol-in-water series (50%, 60%, 70%, 80%, 90%, twice at 95%, and four times at 100%), washed (three times for 10 min) with 100% propylene oxide, and equilibrated into Spurr’s resin (Electron Microscopy Sciences) in a propylene oxide-in-Spurr’s resin series (2:1, 1:1, 1:2, 100% Spurr’s resin). Each equilibration step was carried out rotating for 8 h at room temperature. Heads were placed in molds (Ted Pella) and cured overnight at 70°C. Eyes were sectioned using a Reichert-Jung Ultracut E Microtome. Ultrathin sections were placed on pioloform-coated one-hole slot grids and contrasted with Reynolds lead citrate and 8% uranyl acetate in 50% ethanol. Sections were analyzed on a Philips CM120 electron microscope.
Quantitation of the neurodegenerative phenotype was carried out by scoring the number of morphologically normal photoreceptor neurons in a field of defined size located in the center of the eye. At least four individual eyes and ≥260 ommatidia were scored for each genotype. Ultrathin sections were taken at a constant depth of 25 μm from the surface of the eye. Analysis software (Soft Imaging System Corp.) was used to take multiple overlapping images of the entire eye section at a magnification of 2050×, which were compiled using Adobe Photoshop. Rhabdomeres were considered normal if they did not contain holes and were not condensed, fragmented, or split. P-values were calculated using one-way or two-way ANOVA and Dunnett’s multiple comparison tests or Bonferonni post-tests using Prism 4.0c (Graphpad) software.
Immunofluorescence
For immunofluorescence, eye-antennal imaginal discs were dissected from wandering third instar larvae in 1× PBS and prepared for PH3, BrdU, TUNEL, or acridine orange (Invitrogen) staining as described (Wolff 2002). The primary antibodies used were α-PH3 (1:100 dilution; Cell Signaling Technology), α-Elav (1:500 dilution; Developmental Studies Hybridoma Bank), α-BrdU (1:200 dilution; Becton Dickinson), and rhodamine-conjugated α-DIG (1:200 dilution; Chemicon). The fluorescently conjugated secondary antibodies used were α-mouse FITC (1:200 dilution; Jackson Laboratories), α-rabbit FITC (1:200 dilution; Jackson Laboratories), α-rat rhodamine (1:5000 dilution; Molecular Probes), and α-rat Alexa Fluor 488 (1:200 dilution; Invitrogen). TUNEL assays were performed using the ApopTag Red In Situ Apoptosis Detection Kit (Chemicon). The following changes were made to the Wolff (2002) TUNEL staining protocol: 1× PBS was substituted for 0.1 M sodium phosphate buffer; initial fixation was performed in 5% formaldehyde/1× PBS; eye discs were incubated in stop-reaction mix for 1 h at 37°C; blocking was performed at room temperature for 1 h in a solution of 12% blocking solution (Chemicon)/BSS/0.3% TX-100 (BS/BSS/TX); rhodamine-conjugated α-DIG and α-Elav antibodies were diluted in BS/BSS/TX and eye discs were incubated overnight at 4°C; and α-rat Alexa Fluor 488 was diluted in BS/BSS/TX and eye discs were incubated overnight at 4°C and processed as described by Wolff (2002). The following change was made to the Wolff (2002) acridine orange staining protocol: 1× PBS was substituted for Drosophila Ringer’s solution. Eye-antennal imaginal discs were imaged on a Zeiss Axiovert 200M inverted microscope.
FACS analysis
FACS analysis was performed as described by Neufeld et al. (1998) with the indicated modifications. Thirty to forty pairs of eye-antennal imaginal discs from wandering third instar larvae were dissected in PBS and transferred immediately into 500 μL of TEHP buffer (9× Sigma Trypsin-EDTA, 15 μg/mL Hoechst 33342 in PBS). Eye-antennal imaginal discs were incubated with gentle agitation for 3 h. To aide cell dissociation, discs were gently pipetted up and down 10 times every 20 min. After dissociation, 100 μL of 33 μg/mL propidium iodide in PBS was added to each sample. A Becton Dickinson LSRII was used to analyze the cells. Five-thousand live, single, GFP-positive events were collected for each sample, and the data were analyzed using CellQuest (Becton Dickinson) and ModFit (Verity Software House) software. Propidium iodide was used to discriminate between live and dead cells, and doublets were excluded during data acquisition and analysis. The n-values for each set of experiments are indicated in the figure legends. Different individuals repeated experiments at least two independent times for each genotype. Error bars represent standard error of the mean. P-values were calculated using one-way ANOVA and Tukey’s multiple comparison post-test using Prism 4.0c (Graphpad) software.
Mammalian cell culture
HEK 293T cells were maintained in Eagle’s minimum essential medium (MEM) containing 5% FCS. Antibody suppliers included Genetex (α-ATM, α-HDAC1), Santa Cruz Biotechnology (α-ATM), Rockland (α-pS1981-ATM), Upstate Biotechnology (α-HDAC2), and Sigma (α-Flag M2). For the coimmunoprecipitation experiments, HEK 293T cells were transfected with Flag-HDAC1 or Flag-HDAC2 expression vectors (P. Yao, Duke University), and cell extracts were prepared before or 1 h after irradiation (20 Gy) as described previously (Tibbetts et al. 1999). Cell extracts were immunoprecipitated with 2 mg of the indicated antibodies for 2–4 h prior to analysis by SDS-PAGE and immunoblotting (Shi et al. 2004). Coimmunoprecipitations of endogenous proteins were performed using HEK 293T cell extracts immunoprecipitated with 2 mg of control IgG, HDAC1, HDAC2, or ATM antibodies as described (Tibbetts et al. 1999). Scrambled and HDAC2 siRNA transfections were carried out as described (Dodson and Tibbetts 2004) using SmartPool siRNA mixtures (Dharmacon).
Acknowledgments
We thank Ben August from the University of Wisconsin-Madison Electron Microscopy Facility for sectioning and staining Drosophila eyes for TEM, as well as for training and assistance in the use of the transmission electron microscope. In addition, we thank E. Spalding (Univeristy of Wisconsin-Madison, Botany Department SEM facility; NSF Grant DBI-0421266); G. Boekhoff-Falk, N. Colley, M. Brodsky, R. Carthew, M. Gatti, B. LeMaitre, T. Neufeld, A. DeAntonio, P. Yao, and B. Hay for providing flies, reagents, or technical assistance; G. Boekhoff-Falk, S. Miyamoto, C. Metcalf, and M. Marengo for insights that greatly improved this work; and the A-T Children’s Project for supporting our work on A-T. This work was funded by grants from the National Ataxia Foundation (to D.A.W), the University of Wisconsin-Madison Medical School (to D.A.W.), and the NIH (R01 NS059001 to D.A.W. and R01 GM 067868 to R.S.T.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1639608.
References
- Abraham R.T. Cell cycle checkpoint signaling through ATM and ATR kinases. Genes & Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Abraham R.T. MAPKAP kinase-2: Three’s company at the G2 checkpoint. Mol. Cell. 2005;17:163–164. doi: 10.1016/j.molcel.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Abraham R.T., Tibbetts R.S. Guiding ATM to broken DNA. Science. 2005;308:551–554. doi: 10.1126/science.1112069. [DOI] [PubMed] [Google Scholar]
- Allen D.M., van Praag H., Ray J., Weaver Z., Winrow C.J., Carter T.A., Braquet R., Harrington E., Ried T., Brown K.D., et al. Ataxia telangiectasia mutated is essential during adult neurogenesis. Genes & Dev. 2001;15:554–566. doi: 10.1101/gad.869001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alterman N., Fattal-Valevski A., Moyal L., Crawford T.O., Lederman H.M., Ziv Y., Shiloh Y. Ataxia-telangiectasia: Mild neurological presentation despite null ATM mutation and severe cellular phenotype. Am. J. Med. Genet. 2007;143A:1827–1834. doi: 10.1002/ajmg.a.31853. [DOI] [PubMed] [Google Scholar]
- al-Ubaidi M.R., Hollyfield J.G., Overbeck P.A., Baehr W. Photoreceptor degeneration induced by the expression of simian virus 40 large tumor antigen in the retina of transgenic mice. Proc. Natl. Acad. Sci. 1992;89:1194–1198. doi: 10.1073/pnas.89.4.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiou M.C., Yunis W., Coleman N., Ehlenfeldt R., Clark H.B., Orr H.T., Feddersen R.M. The transcription factor E2F-1 in SV40 T antigen-induced cerebellar Purkinje cell degeneration. Mol. Cell. Neurosci. 1998;12:16–28. doi: 10.1006/mcne.1998.0699. [DOI] [PubMed] [Google Scholar]
- Baker N.E. Patterning signals and proliferation in Drosophila imaginal discs. Curr. Opin. Genet. Dev. 2007;17:287–293. doi: 10.1016/j.gde.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer association. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- Baonza A., Freeman M. Control of cell proliferation in the Drosophila eye by Notch signaling. Dev. Cell. 2005;8:529–539. doi: 10.1016/j.devcel.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Barlow C., Hirotsune S., Paylor R., Liyanage M., Eckhaus M., Collins F., Shiloh Y., Crawley J.N., Ried T., Tagle D. Atm-deficient mice: A paradigm of Ataxia telangiectasia. Cell. 1996;86:159–171. doi: 10.1016/s0092-8674(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Barzilai A., Rotman G., Shiloh Y. ATM deficiency and oxidative stress: A new dimension of defective response to DNA damage. DNA Repair (Amst.) 2002;1:3–25. doi: 10.1016/s1568-7864(01)00007-6. [DOI] [PubMed] [Google Scholar]
- Ben-Levy R., Hooper S., Wilson R., Paterson H.F., Marshall C.J. Nuclear export of the stress-activated protein kinase p38 mediated by its substrate MAPKAP kinase-2. Curr. Biol. 1998;8:1049–1057. doi: 10.1016/s0960-9822(98)70442-7. [DOI] [PubMed] [Google Scholar]
- Bi X., Wei S.-C., Rong Y.S. Telomere protection without a telomerase: The role of ATM and Mre11 in Drosophila telomere maintenance. Curr. Biol. 2004;14:1348–1353. doi: 10.1016/j.cub.2004.06.063. [DOI] [PubMed] [Google Scholar]
- Bi X., Gong M., Srikanta D., Rong Y.S. Drosophila ATM and MRE11 are essential for the G2/M checkpoint induced by low-dose radiation. Genetics. 2005;171:845–847. doi: 10.1534/genetics.105.047720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros R., Dozier C., Ducommun B. The when and whereas of CDC25 phosphatases. Curr. Opin. Cell Biol. 2006;18:185–191. doi: 10.1016/j.ceb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brodsky M.H., Weinert B.T., Tsang G., Rong Y.S., McGinnis N.M., Golic K.G., Rio D.C., Rubin G.M. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 2004;24:1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwald M. Complementation groups: One or more per gene? Nat. Genet. 1995;11:228–230. doi: 10.1038/ng1195-228. [DOI] [PubMed] [Google Scholar]
- Carson C.T., Schwartz R.A., Stracker T.H., Lilley C.E., Lee D.V., Weitzman M.D. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;15:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Nordstrom W., Gish B., Abrams J.M. grim, a novel cell death gene in Drosophila. Genes & Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Chowdhury D., Keogh M.C., Ishii H., Peterson C.L., Buratowski S., Lieberman J. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell. 2005;20:801–809. doi: 10.1016/j.molcel.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Ciapponi L., Cenci G., Ducau J., Flores C., Johnson-Schlitz D., Gorski M.M., Engels W.R., Gatti M. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr. Biol. 2004;14:1360–1366. doi: 10.1016/j.cub.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Clem R.J., Miller L.K. Control of programmed cell death by the baculovirus genes p35 and iap. Mol. Cell. Biol. 1994;14:5212–5222. doi: 10.1128/mcb.14.8.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copani A., Uberti D., Sortino M., Bruno V., Nicoletti F., Memo M. Activation of cell-cycle-associated proteins in neuronal death: A mandatory or dispensable path? Trends Neurosci. 2001;24:25–31. doi: 10.1016/s0166-2236(00)01663-5. [DOI] [PubMed] [Google Scholar]
- Crawford T.O. Ataxia telangiectasia. Semin. Pediatr. Neurol. 1998;5:287–294. doi: 10.1016/s1071-9091(98)80007-7. [DOI] [PubMed] [Google Scholar]
- Davidson F.F., Steller H. Blocking apoptosis prevents blindness in Drosophila retinal degeneration mutants. Nature. 1998;391:587–591. doi: 10.1038/35385. [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. A genome-wide transgenic RNAi library for conditional inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dodson G.E., Tibbetts R.S. DNA replication defects, spontaneous DNA damage, and ATM-dependent checkpoint activation in replication protein A-deficient cells. J. Biol. Chem. 2004;279:34010–34014. doi: 10.1074/jbc.C400242200. [DOI] [PubMed] [Google Scholar]
- Du W., Xie J.-E., Dyson N. Ectopic expression of dE2F and dDP induces cell proliferation and death in the Drosophila eye. EMBO J. 1996;15:3684–3692. [PMC free article] [PubMed] [Google Scholar]
- Dupré A., Boyer-Chatenet L., Gautier J. Two-step activation of ATM by DNA and the Mre11–Rad50–Nbs1 complex. Nat. Struct. Mol. Biol. 2006;13:451–457. doi: 10.1038/nsmb1090. [DOI] [PubMed] [Google Scholar]
- Edgar B.A., O’Farrell P.H. The three postblastoderm cycles of Drosophila embryogenesis are regulated in G2 by string. Cell. 1990;62:469–480. doi: 10.1016/0092-8674(90)90012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., Britton J., de la Cruz A.F., Johnston L.A., Lehman D., Martin-Castellanos C., Prober D. Pattern- and growth-linked cell cycles in Drosophila development. Novartis Found. Symp. 2001;237:3–12. doi: 10.1002/0470846666.ch2. [DOI] [PubMed] [Google Scholar]
- Elson A., Wang Y., Daugherty C.J., Morton C.C., Zhou F., Campos-Torres J., Leder P. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. 1996;93:13084–13089. doi: 10.1073/pnas.93.23.13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falck J., Coates J., Jackson S.P. Conserved modes of recruitment of ATM, ATR and DNA-PKCs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Feddersen R.M., Ehlenfeldt R., Yunis W.S., Clark H.B., Orr H.T. Disrupted cerebellar cortical development and progressive degeneration of Purkinje cells in SV40 T antigen transgenic animals. Neuron. 1992;9:955–966. doi: 10.1016/0896-6273(92)90247-b. [DOI] [PubMed] [Google Scholar]
- Goodarzi A.A., Jonnalagadda J.C., Douglas P., Young D., Ye R., Moorhead G.B., Lees-Miller S.P., Khanna K.K. Autophosphorylation of ataxia-telangiectasia mutated is regulated by protein phosphatase 2A. EMBO J. 2004;23:4451–4461. doi: 10.1038/sj.emboj.7600455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C.Y., Brautigan D.L., Larner J.M. ATM-dependent dissociation of B55 regulatory subunit from nuclear PP2A in response to ionizing radiation. J. Biol. Chem. 2002;277:4839–4844. doi: 10.1074/jbc.M110092200. [DOI] [PubMed] [Google Scholar]
- Hassin-Baer S., Bar-Shira A., Gilad S., Galanty Y., Khosravi R., Lossos A., Giladi N., Weitz R., Ben-Zeev B., Goldhammer Y., et al. Absence of mutations in ATM, the gene responsible for ataxia telangiectasia in patients with cerebellar ataxia. J. Neurol. 1999;246:716–719. doi: 10.1007/s004150050438. [DOI] [PubMed] [Google Scholar]
- Hay B.A., Wolff T., Rubin G.M. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Hay B.A., Wassarman D.A., Rubin G.M. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Heintz N. Cell death and the cell cycle: A relationship between transformation and neurodegeneration. Trends Biochem. Sci. 1993;18:2385–2395. doi: 10.1016/0968-0004(93)90103-t. [DOI] [PubMed] [Google Scholar]
- Herrup K., Busser J.C. The induction of multiple cell cycle events precedes target-related neuronal death. Development. 1995;121:2385–2395. doi: 10.1242/dev.121.8.2385. [DOI] [PubMed] [Google Scholar]
- Herrup K., Yang Y. Cell cycle regulation in the postmitotic neuron: Oxymoron or new biology? Nat. Rev. Neurosci. 2007;8:368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- Hsu C.-D., Whaley M.A., Frazer K., Miller D.A., Mitchell K.A., Adams S.M., O’Tousa J.E. Limited role of developmental programmed cell death pathways in Drosophila norpA retinal degeneration. J. Neurosci. 2004;24:500–507. doi: 10.1523/JNEUROSCI.3328-02.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P. Sensing and repairing DNA double-strand breaks. Carcinogenesis. 2002;23:687–696. doi: 10.1093/carcin/23.5.687. [DOI] [PubMed] [Google Scholar]
- Jowsey P., Morrice N.A., Hastie C.J., McLauchlan H., Toth R., Rouse J. Characterization of the sites of DNA damage-induced 53BP1 phosphorylation catalyzed by ATM and ATR. DNA Repair (Amst.) 2007;6:1536–1544. doi: 10.1016/j.dnarep.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Katzenberger R.J., Marengo M.S., Wassarman D.A. ATM and ATR pathways signal alternative splicing of Drosophila TAF1 pre-mRNA in response to DNA damage. Mol. Cell. Biol. 2006;26:9256–9267. doi: 10.1128/MCB.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana V., Feany M.B. Connecting cell-cycle activation to neurodegeneration in Drosophila. Biochim. Biophys. Acta. 2007;1772:446–456. doi: 10.1016/j.bbadis.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G.D., Choi Y.H., Dimtchev A., Jeong S.J., Dritschilo A., Jung M. Sensing of ionizing radiation-induced DNA damage by ATM through interaction with histone deacetylase. J. Biol. Chem. 1999;274:31127–31130. doi: 10.1074/jbc.274.44.31127. [DOI] [PubMed] [Google Scholar]
- Kranenburg O., ven der Eb A.J., Zantema A. Cyclin D, is an essential mediator of apoptotic neuronal cell death. EMBO J. 1996;15:46–54. [PMC free article] [PubMed] [Google Scholar]
- Kruman I.I., Wersto R.P., Cardozo-Palaez F., Smilenov L., Chan S.L., Chrest F.J., Emokpae R., Gorospe M., Mattson M.P. Cell cycle activation linked to neuronal cell death initiated by DNA damage. Neuron. 2004;41:549–561. doi: 10.1016/s0896-6273(04)00017-0. [DOI] [PubMed] [Google Scholar]
- Lavin M.F., Shiloh Y. The genetic defect in ataxia-telangiectasia. Annu. Rev. Immunol. 1997;15:177–202. doi: 10.1146/annurev.immunol.15.1.177. [DOI] [PubMed] [Google Scholar]
- Lavin M.F., Delia D., Chessa L. ATM and the DNA damage response. EMBO Rep. 2006;7:154–160. doi: 10.1038/sj.embor.7400629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.S., Carthew R.W. Making a better RNAi vector for Drosophila: Use of intron spacers. Methods. 2003;30:322–329. doi: 10.1016/s1046-2023(03)00051-3. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Paull T.T. Direct activation of the ATM protein kinase by the Mre11/Rad50/Nbs1 complex. Science. 2004;304:93–96. doi: 10.1126/science.1091496. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Paull T.T. ATM activation by DNA double-stranded breaks through the Mre11–Rad50–Nbs1 complex. Science. 2005;308:551–554. doi: 10.1126/science.1108297. [DOI] [PubMed] [Google Scholar]
- Liu W., Silverstein A.M., Shu H., Martinez B., Mumby M.C. A functional genomics analysis of the B56 isoforms of Drosophila protein phosphatase 2A. Mol. Cell. Proteomics. 2007;6:319–332. doi: 10.1074/mcp.M600272-MCP200. [DOI] [PubMed] [Google Scholar]
- Lopez-Aviles S., Grande M., Gonzalez M., Helgesen A.-L., Alemany V., Sanchez-Pris M., Bachs O., Millar J.B.A., Aligue R. Inactivation of the CDC25 phosphatase by the stress-activated Srk1 kinase in fission yeast. Mol. Cell. 2005;17:49–59. doi: 10.1016/j.molcel.2004.11.043. [DOI] [PubMed] [Google Scholar]
- Ma Y., Creanga A., Lum L., Beachy P.A. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443:359–363. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- Manke I.A., Nguyen A., Lim D., Stewart M.Q., Elia A.E., Yaffe M.B. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol. Cell. 2005;17:37–48. doi: 10.1016/j.molcel.2004.11.021. [DOI] [PubMed] [Google Scholar]
- Martin D.N., Baehrecke E.H. Caspases function in autophagic programmed cell death in Drosophila. Development. 2004;131:275–284. doi: 10.1242/dev.00933. [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., Huroy K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- McKinnon P.J. ATM and ataxia telangiectasia. EMBO Rep. 2004;5:772–776. doi: 10.1038/sj.embor.7400210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon N.-S., Frolov M.V., Kwon E.-J., Di Stefano L., Dimova D.K., Morris E.J., Taylor-Harding B., White K., Dyson N.J. Drosophila E2F1 has context-specific pro- and antiapoptotic properties during development. Dev. Cell. 2005;9:463–475. doi: 10.1016/j.devcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- Murnane J.P., Painter R.B. Complementation of the defects in DNA synthesis in irradiated and unirradiated ataxia-telangiectasia cells. Proc. Natl. Acad. Sci. 1982;79:1960–1963. doi: 10.1073/pnas.79.6.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T.P., de la Cruz A.F., Johnston L.A., Edgar B.A. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- Oikemus S.R., McGinnis N., Queiroz-Machado J., Tukachinsky H., Saeko T., Sunkel C.E., Brodsky M.H. Drosophila atm.telomere fusion is required for telomeric localization of HP1 and telomere position effect. Genes & Dev. 2004;18:1850–1861. doi: 10.1101/gad.1202504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R., Muller A., Napoli C.A., Selinger D.A., Pikaard C.S., Richards E.J., Bender J., Mount D.W., Jorgensen R.A. Analysis of histone acetyltransferases and histone deacetylase families of Arabidopsis thaliana suggest functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D.S., Farinelli S.E., Green L.A. Inhibitors of cyclin-dependent kinases promote survival of post-mitotic neuronally differentiated PC12 cells and sympathetic neurons. J. Biol. Chem. 1996;271:8161–8169. doi: 10.1074/jbc.271.14.8161. [DOI] [PubMed] [Google Scholar]
- Park Y., Ng C., Datta S. Induction of String rescues the neuroblast proliferation defect in trol mutant animals. Genesis. 2003;36:187–195. doi: 10.1002/gene.10216. [DOI] [PubMed] [Google Scholar]
- Petersen P., Chou D.M., You Z., Hunter T., Walter J.C., Walter G. Protein phosphatase 2A antagonizes ATM and ATR in a Cdk2 and Cdc7-independent DNA damage checkpoint. Mol. Cell. Biol. 2006;26:1997–2011. doi: 10.1128/MCB.26.5.1997-2011.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pile L.A., Wassarman D.A. Chromosomal localization links the SIN3–RPD3 complex to the regulation of chromatin condensation, histone acetylation and gene expression. EMBO J. 2000;19:6131–6140. doi: 10.1093/emboj/19.22.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolig R.L., McKinnon P.J. Linking DNA damage and neurodegeneration. Trends Neurosci. 2000;23:417–424. doi: 10.1016/s0166-2236(00)01625-8. [DOI] [PubMed] [Google Scholar]
- Rubin G.M., Spradling A.C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- Savitsky K., Bar-Shira A., Gilad S., Rotman G., Ziv Y., Vanagaite L., Tagle D.A., Smith S., Uziel T., Sfez S., et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Shi Y., Venkataraman S.L., Dodson G.E., Mabb A.M., LeBlanc S., Tibbetts R.S. Direct regulation of CREB transcriptional activity by ATM in response to genotoxic shock. Proc. Natl. Acad. Sci. 2004;101:5898–5903. doi: 10.1073/pnas.0307718101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Genet. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- Silva E., Tiong S., Pederson M., Homola E., Royou A., Fasulo B., Siriaco G., Campbell S.D. ATM is required for telomere maintenance and chromosomal stability during Drosophila development. Curr. Biol. 2004;14:1341–1347. doi: 10.1016/j.cub.2004.06.056. [DOI] [PubMed] [Google Scholar]
- Soller M., White K. ELAV. Curr. Biol. 2004;14:R53. doi: 10.1016/j.cub.2003.12.041. [DOI] [PubMed] [Google Scholar]
- Song Y.-H. Drosophila melanogaster: A model for the study of DNA damage checkpoint response. Mol. Cells. 2005;19:167–179. [PubMed] [Google Scholar]
- Song Y.-H., Mirey G., Betson M., Haber D.A., Settleman J. The Drosophila ATM ortholog, dATM, mediates the response to ionizing radiation and to spontaneous DNA damage during development. Curr. Biol. 2004;14:1354–1359. doi: 10.1016/j.cub.2004.06.064. [DOI] [PubMed] [Google Scholar]
- Spring K., Cross S., Li C., Watters D., Ben-Senior L., Waring P., Ahangari F., Lu S., Chen P., Misko I., et al. Atm knock-in mice harbor an in-frame deletion corresponding to the human ATM 7636del9 common mutation exhibit a variant phenotype. Cancer Res. 2001;61:4561–4568. [PubMed] [Google Scholar]
- Sun Y., Jiang X., Fernandes N., Price B.D. A role for Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Xu Y., Roy K., Price B.D. DNA damage induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol. Cell. Biol. 2007;27:8502–8509. doi: 10.1128/MCB.01382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts R.S., Brumbaugh K.M., Williams J.M., Sarkaria J.N., Cliby W.A., Shieh S.Y., Tava Y., Prives C., Abraham R.T. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes & Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch M., Bree R.T., Lowndes N.F. The MRN complex: Coordinating and mediating the response to broken chromosomes. EMBO Rep. 2003;4:844–849. doi: 10.1038/sj.embor.embor925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilenchik M.M., Knudson A.G. Endogenous DNA double-strand breaks: Production, fidelity of repair, and induction of cancer. Proc. Natl. Acad. Sci. 2003;100:12871– 12876. doi: 10.1073/pnas.2135498100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang R., Herrup K. E2F1 works as a cell cycle suppressor in mature neurons. J. Neurosci. 2007;27:12555–12564. doi: 10.1523/JNEUROSCI.3681-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters D.J. Oxidative stress in ataxia telangiectasia. Redox Rep. 2003;8:23–29. doi: 10.1179/135100003125001206. [DOI] [PubMed] [Google Scholar]
- Wei Y., Mizzen C.A., Cook R.G., Gorovsky M.A., Allis C.D. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc. Natl. Acad. Sci. 1998;95:7480–7484. doi: 10.1073/pnas.95.13.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K., Tahaoglu E., Steller H. Cell killing by the Drosophila gene reaper. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- Wolff T. Histological techniques for the Drosophila eye. Part I: Larva and pupa. In: Sullivan W., et al., editors. Drosophila protocols. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2002. pp. 201–228. [Google Scholar]
- Wolff T., Ready D.F. Pattern formation in the Drosophila retina. In: Bate M., Martinez Arias A., editors. The development of Drosophila melanogaster. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 1277–1326. [Google Scholar]
- Xu Y., Ashley T., Brainerd E.E., Bronson R.T., Meyn M.S., Baltimore D. Targeted destruction of ATM leads to growth retardation, chromosomal fragmentation during meiosis, immune defects, and thymic lymphoma. Genes & Dev. 1996;10:2411–2422. doi: 10.1101/gad.10.19.2411. [DOI] [PubMed] [Google Scholar]
- Yang L., Baker N.E. Notch activity opposes Ras- induced differentiation during the Second Mitotic Wave of the developing Drosophila eye. BMC Dev. Biol. 2006;6:8. doi: 10.1186/1471-213X-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Herrup K. Loss of neuronal cell cycle control in Ataxia-telangiectasia: A unified disease mechanism. J. Neurosci. 2005;25:2522–2529. doi: 10.1523/JNEUROSCI.4946-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Herrup K. Cell division in the CNS: Protective response or lethal event in post-mitotic neurons? Biochim. Biophys. Acta. 2007;1772:457–466. doi: 10.1016/j.bbadis.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao K.M., White K. Neural specificity of Elav expression: Defining a Drosophila promoter for directing expression to the nervous system. J. Neurochem. 1994;63:41–51. doi: 10.1046/j.1471-4159.1994.63010041.x. [DOI] [PubMed] [Google Scholar]
- You Z., Chahwan C., Bailis J., Hunter T., Russell P. ATM activation and its recruitment to damaged DNA requires binding to the C-terminus of Nbs1. Mol. Cell. Biol. 2005;25:5363–5374. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Lipinski M., Degterev A. Diversity in the mechanisms of neuronal cell death. Neuron. 2003;40:401–413. doi: 10.1016/s0896-6273(03)00601-9. [DOI] [PubMed] [Google Scholar]