Abstract
The Drosophila retinoblastoma family of proteins (RBF1 and RBF2) and their mammalian homologs (pRB, p130, and p107) are best known for their regulation of the G1/S transition via the repression of E2F-dependent transcription. However, RB family members also possess additional functions. Here, we report that rbf1 mutant larvae have extensive defects in chromatin condensation during mitosis. We describe a novel interaction between RBF1 and dCAP-D3, a non-SMC component of the Condensin II complex that links RBF1 to the regulation of chromosome structure. RBF1 physically interacts with dCAP-D3, RBF1 and dCAP-D3 partially colocalize on polytene chromosomes, and RBF1 is required for efficient association of dCAP-D3 with chromatin. dCap-D3 mutants also exhibit chromatin condensation defects, and mutant alleles of dCap-D3 suppress cellular and developmental phenotypes induced by the overexpression of RBF1. Interestingly, this interaction is conserved between flies and humans. The re-expression of pRB into a pRB-deficient human tumor cell line promotes chromatin association of hCAP-D3 in a manner that depends on the LXCXE-binding cleft of pRB. These results uncover an unexpected link between pRB/RBF1 and chromatin condensation, providing a mechanism by which the functional inactivation of RB family members in human tumor cells may contribute to genome instability.
[Keywords: RBF1, RB, CAP-D3, chromatin, condensation, Condensin II]
Rb family proteins (pRB, p130, and p107) are important regulators of the cell cycle (Cobrinik 2005). These proteins repress E2F-dependent transcription, a process that includes both the inhibition of activator E2Fs and the recruitment of corepressor complexes such as NuRD, SWI/SNF, SUV39H1, and hREAM to E2F-dependent promoters (Brehm et al. 1998; Ferreira et al. 1998; Luo et al. 1998; Magnaghi-Jaulin et al. 1998; Nielsen et al. 2001; Trimarchi and Lees 2002; Narita et al. 2003; Gunawardena et al. 2004; Litovchick et al. 2007). In an analogous way, RBF1, a Drosophila homolog of the Rb family, antagonizes the activity of the transcriptional activator dE2F1/dDP. RBF1 and the related protein RBF2 also combine with dE2F2, dDP, and other components of the dREAM/Myb–Muv complex to repress transcription of E2F-regulated genes (Du et al. 1996; Stevaux et al. 2002; Dimova et al. 2003; Korenjak et al. 2004; Lewis et al. 2004; Stevaux et al. 2005). The functions of Rb family members are vitally important for the control of cell proliferation during normal development and are compromised in human disease. Most significantly, the mutation of RB1 is the rate-limiting step in development of retinoblastoma. Mutation or loss of RB1 also occurs in 80% of small-cell lung cancer and at a lower level in a variety of other cancers, including non-small-cell lung cancer, pancreatic cancer, breast cancer, glioblastoma multiforme, T-cell acute lymphoblastic leukemia, and mantle cell lymphomas (Hanahan and Weinberg 2000; Sherr and McCormick 2002).
In normal cells, pRB activity is regulated by Cyclin/CDK complexes through the phosphorylation of residues in its C-terminal tail. These modifications cause the C terminus of pRB to fold toward the middle of the protein, disrupting the E2F-binding site and blocking the binding of corepressor complexes to the LXCXE cleft (Rubin et al. 2005). This process, which begins in late G1, inactivates pRB and permits a broad program of E2F-dependent gene expression that supports cell proliferation. pRB is dephosphorylated in mid- to late M phase, a change that appears to involve multiple phosphatases, including PP1 (Rubin et al. 2001). The C termini of p107, p130, RBF1, and RBF2 also contain clusters of consensus Cyclin/CDK phosphorylation sites, and are thought to be regulated in a manner resembling pRB (Du et al. 1996; Stevaux et al. 2002).
Although E2F is the best known pRB-associated protein, there is very clear evidence that pRB has additional targets and performs multiple functions. pRB has been reported to physically interact with a diverse assortment of proteins, to inhibit transcription from Pol I-, Pol II-, and Pol III-dependent promoters, and it has been implicated in both the repression and activation of gene expression (Morris and Dyson 2001). Mice carrying mutant alleles of Rb, p107, and p130 have complex phenotypes that are not fully understood, but include changes in both E2F-dependent and E2F-independent functions (Classon and Harlow 2002). Cells derived from these mutant animals have been noted to possess abnormalities that have yet to be explained. For example, in mitotic prophase, triple knockout mouse embryonic fibroblasts (TKO MEFs) have butterfly chromosomes that are balloon-like in appearance (Gonzalo et al. 2005). Fibroblasts cultured from Rb−/− mice, and more specifically from animals that carry a point mutation in the LXCXE-binding cleft of Rb (RbΔLXCXE), display aneuploidy and exhibit anaphase defects such as chromosome bridges (Herrera et al. 1996; Mayhew et al. 2004; Iovino et al. 2006; Isaac et al. 2006). It has also been noted that Rb−/− or TKO MEFs have relaxed chromatin that is very susceptible to nuclease digestion, suggesting that removing Rb family members may have a general effect on chromatin structure (Herrera et al. 1996).
The Condensin complexes, Condensin I and Condensin II, are necessary for the initial condensation and stability of chromatin during early mitotic prophase in organisms that range from yeast to humans (Nasmyth and Haering 2005). Each Condensin complex contains two SMC subunits (SMC4 and SMC2) that heterodimerize, thereby forming an active ATPase. SMC2/SMC4 heterodimers bind to a kleisin subunit, which in turn binds to two HEAT repeat proteins (Aono et al. 2002; Nasmyth and Haering 2005; Onn et al. 2007). While Condensin I and Condensin II both contain SMC2 and SMC4, they differ in their non-SMC proteins, with Condensin I containing CAP-H, CAP-G, and CAP-D2, and Condensin II containing CAP-H2, CAP-G2, and CAP-D3. SMC2/SMC4 heterodimers can bind to DNA and hydrolyze ATP to constrain positive supercoils, and this is dependent on the presence of non-SMC proteins (Bazett-Jones et al. 2002; Hirano 2002).
Condensin I and Condensin II complexes associate independently with chromatin and localize to different regions of the chromosome (Ono et al. 2003, 2004). The depletion of Condensin I and Condensin II subunits from Xenopus extracts or by siRNA treatment of mammalian cells leads to defects in chromosome condensation. In these studies, the selective targeting of Condensin I- or Condensin II-specific components had different consequences, indicating that the two complexes most likely perform distinct functions. The chromosomes formed under the depleted conditions lack the normal degree of rigidity, and this has led to the suggestion that Condensins may be most important for chromosome rigidity (Hirota et al. 2004; Ono et al. 2004).
Chromatin condensation is an important part of the processes that facilitate the resolution of sister chromatids in mitosis. Genetic studies in several different model organisms have shown that the mutation of either SMC or non-SMC Condensin subunits leads to inefficient resolution of chromatids, causing anaphase defects such as lagging chromosomes and chromosome bridges (Steffensen et al. 2001; Dej et al. 2004; Oliveira et al. 2005; Savvidou et al. 2005; Lam et al. 2006).
Non-SMC subunits have been shown to enhance the association of SMC heterodimers with DNA (Kimura et al. 1998, 2001) but the parameters that control the loading of Condensin subunits are not well understood. The factors that are responsible for the selective recruitment of Condensin I and Condensin II complexes to different regions of the chromosome have yet to be identified. In this study we show that the mutation of rbf1 causes mitotic defects including chromatin hypocondensation and anaphase defects. Through genetic and biochemical methods, we describe an interaction between RBF1 and the Condensin II subunit dCAP-D3 that promotes the localization of dCAP-D3 to DNA. RBF1 interacts specifically with dCAP-D3, not with dCAP-D2, and the results suggest that RBF1 is important for the targeting of Condensin II complexes. This interaction between RBF1 and dCAP-D3 reveals a novel, E2F-independent function of RB family proteins in cell cycle progression.
Results
RBF1 is required for normal chromatin condensation
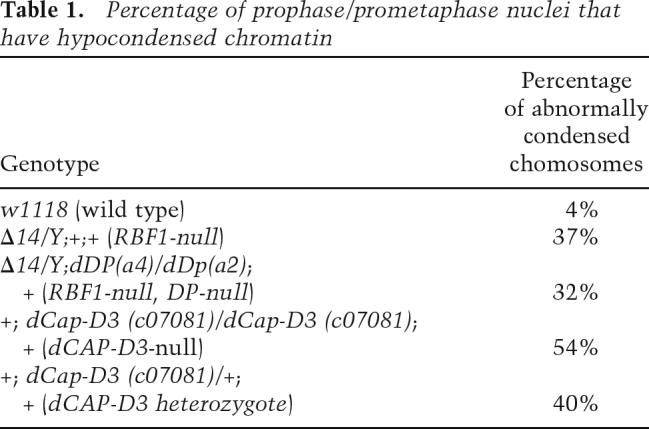
Previous studies of RBF1 have focused on its role in the G0/G1 phase of the cell cycle and the control of the G1-to-S transition. Far less attention has been paid to the possibility that RBF1 may influence progression through later stages of the cell cycle. Neuroblast squashes are a Drosophila technique that allows visualization of all phases of mitosis within a population of cells, and we used this method to examine mitotic cells of rbf1 mutant (Δ14 allele) larvae. Chromatin was detected using DAPI, a dye that preferentially stains condensed, heterochromatic DNA. Chromatin condenses uniformly during mitosis in wild-type cells. We noticed that the chromatin in mitotic cells of rbf1 mutants was highly abnormal in appearance. The greatest difference was seen in early prophase/prometaphase, where rbf1 mutant chromosomes are often long and spindly and contain regions of condensed chromatin that are interspersed with hypocondensed regions (Fig. 1A). This was in stark contrast to the uniformly condensed chromatin visualized in prophase/prometaphase neuroblast squashes of wild-type larvae (Fig. 1A; Supplemental Fig. 1A). Approximately 37% of prophase/prometaphase nuclei in neuroblast squashes of rbf1 mutant larvae had abnormally condensed chromatin (Table 1). rbf1 mutant cells were also observed with fused or broken chromosomes, with anaphase bridges in which chromosomes failed to separate normally, and with nonparallel anaphase separation (Fig. 1B). Similar defects were seen with two additional mutant alleles of rbf1 (rbf1Cas21 and rbf1SLS15) (Fig. 1; data not shown), and were also evident at a lower frequency in animals carrying the hypomorphic allele, rbf120a. Thus, the loss of RBF1 leads to multiple mitotic defects, including abnormal chromatin condensation.
Figure 1.
rbf1 mutants have defects in chromatin condensation. (A) Prophase/prometaphase spreads of neuroblast squashes from wild-type, rbf1 hypomorphic (rbf1120a), and rbf1-null (rbf1Δ14) Drosophila late second instar larvae contain regions of hypocondensed chromatin (indicated by the yellow arrows) as compared with the more condensed regions of chromatin within the same spread (indicated by the blue arrows). Note the uneven DAPI staining and regions of chromatin that appear spindly and poorly organized. (B) Anaphase spreads of late second instar larvae of the same genotypes as in A illustrating chromosome bridges and nonparallel separation. (C) Condensation defects are present in prophase/prometaphase spreads of neuroblast squashes from late second instar larvae mutant for both rbf1 and dDP (dDPa4/a2). Prometaphase spreads of neuroblast squashes from dDPa4/a2 mutant, de2f276Q1.21 mutant, and de2f276Q1.21, de2f1rm729 double mutant larvae have primarily uniform chromatin condensation.
Table 1.
Percentage of prophase/prometaphase nuclei that have hypocondensed chromatin
The best-known function of RBF1 is its ability to repress dE2F-dependent transcription, a role that involves two distinct activities. RBF1 can bind to dE2F1/dDP complexes, preventing them from activating transcription, and, additionally, RBF1 can associate with dE2F2/dDP in complexes that actively repress transcription. Interestingly, the abnormal chromatin condensation evident in rbf1 mutants was not seen in neuroblast squashes of de2f2 or dDP mutants or in de2f1, de2f2 double mutants, suggesting that this phenotype is not caused by the loss of dE2F2/dDP/RBF1 repressor complexes (Fig. 1C). Abnormal chromatin condensation was still evident in neuroblast squashes from animals that were mutant for both rbf1 and dDP, showing that this change does not require dE2F/dDP-activated transcription (Fig. 1C). Thus, the abnormally condensed chromatin that is seen in rbf1 mutant animals reflects a function of RBF1 that can be separated from both of its previously studied roles in E2F regulation.
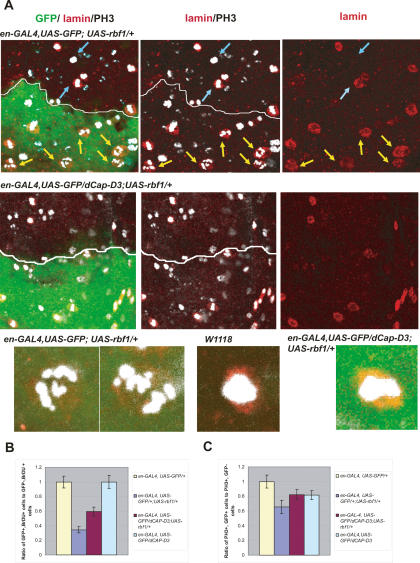
To investigate the link between RBF1 and chromatin condensation, we took advantage of a genetic assay to score functional interactions with rbf1. ptc-GAL4; UAS-rbf1 allows RBF1 to be expressed specifically in a subset of cells in the developing wing. Elevated expression of RBF1 from the ptc-GAL4 driver interferes with cell proliferation in cells that will form the L3/L4 intervein region of the adult wing (Supplemental Fig. 2A). Analysis of imaginal wing discs confirmed that elevated levels of RBF1 reduces the number of cells that incorporate bromodeoxyuridine (BrdU) or that stain with phosphorylated histone-3 (phospho-H3, a marker of condensed chromatin), and in ptc-GAL4; UAS-rbf1 flies this ultimately narrows the L3/L4 intervein distance in the adult wing (Fig. 2B,C; Supplemental Fig. 2A). The change in intervein distance is easy to quantify, and we used it to score the effects of various alleles on the ability of RBF1 to reduce cell proliferation. As expected, the coexpression of Cyclin E completely suppressed the activity of RBF1 in this assay (Table 2; Supplemental Fig. 2A,B).
Figure 2.
RBF1 overexpression causes the appearance of nuclei with hypercondensed chromatin, an effect that is suppressed by halving the dosage of dCap-D3. (A) Immunostaining of Drosophila third instar larval wing discs engineered to express RBF1, and GFP, under the control of the engrailed-GAL4 (en-GAL4) driver. Condensed DNA was visualized with an antibody to phospho-H3. The presence of an intact nuclear membrane was visualized with an antibody to lamin-β. The region of the disc expressing RBF1 contains nuclei with hypercondensed DNA (yellow arrows) that differ from the phospho-H3-positive DNA in adjacent wild-type tissue (blue arrows). Images of hypercondensed and wild-type DNA were magnified in Photoshop and are shown in the bottom row. (B,C) Overexpression of RBF1 reduces the overall number of BrdU-positive cells (B) and phospho-H3-positive cells (C) in the wing disc, in comparison with driver controls, and these effects are partly rescued by halving the dosage of dCap-D3. BrdU-positive (B) and phospho-H3-positive (C) cells were counted in GFP-positive and -negative areas in Drosophila third instar larval wing discs of the genotypes indicated in the legend on the right. Ratios were normalized against the wild-type control.
Table 2.
Heterozygous mutation of non-SMC Condensin II subunits suppresses an RBF1 overexpression phenotype
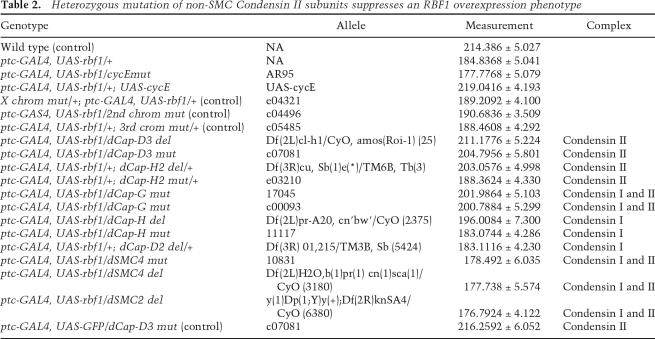
ptc-GAL4; UAS-rbf1 was crossed to stocks carrying mutant alleles of genes encoding components of Condensin I and Condensin II complexes, two protein complexes that are important for chromatin condensation. The effect of RBF1 expression on the L3/L4 intervein region was suppressed by the heterozygous mutation of dCap-D3, dCap-H2, or dCap-G, three genes that encode the non-SMC subunits of the Condensin II complex (Table 2; Supplemental Fig. 2B). The strongest interaction was seen with two independent alleles affecting dCap-D3: c07081, an insertion in dCap-D3, and Δ25 (Df(2L)pr-A20,cn′bw′/CyO), a deletion that removes dCap-D3. In contrast, the ptc-GAL4; UAS-rbf1 phenotype was unaffected by mutant alleles of Cap-H and Cap-D2, which encode non-SMC subunits of the Condensin I complex, or by heterozygous mutations of randomly selected genes on the X, second, or third chromosomes. Note that dCAP-G is a common component of both Condensin 1 and Condensin II complexes in Drosophila. Unlike alleles of genes encoding the non-SMC subunits on the Condensin II complex, mutant alleles of dSMC2 and dSMC4 failed to suppress the ptc-GAL4; UAS-rbf1 phenotype. Negative data in this type of assay are difficult to interpret because halving gene dosage may or may not be rate limiting for gene function. Nevertheless, the interactions that were found demonstrate that the levels of the non-SMC subunits of the Condensin II complex are important for this RBF1-dependent phenotype.
To examine the interaction between RBF1 and dCAP-D3 in more detail, we used engrailed-GAL4 (en-GAL4) to direct the expression of UAS-rbf1 in the posterior compartment of developing wing discs (Fig. 2, marked with UAS-GFP). While the elevated expression of RBF1 reduced the number of cells that stain with phospho-H3, we noticed that many of the phospho-H3-positive nuclei that remained had an unusual appearance. These nuclei contained chromatin that was highly condensed, to the extent that individual chromosomes were apparent (Fig. 2A, yellow arrows), and this was in stark contrast to the more globular phospho-H3-positive nuclei present in the adjacent wild-type tissue (Fig. 2A, blue arrows) and in tissue that expressed only the engrailed driver (Supplemental Fig. 3). The phospho-H3-positive nuclei that overexpressed RBF1 also stained with antibodies to lamin β (Fig. 2A), demonstrating that the hypercondensed chromatin was contained within a single nucleus. These cells were TUNEL-negative, indicating that chromatin hypercondensation was not due to apoptosis (data not shown). Interestingly, RBF1’s ability to induce chromatin hypercondensation was completely suppressed by the heterozygous mutation of dCap-D3 (Fig. 2A).
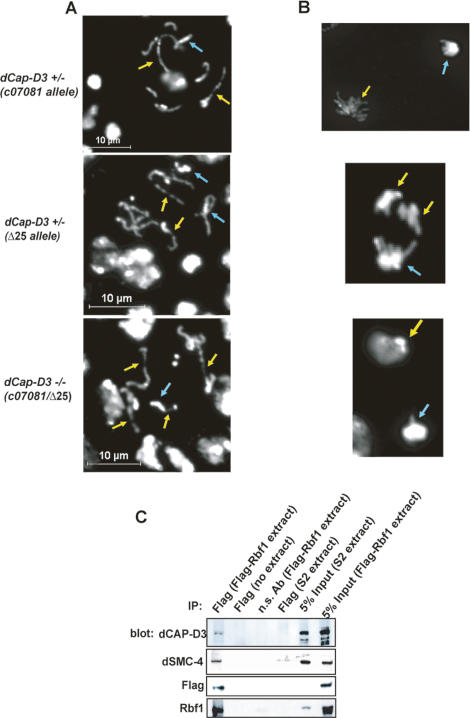
Neuroblast squashes prepared from larvae transheterozygous for mutant alleles of dCap-D3 displayed chromosome condensation defects that were very similar to those observed in rbf1 mutant larvae (Fig. 3A,B). Mitotic cells with abnormally condensed chromatin were also observed in larvae heterozygous for dCap-D3, albeit at a lower frequency than the homozygous mutants (Table 1). Larvae that were null for both rbf1 and dCap-D3 did not develop to a point where neuroblast squashes could be analyzed. The fact that rbf1 and dCap-D3 mutants have similar defects in chromosome condensation and the finding that RBF1-induced phenotypes can be suppressed by reducing the dose of dCap-D3 suggests that RBF1 cooperates with dCAP-D3, directly or indirectly, to promote normal chromatin condensation.
Figure 3.
dCAP-D3 physically interacts with RBF1 and is necessary for uniform chromatin condensation. (A) Prophase/prometaphase spreads of neuroblast squashes from Drosophila third instar larvae heterozygous or null for dCap-D3 show chromatin hypocondensation phenotypes that are very similar to those seen in rbf1 mutants (Fig. 1). Yellow arrows mark regions of chromosomes that are hypocondensed. Blue arrows mark regions of chromosomes that are more condensed. (B) Anaphase spreads of neuroblast squashes from Drosophila larvae of the genotypes described in A. Yellow arrows indicate the group of chromosomes that are hypocondensed compared with the other group of chromosomes in the anaphase pair. (C) Condensin II subunits coimmunprecipitate with RBF1. Immunoblotting was performed using antibodies to Condensin II subunits dCAP-D3 and dSMC4. Extracts of SL2 cells, or SL2 cells stably expressing Flag-RBF1, were immunopreciptated with antibody to Flag-RBF1 or a nonspecific control antibody. Note that dCAP-D3 and dSMC4 coimmunopreciptated with Flag-RBF1; no signal was seen using either the control antibody or control lysates.
RBF1 and dCAP-D3 physically interact and colocalize on polytene chromosomes
Given this genetic data, we asked whether RBF1 and dCAP-D3 physically interact with one another. Using a SL2 cell line stably transfected with a Flag-RBF1 expression construct, we found that dCAP-D3 and dSMC4 coimmunoprecipitate specifically with RBF1 (Fig. 3C). Consistent with the genetic data, dCAP-D2, a component of the Condensin I complex, was not detected in RBF1 immunoprecipitates (data not shown).
At first glance, an interaction between RBF1, a protein that is thought to be active primarily in G1, and components of Condensin II, a complex that promotes chromosome condensation during mitosis, might seem unexpected. However, previous studies have shown that Condensin II is present in the nucleus of mammalian cells during interphase, raising the possibility that this might also be true in Drosophila and that CAP-D3 and RBF1 might physically interact earlier in the cell cycle. As a test of the potential link between RBF1 and Condensin II, we used polytene chromosomes to visualize the genome-wide distribution of RBF1 and dCAP-D3 on chromatin of interphase cells.
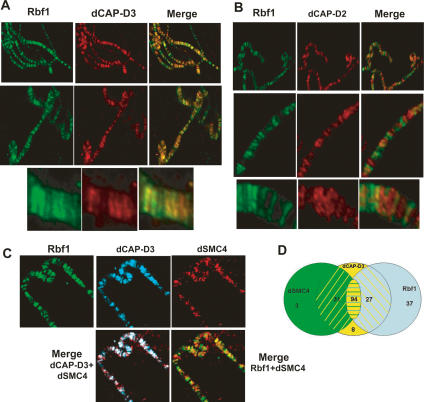
Antibodies to RBF1, dCAP-D3, and dCAP-D2 each stain multiple bands on polytene chromosomes. In all three cases, the banding patterns are largely excluded from regions that stain strongly with DAPI, indicating that these proteins localize primarily to areas devoid of constituitive heterochromatin (Supplemental Fig. 4). Previous studies in humans and Xenopus have shown that the specific components of Condensin I and Condensin II complexes localize to distinct regions of the chromosome (Ono et al. 2003). Coimmunostaining experiments with antibodies that were specific for RBF1 and dCAP-D3 revealed a remarkable colocalization of these proteins on polytene chromosomes (Fig. 4A). While this colocalization was not complete for either protein, the majority of the bands stained positive for both dCAP-D3 and RBF1. Consistent with our genetic data (Fig. 1), the colocalization of RBF1 and dCAP-D3 was still seen on polytene squashes from dDP-null larvae (Supplemental Fig. 4B), confirming that the localization of these proteins is independent of dE2F/dDP complexes. Unlike the experiments with antibodies to dCAP-D3, we saw little or no overlap between the sites stained by antibodies to RBF1 and dCAP-D2 (Fig. 4B). This observation agrees with the results of both genetic and biochemical experiments suggesting that RBF1 interacts specifically with Condensin II components and not with Condensin I components (Table 2; data not shown). Similar experiments with antibodies to dSMC4, dCAP-D3, and RBF1 showed a significant degree of overlap between dCAP-D3 and dSMC4 and between all three proteins. Overlap between RBF1 and dSMC4 was only apparent at bands that also stained positive for dCAP-D3 protein (Fig. 4C,D).
Figure 4.
RBF1 colocalizes on polytene chromatin with dCAP-D3 but not with dCAP-D2. (A) Polytene chromatin spreads from wild-type Drosophila third instar larvae were immunostained with antibodies to dCAP-D3 (red) and RBF1 (green). The partial colocalization of the two proteins is shown by the yellow bands in the merged image. (B) RBF1 and dCAP-D2 localize to different regions. Polytene chromatin spreads from wild-type Drosophila larvae were immunostained with antibodies to dCAP-D2 (red) and RBF1 (green). (C) dSMC4 displays partial overlap with dCAP-D3 and RBF1 on polytene chromatin. Chromatin spreads from wild-type larvae were immunostained with antibodies to dCAP-D3 (blue), dSMC4 (red), and RBF1 (green). (D) A Venn diagram depicting the number of chromatin bands out of a total of 200 randomly selected bands that stained for RBF1 and/or dCAP-D3 and/or dSMC4.
In summary, these results show that RBF1 and dCAP-D3 physically interact and localize to similar sites on chromatin. This extensive pattern of colocalization provides a rational explanation for the strong genetic interactions seen between rbf1 and dCap-D3.
RBF1 promotes the association of dCAP-D3 with chromatin, and this functional interaction is conserved in human cells
We used mutant alleles and antibodies to dCAP-D3 and RBF1 to investigate the functional relationship between these proteins. RBF1 antibodies gave a robust banding pattern on polytene chromosomes prepared from dCap-D3 mutant larvae, indicating that dCAP-D3 is not needed for RBF1 to localize to DNA (Supplemental Fig. 4B). In contrast, the staining obtained with antibodies to dCAP-D3 was greatly diminished on polytene chromosomes prepared from rbf1-null larvae (Fig. 5A; Supplemental Fig. 4C). The few dCAP-D3 bands that do form in the absence of RBF1 are thinner and less organized than those seen in the wild-type polytene chromatin. These results suggest that RBF1 promotes the chromatin association of dCAP-D3. To test whether a similar change occurs in dividing cells, we immunostained neuroblast squashes from wild-type and rbf1-null larvae. In wild-type cells, accumulation of dCAP-D3 on chromatin is evident in late anaphase/telophase cells (Fig. 5B). In contrast, in rbf1 mutant cells, dCAP-D3 no longer localizes to mitotic DNA (Fig. 5B, right panel). Similar experiments using antibodies to dCAP-D2 showed no difference in staining between wild-type and rbf1-null chromosomes (data not shown). To quantify the change in the amount of chromatin-associated dCAP-D3 in the presence or absence of RBF1, we treated SL2 cells with dsRNA to RBF1 or to the white gene, as a control (Supplemental Fig. 5B). The results showed an ∼45% decrease in dCAP-D3 protein associated with chromatin in the insoluble chromatin fraction upon loss of RBF1. Taken together, these results indicate that RBF1 recruits dCAP-D3 to chromatin and suggest that the chromosome condensation defects seen in rbf1 mutant animals may be due, at least in part, to reduced dCAP-D3 or Condensin II activity.
Figure 5.
dCAP-D3 localization to DNA is strongly reduced in rbf1 mutants. (A) Polytene chromatin spreads from wild-type and rbf1-null larvae were immunostained with an antibody to dCAP-D3 (red) and with DAPI (white). Yellow arrows indicate DAPI-stained chromatin bands. (B) Anaphase spreads of neuroblast squashes from Drosophila late second instar larvae immunostained with an antibody to dCAP-D3 (red) and with DAPI (white). Wild-type spreads are shown on the left while rbf1-null spreads are shown on the right. Note that dCAP-D3 is enriched on chromatin in wild-type cells (blue arrows) but not in the rbf1 mutants (yellow arrows).
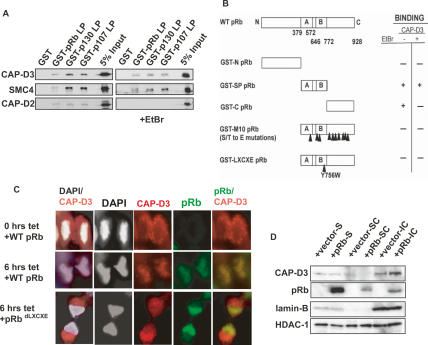
Components of the Condensin II complex are conserved in many higher eukaryotes, including humans. To ask whether the functional connection between RBF1 and dCAP-D3 is also conserved, we first tested fusion proteins containing each of the three human Rb family members (pRB, p130, and p107) in in vitro binding assays. All three RB family members bound hCAP-D3 in cell extracts and pulled down hSMC4 as well (Fig. 6A). A small amount of hCAP-D2 binding was also detected in these assays, but this was most likely an indirect association mediated through DNA, as it was eliminated by treating the extracts with ethidium bromide, whereas the association with hCAP-D3 and hSMC4 was unaffected by this treatment. Domain mapping experiments showed that the small pocket domain of pRB (amino acids 379–792) was sufficient for the DNA-independent association with hCAP-D3 (Fig. 6B; data not shown). Interestingly this interaction was disrupted by point mutations in the LXCXE-binding cleft of pRB and by mutations in pRB that mimicked phosphorylation (Barrientes et al. 2000).
Figure 6.
Rb family members associate with hCAP-D3 and promote its localization to DNA in human cells. (A) pRB, p107, and p130 fusion proteins bind to the Condensin II subunits hCAP-D3 and hSMC4, independent of DNA. GST-binding assays were performed using nuclear extracts from confluent IMR90 cells, in either the presence or absence of ethidium bromide (EtBr). Immunoblotting was performed using antibodies to human Condensin II subunits hCAP-D3 and hSMC4 or to the human Condensin I subunit hCAP-D2. (B) The binding of hCAP-D3 to pRB is abrogated by mutations that disrupt the LXCXE-binding cleft of pRB or that mimick phosphorylation. Wild-type and mutated GST-tagged fragments of pRB assayed as described in A. A positive interaction between proteins is indicated by a plus sign (+), while the absence of an interaction is indicated by a minus sign (−). Fragments used included an N-terminal fusion protein (GST-N-pRB, amino acids 1–379), a small pocket fusion protein (GST-SP-pRB, amino acids 379–772), a C-terminal fusion protein (GST-C-pRB, amino acids 772– 928), a mutant (GST-M10 pRB) (Barrientes et al. 2000) that encompassed amino acids 379–928 and contained S/T to E mutations in the 10 C-terminal Cyclin/CDK phosphorylation sites, and GST-ΔLXCXE, which encompassed amino acids 379–928 and contained a mutation at Y756. Binding assays were performed in the presence or absence of EtBr to identify interactions that might be mediated via DNA. (C) The re-expression of pRB in SAOS-2 cells relocalizes hCAP-D3 to chromatin in late anaphase/telophase. Cells expressing tetracycline-inducible pRB (wild-type or LXCXE cleft mutant) were immunostained with antibodies to pRB (green) and hCAP-D3 (red) 6 h after induction. DNA was stained with DAPI (white). (D) Chromatin fractionation of lysates from SAOS-2 cells treated as described in C showing that pRB expression increases the amount of hCAP-D3 present in the insoluble chromatin pellet (IC). (S) Soluble cellular protein fraction; (SC) soluble chromatin fraction. Immunoblotting was performed with antibodies to hCAP-D3 and pRB. Lamin-B was used as a loading control for the insoluble fraction and HDAC1 served as a control for overall loading.
As a further test, we asked whether the re-expression of pRB into human SAOS-2 cells, an osteosarcoma cell line that is homozygous mutant for pRB, would alter the localization of hCAP-D3. For these experiments we used SAOS-2 cells carrying a Tet-inducible form of pRB. This allowed us to control the time of pRB expression, and we compared the localization of hCAP-D3 in cells before pRB expression with cells 6 h after induction, at a time before the population of pRB-expressing cells has begun to accumulate in G1 (Binne et al. 2007). The expression of pRB caused a change in the localization of hCAP-D3 that was most evident in cells at late anaphase/early telophase (Fig. 6C). At this stage in the cell cycle, the nuclear envelope is disrupted and nuclear proteins are free to disperse through the cytoplasm. This is also the phase in the cell cycle where pRB normally becomes dephosphorylated and was the earliest point at which we could detect pRB association with chromatin. When no pRB protein was present in the cells (0 h tet), hCAP-D3 was mostly present in the cytoplasm, with only a small amount present on chromatin in late anaphase/early telophase. However, in cells collected 6 h after pRB expression, hCAP-D3 localized primarily on chromatin, together with pRB. As a control, a mutant form of pRB that is disrupted in the LXCXE-binding cleft (RbΔLXCXE) failed to cause this redistribution of hCAP-D3 (Fig. 6C). The ability of pRB to promote the localization of hCAP-D3 to DNA was confirmed by chromatin fractionation experiments (Fig. 6D). SAOS-2 cells expressing either a control vector or Tet-inducible wild-type pRB were induced with tetracycline for 6 h and then fractionated into soluble and insoluble chromatin fractions. In SAOS-2 cells carrying the control vector alone, hCAP-D3 protein was found in the soluble, soluble chromatin, and insoluble chromatin fractions. Following pRB induction, the overall level of hCAP-D3 was unchanged, but hCAP-D3 was no longer present in the soluble chromatin fraction and increased levels were detected in the insoluble pellet. Taken together, these results show that a physical interaction between RB family members and the Condensin II subunit CAP-D3 has been conserved from flies to humans and that in both species RB family members promote the tight chromatin association of CAP-D3.
Discussion
Rbf1 interacts specifically with dCAP-D3/Condensin II
Previous studies have shown that non-SMC proteins are important for the function of Condensin complexes (Hirano 2005; Nasmyth and Haering 2005). While the SMC proteins contain the ATPase domains that are essential for DNA supercoiling, non-SMC proteins like CAP-D2 and CAP-D3 are thought to help target Condensins to chromatin. Condensin I and Condensin II complexes are known to be selectively recruited to different chromosomal locations (Ono et al. 2004; Savvidou et al. 2005), but the mechanisms responsible for specificity have been elusive. The results described here show that RB- family members are important for the chromatin association by dCAP-D3, a specific component of the Condensin II complex. One of the most striking features of this data is the specificity of the interactions. RBF1 interacts consistently with dCAP-D3 but not with dCAP-D2. This specificity is illustrated by the polytene staining experiments showing that RBF1 colocalizes with dCAP-D3 but not with dCAP-D2 (Fig. 4). A similar specificity was observed in the genetic assays, in binding experiments, and in immunostaining experiments and was seen in experiments in both flies and human cells (Table 2; Fig. 6; Supplemental Fig. 2; data not shown).
While the lack of interaction with dCAP-D2 provides a valuable negative control, it also suggests that there is an important distinction between the two Condensin subunits and, by inference, between the two Condensin complexes. When tested in Xenopus extracts, the depletion of Condensin I resulted in decondensation of sperm chromatin with no chromosome formation while depletion of Condensin II primarily affected the morphology of chromosomes, resulting in kinked chromosomes that lacked the normal degree of rigidity (Ono et al. 2003). These differences indicate that Condensin I and Condensin II play different roles in the organization of chromosome structure, but precisely how the functions of these two complexes differ, or how their functions are integrated, is not well understood (for review, see Hirano 2005). One idea, suggested by studies in mammalian cells showing that Condensin II associates with chromatin prior to nuclear envelope breakdown (NEB), whereas Condensin I complexes are excluded from the nucleus and only gain access to chromatin following NEB, is that Condensin II complexes perform the initial compaction of the chromosome in prophase, and that Condensin I helps to complete the process (Ono et al. 2004). However, in Drosophila, both Condensin I and II proteins have been found to localize with chromatin prior to NEB (Savvidou et al. 2005; Gerlich et al. 2006; Oliveira et al. 2007) and this temporal distinction is less certain. The fact that the two Condensin complexes localize to distinct regions of euchromatin suggests that they determine the architecture of different subdomains. If this model is correct, then this implies that RB family members are important for the structural organization of particular regions of the chromosome. This may explain why the abnormally condensed chromosomes seen in rbf1 mutant animals contain interspersed segments of condensed and hypocondensed chromatin. In a more general sense, this link between RB proteins and chromosome organization suggests that RB family members are poised to control global changes in chromatin architecture, a position that may help them to act as global regulators of proliferation and differentiation.
We note that several studies have described roles for Condensins outside of mitosis. The Caenorhabditis elegans SMC4 homolog DPY-27 is required for the regulation of the dosage compensation complex and the transcriptional repression of the autosomal male sex determination gene, her-1 (Chuang et al. 1994; Chu et al. 2002). Non-SMC subunits, which have been shown to form complexes independent of the SMC heterodimer, can also effect gene expression. dCAP-G, (a member of both Condensin I and II) has been shown to effect position effect variegation (PEV), suggesting that it may have a role in maintenance of heterochromatin (Dej et al. 2004; Cobbe et al. 2006). An interaction between murine CAP-G2 (MTB) and the SCL and E12 transcription factors allows MTB to repress transcription during erythoid cell development (Xu et al. 2006). Recently, a kleisin β mutation in mice was shown to be responsible for defects in T-lymphocyte differentiation (Gosling et al. 2007). It is possible, therefore, that the interaction between RB family members and CAP-D3 protein may be used, in part, to control gene expression, and that the changes in chromosome condensation seen when RBF1 levels are altered may be a consequence of too little/too much chromatin-associated CAP-D3 that was initially recruited for a different purpose, such as transcriptional regulation.
Interestingly, we find that mutant alleles of dCap-D3 suppress PEV (Supplemental Fig. 6), supporting the idea that this complex can affect chromatin states. dCAP-D3 and RBF1 do not colocalize with bands of constituitive heterochromatin, which stain strongly with DAPI, but are found in the lighter DAPI-stained or non-DAPI-stained regions (Supplemental Fig. 4), raising the possibility that they act together in areas of repressed euchromatin or facultative heterochromatin. Clearly, further studies are needed to identify the regions of dCAP-D3 that are important for this interaction and to identify the chromatin elements that are bound by dCAP-D3/RBF1.
Several of our results raise the intriguing possibility that RBF1 may target a subpopulation of dCAP-D3 that acts independently of dSMC proteins. It is striking that the UAS-RBF1 phenotype was not modified by mutant alleles of dsmc2 or dsmc4 (Table 2). Moreover, antibodies to dSMC4 failed to stain several bands on polytene chromosomes that were costained with antibodies to dCAP-D3 and RBF1 (Fig. 4), and, while we saw a clear decrease in the levels of dCAP-D3 associated with chromatin in the absence of RBF1, we failed to detect a clear reduction in the level of dSMC4 (Supplemental Fig. 5). Possibly, dCAP-D3 may exist in several different complexes, some of which contain dSMC proteins and some that do not, and the activity that is targeted by RBF1 may be different from the traditional view of Condensin complexes. Alternatively, since dSMC2 and dSMC4 are components of both Condensin I and Condensin II, one could argue that the lack of interaction with dsmc2 or dsmc4 and the failure to see a clear reduction in dSMC4 levels when RBF1 was removed are simply due to redundancy between the two Condensin complexes. The overlap between dSMC4 and dCAP-D3 staining on polytene chromosomes was surprisingly high (Fig. 4), and it is possible that the dSMC4 antibody marks some Condensin complexes better than others. Since dSMC4 coimmunoprecipitates with RBF1 (Fig. 3), since SMC4 binds to Rb, p107, and p130 fusion proteins in vitro (Fig. 6), since many of the dCAP-D3/RBF bands on the polytene chromosomes also costain with dSMC4 (Fig. 4), and mutant alleles of dCap-H2 and dCap-G also modified the UAS-rbf1 phenotype, it seems likely that this activity of RBF1 involves its interaction with multiple components of the Condensin II complex. However, further studies are needed to fully characterize the RBF1/dCAP-D3 complexes and to determine which of the Condensin II proteins are important for this activity.
An E2F/DP-independent function of RBF1 is required for normal chromatin condensation
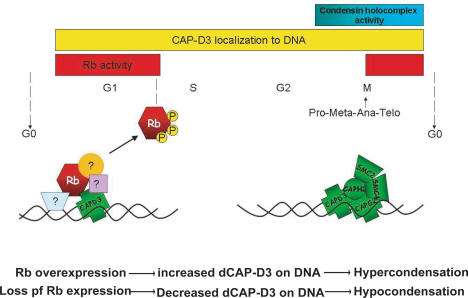
Analysis of polytene chromosomes shows that RBF1 is needed for chromatin localization of CAP-D3 in interphase cells. In experiments carried out in human cells, the earliest point in the cell cycle when we could detect the ability of pRB to promote chromatin association by dCAP-D3 was late anaphase, the point in the cell cycle where pRB becomes dephosphorylated and associates tightly with chromatin. This, together with the observation that recombinant RB polypeptides associate with hCAP-D3 and the finding that the interaction between RB and hCAP-D3 was disrupted by mutations that mimic phosphorylation, leads us to propose that RBF1/RB proteins recruit CAP-D3 to DNA at the times when they are traditionally thought to act. While we cannot exclude the possibility that a subpopulation of RBF1 interacts with dCAP-D3 later in the cell cycle, the changes in chromatin condensation in rbf1 mutants are most likely to be a consequence of reduced dCAP-D3 localization earlier in the cell cycle (Fig. 7).
Figure 7.
RB family members interact with CAP-D3, promoting its association with chromatin. Changing the levels or activity of RB family proteins alters the level of dCAP-D3 associated with chromatin, altering the efficiency of chromatin condensation. The analysis of polytene spreads shows that RBF1 and dCAP-D3 colocalize on the chromatin of interphase cells and that RBF1 is needed for the efficient localization of dCAP-D3 to DNA. In proliferating SAOS-2 cells, the recruitment of hCAP-D3 to chromatin by pRB was evident at late anaphase/telophase, at the time where pRB is activated by dephosphorylation (see Discussion for details).
The best-known functions of the RB family proteins in both mammalian cells and Drosophila involve interactions with E2F transcription factors. However neuroblast squashes of rbf1/dDP mutants and either dE2F2 or dDP-null larvae (Fig. 1C) show that the hypocondensation caused by the loss of RBF1 does not require dE2F/dDP activity and cannot be generated by simply inactivating dE2F/dDP complexes. In agreement with this, we find that the striking colocalization of RBF1 and dCAP-D3 is maintained on polytene squashes from dDP-null larvae (Supplemental Fig. 4B). This does not preclude the possibility that dCAP-D3 and RBF1 complex may interact in the vicinity of dE2F-regulated promoters, or that the dCAP-D3/RBF1 interaction may be influenced by changes in E2F activity. Clearly though, these results show that RBF1 recruits dCAP-D3 to chromatin in a manner that does not depend on the conventional model of E2F/DP DNA-binding activity. Many different transcription factors have been shown to interact with RB family members, and there is no shortage of possibilities for such an E2F-independent function. Intriguingly, DNMT3b and HDAC1, which have been linked previously to pRB, have also been found to associate with both SMC4 and SMC2 (Geiman et al. 2004). This raises the possibility that the interaction between Condensin II proteins and RBF1 may involve one or more of the chromatin-associated complexes that have been connected previously to pRB.
Links between Condensin II and cancer
The results described here provide a very simple explanation for the aneuploidy and anaphase defects observed in Rb−/− and RbΔLXCXE MEFs (Herrera et al. 1996; Mayhew et al. 2004; Iovino et al. 2006; Isaac et al. 2006). Since the inactivation of Condensin II complexes causes global changes in chromatin structure, these findings may also explain the balloon-like, butterfly chromosomes described in TKO MEFs (Gonzalo et al. 2005) or the changes in chromatin structure previously noted in Rb−/− MEFs (Herrera et al. 1996).
pRB is one of the most well-studied tumor suppressor proteins and is mutated or inactivated in many types of human cancer. The idea that RB family members are required for normal Condensin II function provides a simple route through which the functional inactivation of RB family proteins, either by deletion, phosphorylation, or by viral proteins that bind to the LXCXE-binding cleft, will promote genomic instability. Indeed, there are several independent lines of evidence implicating Condensin activity in tumorigenesis. Mutations in Condensin subunits SMC2 and SMC4 have been identified in cases of pyothorax-associated lymphoma (Ham et al. 2007). Strikingly, a recent study of 102 early-onset breast cancer patients showed that 53% of cases had loss of heterozygosity (LOH) occurring in the region that includes the hCAP-D3 locus (Gentile et al. 2001). This LOH also correlated with higher tumor grade and a more unfavorable prognosis. These observations seem particularly significant when one considers that heterozygosity for dCAP-D3 was not only sufficient to suppress RBF1-induced phenotypes but also caused visible defects in chromatin condensation and segregation (Figs. 2 3). Given the evidence that RB family members promote CAP-D3 association with chromatin and the evidence that LOH at the CAP-D3 locus occurs frequently in breast cancer, we suggest that Condensin II function is likely to be compromised in many different human tumor cells.
Materials and methods
Drosophila stocks
Unless otherwise indicated, fly strains were obtained from Bloomington Stock Center, the Harvard Medical School Exelixis collection, or generated in the laboratory. All fly stocks and crosses were maintained at 25°C. Tp(3;Y)BL2 flies were a generous gift from Joe Eissenberg (St. Louis State University) and en-GAL4, UAS-GFP flies were a generous gift of Laurel Raftery (MGH Cancer Center, Charlestown, MA). The ptc-GAL4 phenotypes were from female progeny, while all other phenotypes described were from male progeny.
Cell lines and constructs
IMR90 primary human fibroblasts were maintained in Dulbecco’s Minimal Essential Media (DMEM, Cellgro/Mediatech) supplemented with 10% fetal bovine serum (FBS) (Gibco-Invitrogen), 1% Pen-Strep (Gibco-Invitrogen), and 1% L-glutamine (Gibco-Invitrogen). Inducible SAOS-2 cell lines were described previously (Binne et al. 2007) and were also maintained in this media with a drug selection of 2.5 mg/mL Blastocydin (Invitrogen). Flag-Rbf1-transfected Drosophila Schneider line 2 (SL2) cells were described previously (Korenjak et al. 2004) and were maintained in Schneider’s Insect Medium (Sigma) with 10% FBS and 1% Pen-Strep.
Antibodies
Primary antibodies included rabbit anti-dCAP-D3 and rabbit anti-dCAP-D2 (1:50 for immunostaining of polytene chromatin and 1:100 for immunostaining of other larval tissues, 1:500 immunoblotting), both gifts of M. Heck (Edinburgh, UK). Mouse anti-Rbf1 was used at a dilution of 1:25 for polytene chromatin immunostaining and 1:5 for coimmunoprecipitation (co-IP)/immunoblotting. Sheep anti-dSMC4 was used at 1:50 for polytene chromatin immunostaining and 1:500 for immunoblotting (gift of C. Sunkel, Porto, Portugal). Mouse anti-Flag M2 was used in co-IP experiments at 1:100 and in immunoblotting experiments at 1:2000 (Sigma). Rabbit anti-hCAP-D3, rabbit anti-hCAP-D2, and rabbit anti-hSMC4 were all used for immunoblotting at 1:1000 and for immunostaining at 1:200 (Bethyl). Mouse anti-pRb was used for immunostaining at 1:100 (BD Pharmingen). Rabbit anti-phospho-H3 was used at a dilution of 1:500 (Upstate Cell Signaling Solutions). The monoclonal Drosophila lamin antibody was used at 1:200 and was obtained from the Developmental Studies Hybridoma Bank. Secondary antibodies included the appropriate anti-species Alexa 488, anti-species Cy3, and anti-species Cy5 used at 1:500 (Jackson Immunoresearch).
Neuroblast squashes and immunostaining of Drosophila larval tissues
Drosophila larval brains were dissected into 0.7% NaCl and were fixed in 45% acetic acid/H2O for 3 min at room temperature. Brains were then placed on a coverslip (Fisher Scientific) and a Poly-L lysine-coated slide (Polysciences, Inc.) was placed over the coverslip. With a paper towel placed between thumb and coverslip, pressure was applied to the top of the coverslip, squashing the brains between the coverslip and slide. Slides were briefly frozen in liquid nitrogen and then placed in a dry-ice/methanol bath for 10 min. Slides were washed in 0.7% NaCl for 5 min at room temperature and then stained with 2 × 10−4 mg/mL DAPI for 5 min at room temperature. This was followed by another 0.7% NaCl wash, and the slides were air dried and mounted in Vectashield (Vector Laboratories). If immunostaining was performed on the neuroblast squashes, the same protocol was followed with a few changes. Brains were instead fixed in a solution of 2% formaldehyde, 2% Triton X-100, and 1× phosphate-buffered saline (PBS) for 2 min, followed by 6 min in a solution of 2% formaldehyde and 45% acetic acid. The incubation in methanol was omitted, and instead slides were washed in PBT (1× PBS, 0.1% Triton X-100) for 30 min and then blocked for 1 h in PBT + 5% bovine serum albumin (BSA). Slides were incubated overnight at 4°C in blocking solution containing the appropriate dilutions of primary antibody. Three 15-min, room-temperature washes in PBT were followed by incubation in blocking solution with the appropriate dilution of secondary antibody for 2 h at room temperature. Finally, slides were washed three times in PBT at room temperature, air dried, and mounted in Vectashield. Immunostaining of all other Drosophila larval tissues was performed as described above; however, fixation was for 20 min at room temperature in a solution of 4% formaldehyde/PBT. Prophase/prometaphase nuclei were identified as mitotic cells in which chromosomes were partially or fully condensed and had not yet aligned on the spindle (illustrated in the metaphase cells shown in Supplemental Fig. 1B).
TUNEL labeling
Discs were dissected in PBS/0.3% Triton X-100 and fixed in 4% paraformaldehyde in PBS/0.3% Triton X-100 for 1 h at room temperature. After rinsing twice in PBS/0.3% Triton X-100, discs were permeabilized in PBS/0.3% Triton X-100 for 30 min at 37°C. The discs were blocked in 1× TUNEL buffer, and 100 μL of TUNEL enzymatic reaction was added as per the manufacturer’s protocol (Roche) for 1 h at 37°C. Three 15-min, room-temperature washes in PBS/0.3% Triton X-100 were performed and discs were mounted with Vectashield.
BrdU labeling
BrdU labeling was performed as described in Moon et al. (2006).
Polytene chromatin staining
Polytene squashes and stainings were performed using a standard protocol (Sullivan et al. 2000).
Co-IP, immunoblotting, and GST pull-down analysis
Co-IPs, GST pull-downs, and Western blots were performed as described previously (Binne et al. 2007). The GST-N-terminal fusion construct was a gift of Jonathan Horowitz (Raleigh, NC) and the GST-Rb phosphorylation mutant fusion construct was a gift of David Goodrich (Houston, TX).
Immunofluorescent staining of mammalian cell
SAOS-2 cells were grown on coverslips, washed once with 1× PBS and fixed in 4% paraformaldehyde for 10 min at room temperature. Coverslips were washed three times in PBS at room temperature. Cells were permeabilized in PBS/0.2% Triton X-100 for 5 min on ice, followed by three 5-min room temperature washes in PBS. Coverslips were incubated in blocking buffer (0.5% NP-40, 1% BSA in PBS) for 30 min at room temperature and then transferred to a humid chamber where they were incubated with primary antibody diluted in blocking buffer overnight at 4°C. Coverslips were then washed three times in PBS for 5 min at room temperature and incubated for 1 h at room temperature with the secondary antibodies diluted in blocking buffer. This was followed by three washes in PBS, DAPI staining for 5 min at room temperature, and mounting in Vectashield.
Chromatin fractionation
SAOS-2 cells were grown to 75% confluence and induced with tetracycline for 6 h, after which they were trypsinized, washed, and counted for aliquotting of 1 × 107 cells per eppendorf. Cells were resuspended in 200 μL of CSK + buffer (10 mM PIPES at pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 0.5% Triton X-100) plus protease inhibitors. Tubes were incubated on ice for 20 min before being centrifuged at 4000 rpm for 5 min. The supernatant was collected and saved for the soluble fraction. Pellets were resuspended in 200 μL of CSK + buffer for 20 min on ice and centrifuged as before, and supernatant was removed. Pellets were resuspended in 40 μL of DNaseI buffer with protease inhibitors (400 mM Tris-HCl at pH 8.0, 100 mM MgSO4, 10 mM CaCl2) for 2 min and then centrifuged as before. Pellets were incubated in 40 μL of DNase I buffer and 20 μL of RQ1 RNase-Free DNase I (Promega) for 25 min at room temperature. Supernatant was collected for the soluble chromatin fraction, and the insoluble chromatin pellet was resuspended in CSK + buffer.
dsRNA treatment of SL2 cells
These experiments were performed as described in Dimova et al. (2003). Treatment of cells with dsRNA lasted 8 d with a media change and readdition of dsRNAs on the fourth day. SL2 cells were then chromatin-fractionated as described above. Western blots using equal amounts of protein from each fraction were performed, and bands were quantified using a Bio-Rad Transilluminator and the Bio-Rad Quantity One software program.
Position effect variegation analysis
These experiments used the Drosophila Tp(3;Y)BL2 stock and were performed as described previously (Lu et al. 1998).
Microscopy
Bright-field microscopy and some fluorescence microscopy were carried out on a Nikon Eclipse 90i microscope. Fluorescence microscopy was also carried out using the Bio-Rad MRC-1000 confocal on a Leica DMRBE microscope excited by a 488-nm excitation line and a 522DF32 emission filter (green), a 568-nm excitation line and a 605DF32 emission filter (red), or a 647-nm excitation line and a 680DF32 emission filter (far red). All images of Drosophila larval tissues were taken using the 60× lens while images of human cells were taken using the 40× lens. Images were captured with the LaserSharp 2000 computer program. Adobe Photoshop and Adobe Illustrator were used to generate the figures of this study.
Acknowledgments
We thank Joe Eissenberg, David Goodrich, Margarete Heck, Jonathan Horowitz, Laurel Raftery, and Claudio Sunkel for reagents used in this study. We thank Spiros Artavanis-Tsakonas and the Harvard/Exelixis Collection for the use of multiple Drosophila stocks. The monoclonal Drosophila lamin antibody developed by Paul A. Fisher was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Science, Iowa City, IA. We also thank the members of the Dyson laboratory for advice and technical assistance. M.S.L. is supported by a Leukemia and Lymphoma Society Career Development Fellowship Award. This study was supported by NIH grants GM53203 and CA64402 (to N.D.).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1631508.
References
- Aono N., Sutani T., Tomonaga T., Mochida S., Yanagida M. Cnd2 has dual roles in mitotic condensation and interphase. Nature. 2002;417:197–202. doi: 10.1038/417197a. [DOI] [PubMed] [Google Scholar]
- Barrientes S., Cooke C., Goodrich D.W. Glutamic acid mutagenesis of retinoblastoma protein phosphorylation sites has diverse effects on function. Oncogene. 2000;19:562–570. doi: 10.1038/sj.onc.1203332. [DOI] [PubMed] [Google Scholar]
- Bazett-Jones D.P., Kimura K., Hirano T. Efficient supercoiling of DNA by a single condensin complex as revealed by electron spectroscopic imaging. Mol. Cell. 2002;9:1183–1190. doi: 10.1016/s1097-2765(02)00546-4. [DOI] [PubMed] [Google Scholar]
- Binne U.K., Classon M.K., Dick F.A., Wei W., Rape M., Kaelin W.G., Naar A.M., Dyson N.J. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat. Cell Biol. 2007;9:225–232. doi: 10.1038/ncb1532. [DOI] [PubMed] [Google Scholar]
- Brehm A., Miska E.A., McCance D.J., Reid J.L., Bannister A.J., Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Chu D.S., Dawes H.E., Lieb J.D., Chan R.C., Kuo A.F., Meyer B.J. A molecular link between gene-specific and chromosome-wide transcriptional repression. Genes & Dev. 2002;16:796–805. doi: 10.1101/gad.972702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang P.T., Albertson D.G., Meyer B.J. DPY-27: A chromosome condensation protein homolog that regulates C. elegansdosage compensation through association with the X chromosome. Cell. 1994;79:459–474. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- Classon M., Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer. 2002;2:910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- Cobbe N., Savvidou E., Heck M.M. Diverse mitotic and interphase functions of condensins in Drosophila. Genetics. 2006;172:991–1008. doi: 10.1534/genetics.105.050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- Dej K.J., Ahn C., Orr-Weaver T.L. Mutations in the Drosophila condensin subunit dCAP-G: Defining the role of condensin for chromosome condensation in mitosis and gene expression in interphase. Genetics. 2004;168:895–906. doi: 10.1534/genetics.104.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova D.K., Stevaux O., Frolov M.V., Dyson N.J. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes & Dev. 2003;17:2308–2320. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W., Vidal M., Xie J.E., Dyson N. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes & Dev. 1996;10:1206–1218. doi: 10.1101/gad.10.10.1206. [DOI] [PubMed] [Google Scholar]
- Ferreira R., Magnaghi-Jaulin L., Robin P., Harel-Bellan A., Trouche D. The three members of the pocket proteins family share the ability to repress E2F activity through recruitment of a histone deacetylase. Proc. Natl. Acad. Sci. 1998;95:10493–10498. doi: 10.1073/pnas.95.18.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman T.M., Sankpal U.T., Robertson A.K., Chen Y., Mazumdar M., Heale J.T., Schmiesing J.A., Kim W., Yokomori K., Zhao Y., et al. Isolation and characterization of a novel DNA methyltransferase complex linking DNMT3B with components of the mitotic chromosome condensation machinery. Nucleic Acids Res. 2004;32:2716–2729. doi: 10.1093/nar/gkh589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentile M., Wiman A., Thorstenson S., Loman N., Borg A., Wingren S. Deletion mapping of chromosome segment 11q24-q25, exhibiting extensive allelic loss in early onset breast cancer. Int. J. Cancer. 2001;92:208–213. doi: 10.1002/1097-0215(200102)9999:9999<::aid-ijc1169>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Gerlich D., Hirota T., Koch B., Peters J.M., Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Gonzalo S., Garcia-Cao M., Fraga M.F., Schotta G., Peters A.H., Cotter S.E., Eguia R., Dean D.C., Esteller M., Jenuwein T., et al. Role of the RB1 family in stabilizing histone methylation at constitutive heterochromatin. Nat. Cell Biol. 2005;7:420–428. doi: 10.1038/ncb1235. [DOI] [PubMed] [Google Scholar]
- Gosling K.M., Makaroff L.E., Theodoratos A., Kim Y.H., Whittle B., Rui L., Wu H., Hong N.A., Kennedy G.C., Fritz J.A., et al. A mutation in a chromosome condensin II subunit, kleisin β, specifically disrupts T cell development. Proc. Natl. Acad. Sci. 2007;104:12445–12450. doi: 10.1073/pnas.0704870104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena R.W., Siddiqui H., Solomon D.A., Mayhew C.N., Held J., Angus S.P., Knudsen E.S. Hierarchical requirement of SWI/SNF in retinoblastoma tumor suppressor-mediated repression of Plk1. J. Biol. Chem. 2004;279:29278–29285. doi: 10.1074/jbc.M400395200. [DOI] [PubMed] [Google Scholar]
- Ham M.F., Takakuwa T., Rahadiani N., Tresnasari K., Nakajima H., Aozasa K. Condensin mutations and abnormal chromosomal structures in pyothorax-associated lymphoma. Cancer Sci. 2007;98:1041–1047. doi: 10.1111/j.1349-7006.2007.00500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Herrera R.E., Chen F., Weinberg R.A. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts. Proc. Natl. Acad. Sci. 1996;93:11510–11515. doi: 10.1073/pnas.93.21.11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. The ABCs of SMC proteins: Two-armed ATPases for chromosome condensation, cohesion, and repair. Genes & Dev. 2002;16:399–414. doi: 10.1101/gad.955102. [DOI] [PubMed] [Google Scholar]
- Hirano T. Condensins: Organizing and segregating the genome. Curr. Biol. 2005;15:R265–R275. doi: 10.1016/j.cub.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Hirota T., Gerlich D., Koch B., Ellenberg J., Peters J.M. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell Sci. 2004;117:6435–6445. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- Iovino F., Lentini L., Amato A., Di Leonardo A. RB acute loss induces centrosome amplification and aneuploidy in murine primary fibroblasts. Mol. Cancer. 2006;5:38. doi: 10.1186/1476-4598-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac C.E., Francis S.M., Martens A.L., Julian L.M., Seifried L.A., Erdmann N., Binne U.K., Harrington L., Sicinski P., Berube N.G., et al. The retinoblastoma protein regulates pericentric heterochromatin. Mol. Cell. Biol. 2006;26:3659–3671. doi: 10.1128/MCB.26.9.3659-3671.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Hirano M., Kobayashi R., Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Kimura K., Cuvier O., Hirano T. Chromosome condensation by a human condensin complex in Xenopus egg extracts. J. Biol. Chem. 2001;276:5417–5420. doi: 10.1074/jbc.C000873200. [DOI] [PubMed] [Google Scholar]
- Korenjak M., Taylor-Harding B., Binne U.K., Satterlee J.S., Stevaux O., Aasland R., White-Cooper H., Dyson N., Brehm A. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- Lam W.W., Peterson E.A., Yeung M., Lavoie B.D. Condensin is required for chromosome arm cohesion during mitosis. Genes & Dev. 2006;20:2973–2984. doi: 10.1101/gad.1468806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.W., Beall E.L., Fleischer T.C., Georlette D., Link A.J., Botchan M.R. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes & Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L., Sadasivam S., Florens L., Zhu X., Swanson S.K., Velmurugan S., Chen R., Washburn M.P., Liu X.S., DeCaprio J.A. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell. 2007;26:539–551. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Lu B.Y., Ma J., Eissenberg J.C. Developmental regulation of heterochromatin-mediated gene silencing in Drosophila. Development. 1998;125:2223–2234. doi: 10.1242/dev.125.12.2223. [DOI] [PubMed] [Google Scholar]
- Luo R.X., Postigo A.A., Dean D.C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L., Groisman R., Naguibneva I., Robin P., Lorain S., Le Villain J.P., Troalen F., Trouche D., Harel-Bellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- Mayhew C.N., Perkin L.M., Zhang X., Sage J., Jacks T., Knudsen E.S. Discrete signaling pathways participate in RB-dependent responses to chemotherapeutic agents. Oncogene. 2004;23:4107–4120. doi: 10.1038/sj.onc.1207503. [DOI] [PubMed] [Google Scholar]
- Moon N.-S., Di Stefano L., Dyson N. A gradient of epidermal growth factor receptor signaling determines the sensitivity of rbf1 mutant cells to E2F-dependent apoptosis. Mol. Cell. Biol. 2006;26:7601–7615. doi: 10.1128/MCB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris E.J., Dyson N.J. Retinoblastoma protein partners. Adv. Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- Narita M., Nunez S., Heard E., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., Lowe S.W. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Nielsen S.J., Schneider R., Bauer U.M., Bannister A.J., Morrison A., O’Carroll D., Firestein R., Cleary M., Jenuwein T., Herrera R.E., et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Oliveira R.A., Coelho P.A., Sunkel C.E. The condensin I subunit Barren/CAP-H is essential for the structural integrity of centromeric heterochromatin during mitosis. Mol. Cell. Biol. 2005;25:8971–8984. doi: 10.1128/MCB.25.20.8971-8984.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R.A., Heidmann S., Sunkel C.E. Condensin I binds chromatin early in prophase and displays a highly dynamic association with Drosophila mitotic chromosomes. Chromosoma. 2007;116:259–274. doi: 10.1007/s00412-007-0097-5. [DOI] [PubMed] [Google Scholar]
- Onn I., Aono N., Hirano M., Hirano T. Reconstitution and subunit geometry of human condensin complexes. EMBO J. 2007;26:1024–1034. doi: 10.1038/sj.emboj.7601562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F., Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Ono T., Fang Y., Spector D.L., Hirano T. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell. 2004;15:3296–3308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin E., Mittnacht S., Villa-Moruzzi E., Ludlow J.W. Site-specific and temporally-regulated retinoblastoma protein dephosphorylation by protein phosphatase type 1. Oncogene. 2001;20:3776–3785. doi: 10.1038/sj.onc.1204518. [DOI] [PubMed] [Google Scholar]
- Rubin S.M., Gall A.L., Zheng N., Pavletich N.P. Structure of the Rb C-terminal domain bound to E2F1-DP1: A mechanism for phosphorylation-induced E2F release. Cell. 2005;123:1093–1106. doi: 10.1016/j.cell.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Savvidou E., Cobbe N., Steffensen S., Cotterill S., Heck M.M. Drosophila CAP-D2 is required for condensin complex stability and resolution of sister chromatids. J. Cell Sci. 2005;118:2529–2543. doi: 10.1242/jcs.02392. [DOI] [PubMed] [Google Scholar]
- Sherr C.J., McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- Steffensen S., Coelho P.A., Cobbe N., Vass S., Costa M., Hassan B., Prokopenko S.N., Bellen H., Heck M.M., Sunkel C.E. A role for Drosophila SMC4 in the resolution of sister chromatids in mitosis. Curr. Biol. 2001;11:295–307. doi: 10.1016/s0960-9822(01)00096-3. [DOI] [PubMed] [Google Scholar]
- Stevaux O., Dimova D., Frolov M.V., Taylor-Harding B., Morris E., Dyson N. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 2002;21:4927–4937. doi: 10.1093/emboj/cdf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevaux O., Dimova D.K., Ji J.Y., Moon N.S., Frolov M.V., Dyson N.J. Retinoblastoma family 2 is required in vivo for the tissue-specific repression of dE2F2 target genes. Cell Cycle. 2005;4:1272–1280. doi: 10.4161/cc.4.9.1982. [DOI] [PubMed] [Google Scholar]
- Sullivan W., Ashburner M., Hawley R.S. Drosophila Protocols. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2000. [Google Scholar]
- Trimarchi J.M., Lees J.A. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- Xu Y., Leung C.G., Lee D.C., Kennedy B.K., Crispino J.D. MTB, the murine homolog of condensin II subunit CAP-G2, represses transcription and promotes erythroid cell differentiation. Leukemia. 2006;20:1261–1269. doi: 10.1038/sj.leu.2404252. [DOI] [PubMed] [Google Scholar]