Abstract
Translational repression during mRNA transport is essential for spatial restriction of protein production. In the yeast Saccharomyces cerevisae, silencing of ASH1 mRNA before it is localized to the bud cortex in late anaphase is critical for asymmetric segregation of Ash1p to the daughter cell nucleus. Puf6p, an ASH1 mRNA-binding protein, has been implicated in this process as a translational repressor, but the underlying mechanism is unknown. Here, we used yeast extract-based in vitro translation assays, which recapitulate translation and phosphorylation, to characterize the mechanism of Puf6p-mediated translational regulation. We report that Puf6p interferes with the conversion of the 48S complex to the 80S complex during initiation, and this repression by Puf6p is mediated through the general translation factor eIF5B (Fun12p in S. cerevisiae). Puf6p interacts with Fun12p via the PUF domain, and this interaction is RNA-dependent and essential for translational repression by Puf6p. This repression is relieved by phosphorylation of the N-terminal region of Puf6p mediated by protein kinase CK2 (casein kinase II). Inhibition of phosphorylation at Ser31, Ser34, and Ser35 of Puf6p increases its translational repression and results in ASH1 mRNA delocalization. Our results indicate that Puf6p suppresses the translation initiation of ASH1 mRNA via interaction with Fun12p during its transport, and this repression can be released by CK2 phosphorylation in the N-terminal region of Puf6p when the mRNA reaches the bud tip.
[Keywords: Translational regulation, RNA localization, RNA transport]
RNA localization is a fundamental mechanism to restrict protein expression to a specific region in the cell, vital to the establishment of cellular polarity and determination of cell fate (St Johnston 2005; Corral-Debrinski 2007; Du et al. 2007). In budding yeast Saccharomyces cerevisiae, ASH1 mRNA localization is required for mating-type switching. The ASH1 transcripts are localized at the bud cortex in late anaphase, which confines the Ash1 protein to the daughter cell nucleus (Long et al. 1997; Takizawa et al. 1997). ASH1 mRNA localization is achieved by active transport along actin bundles (Long et al. 1997; Takizawa et al. 1997) by a core localization complex (the “locasome”) consisting of proteins She1/Myo4, She2, and She3 (Chartrand et al. 2001; Kwon and Schnapp 2001; Darzacq et al. 2003). She2p is the primary RNA-binding protein that recognizes four cis localization elements (E1, E2A, E2B, and E3) on the ASH1 transcript (Chartrand et al. 1999). She2p recruits Myo4p, a type V myosin, to the ASH1 mRNA via the adaptor protein She3p (Bohl et al. 2000; Long et al. 2000; Takizawa and Vale 2000). ASH1 mRNA localization in budding yeast serves as a model to study RNA transport and localization in mammals and other species (Darzacq et al. 2003; St Johnston 2005).
To achieve spatial and temporal regulation of ASH1 expression, translational repression is coordinated during RNA transport to prevent premature protein synthesis. Both cis- and trans-factors have been shown to play critical roles in translational repression during ASH1 mRNA transport. There are four elements in the coding region of ASH1 mRNA that have been proposed to slow down translation during mRNA transport and prevent premature translation of ASH1 (Chartrand et al. 1999, 2002). Two RNA-binding proteins, Khd1p and Puf6p, have been identified that are required for the localization and translation of ASH1 mRNA (Irie et al. 2002; Gu et al. 2004; Paquin et al. 2007). Release of translational repression is needed once ASH1 mRNA localizes and has been implicated in proper ASH1 mRNA anchoring at the bud tip (Gonzalez et al. 1999; Irie et al. 2002). A casein kinase I (CK1) protein kinase-mediated release of the translational control by Khd1p has been identified recently (Paquin et al. 2007). The mechanism by which Puf6p functions as a translational repressor and how this repression is released remain elusive.
In this study, we examine the role of Puf6p, a PUF protein, in regulating translation of ASH1 mRNA. We show that Puf6p represses translation by interfering with the conversion of the 48S complex to 80S, and that this repression is mediated through the general translation initiation factor eIF5B/Fun12p. Both the N-terminal region and the PUF domain of Puf6p are required for Puf6p repression activity. Casein kinase II (CK2) phosphorylation sites on Puf6p have been identified in the N-terminal region, and CK2 phosphorylation reduces Puf6p repression activity. CK2 localizes to the bud tip before ASH1 expression. These results suggest a mechanism of translational repression by Puf6p involving Fun12p and a spatially controlled phosphorylation step to relieve it.
Results
Puf6p interferes with 80S assembly in translation initiation
To investigate the mechanism of translation regulation by Puf6p, we developed an in vitro translation assay using cell-free yeast extracts. We constructed a reporter mRNA with the coding sequence for renilla luciferase and a 3′ untranslated region (UTR) containing the E3 element of ASH1 mRNA (15 nucleotides [nt] of the coding sequence of ASH1 mRNA and 121 nt of the 3′UTR) that has been shown to be recognized by Puf6p (Fig. 1A; Gu et al. 2004). A similar construct lacking the E3 element (R-luc) serves as a control for specificity. We incubated the in vitro synthesized mRNA with yeast extracts containing 35S methionine and verified the synthesis of renilla protein (data not shown). We preincubated this reporter mRNA with recombinant Puf6 and measured its in vitro translation. In the presence of Puf6, considerably less protein was produced using R-luc-E3 mRNA (Fig. 1B), by 82% and 94% at protein-to-RNA molar ratios of 20 and 100, respectively. The renilla luciferase synthesized from R-luc RNA without the Puf6p-binding site (E3 element) did not show a dosage-dependent decrease in the presence of Puf6. Thus, Puf6 represses translation in a sequence-specific fashion in yeast extracts, consistent with the results obtained using rabbit reticulocyte lysates (Gu et al. 2004).
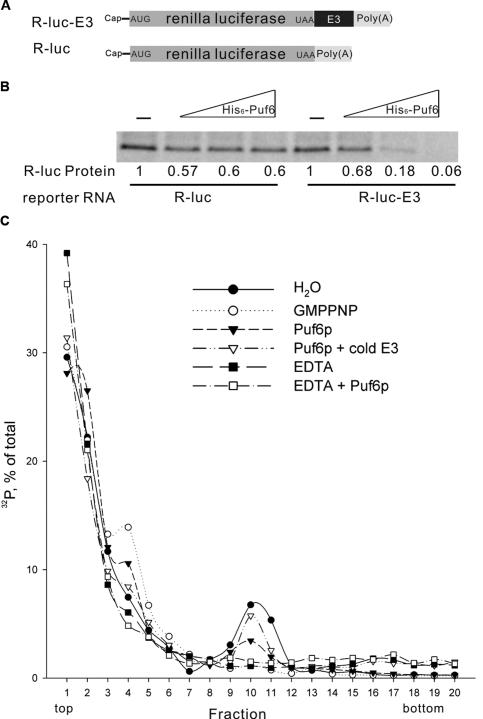
Figure 1.
Puf6p is an E3-dependent translational repressor that blocks 80S complex assembly. (A) Schematic representation of the mRNA constructs for in vitro translation assays. E3 is the sequence in ASH1 mRNA containing binding sites for Puf6p. (B) Autoradiaograph of renilla luciferase (35S-methionine-labeled) translated in yeast extracts. R-luc or R-luc-E3 mRNA (17 nM) was incubated with full-length His6-Puf6 at protein-to-RNA molar ratios of 0, 5, 20, and 100. Numbers indicate the protein levels quantified using a PhosphorImager and normalized to no Puf6 added. There is a slight nonspecific effect of His6-Puf6 with R-luc. (C) Sucrose gradient profile of 32P-labeled miniORF-E3 RNA. RNA was preincubated with H2O (filled circles), 2 mM GMPPNP (open circles), full-length His6-Puf6 (filled triangles), or full-length His6-Puf6 with 3 μg of competitor E3 RNA (open triangles); then incubated with cycloheximide-treated yeast extract; and resolved by centrifugation in 7%–47% linear sucrose gradients. After fractionation, radioactivity was monitored, expressed as percentage of total counts recovered, and plotted against the fraction number. To dissociate ribosomes, 5 mM EDTA was supplemented in yeast extract without (filled squares) or with (open squares) full-length His6-Puf6.
Translation initiation is the rate-limiting step in translation and serves as a target for translational regulation (Dever 2002). To dissect the mechanism of Puf6p inhibition, we analyzed the distribution of translation complexes by sucrose density gradient. To increase the resolution, we constructed a short coding region followed by the E3 element of ASH1, 179 nt in total (Trachsel et al. 1977). Cycloheximide was used to inhibit translation elongation. We found that the labeled E3 RNA under control conditions sedimented at fractions 9–11 (Fig. 1C, filled circles), which corresponded to the 80S complex from the absorbance profile (Supplemental Fig. 1), indicating that the reporter was translated. In the presence of GMP-PNP, a nonhydrolyzable analog of GTP that blocks 60S subunit joining (Lee et al. 2002), the E3 RNA formed a complex sedimenting in fractions 3–5 that corresponded to the 48S complex (Fig. 1C, open circles). The decrease of the 80S complex and concomitant increase of the 48S complex indicated that GMP-PNP allowed translation initiation to proceed to the 48S complex but efficiently blocked 60S joining. Incubation of the E3 RNA with recombinant Puf6 resulted in an increase in the 48S complex and a decrease in the 80S complex similar to results obtained with GMP-PNP (Fig. 1C, filled triangles). This increase of the 48S peak was reproducible using different gradients (see the Materials and Methods). This suggested that assembly of the 80S complex but not the formation of the 48S complex was affected by Puf6p. Inhibition by Puf6 was specific, as competition with cold E3 RNA led to a recovery of the 80S complex preformed by the labeled E3 (Fig. 1C, open triangles). In the presence of EDTA, RNA–ribosomal complexes were not detected, independent of Puf6 (Fig. 1C, open and filled squares), consistent with the essential role of Mg2+ in RNA–ribosomal complex formation. These results suggest that Puf6p blocked 80S complex assembly but not 48S complex formation during translation initiation, which may result from its effect on 60S joining.
Fun12p associates with Puf6p and is required for ASH1 translation and localization
General translation initiation factors are common targets for translational regulation. Puf6p was found to interact with Fun12p (eIF5B in yeast), which assists 60S subunit joining (Gavin et al. 2002; Lee et al. 2002; Collins et al. 2007). To evaluate the possibility that Puf6p could repress translation initiation by suppressing Fun12p, we first tested the interaction between Puf6p and Fun12p by coimmunoprecipitation (co-IP) using yeast extracts from cells expressing both Fun12p-HA and Puf6p-Tap. Fun12p-HA coprecipitated from cells expressing Puf6p-Tap but did not from cells with untagged Puf6p using Tap purification (Fig. 2A). In the reciprocal co-IP, we found Puf6p-HA coprecipitated with Fun12p-Tap but not with untagged Fun12p (Fig. 2B). This interaction is RNA-dependent, as the association between Puf6p and Fun12p was significantly decreased after RNase treatment (Supplemental Fig. 2A).
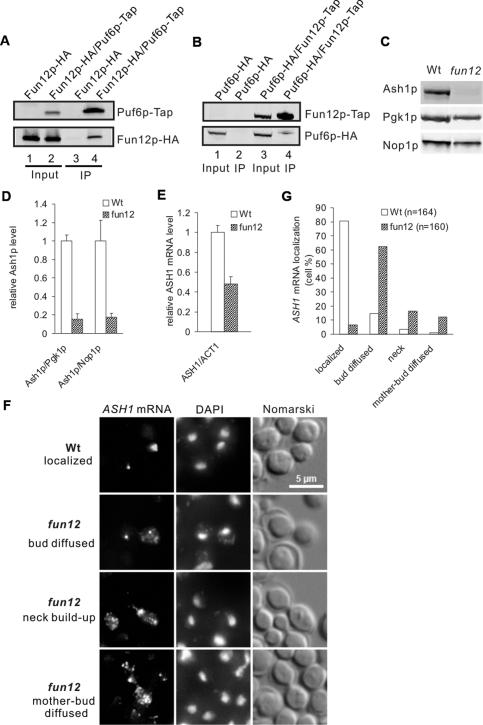
Figure 2.
Fun12p associates with Puf6p and is required for ASH1 mRNA translation and localization. (A) Co-IP with protein A tag (Tap) and IgG-coated beads. Proteins in yeast extract expressing Fun12p-HA (lanes 1,3) or Puf6p-Tap/Fun12p-HA (lanes 2,4) were affinity-purified on matrix-bound IgG. Eluates (lanes 3,4) and 16% total inputs (lanes 1,2) were analyzed by Western blotting with anti-HA antibody. (B) Reciprocal co-IP of A. Proteins in extract expressing Puf6p-HA (lanes 1,2) or Fun12p-Tap/Puf6p-HA (lanes 3,4) were affinity-purified on matrix-bound IgG. Eluates (lanes 2,4) and 16% total inputs (lanes 1,3) were analyzed by Western blotting with anti-HA antibody. (C) Proteins in yeast extract from cells expressing wild-type FUN12 (Wt) or with FUN12 deletion (fun12) were analyzed by Western blotting. Extract from equal amount of cells were loaded. Nine repeats of the c-myc peptide sequence were inserted at the C terminus of Ash1p in each strain. Ash1p, Pgk1p, and Nop1p were detected by anti-Myc, anti-Pgk1, and anti-Nop1 antibodies, respectively. (D) Ash1 protein levels quantified by Western blotting and normalized by Pgk1p or Nop1p. Extracts from three individual clones of each strain were analyzed. (E) ASH1 mRNA levels quantified by real-time PCR and normalized to ACT1 mRNA. mRNA extracted from three individual clones of each strain was analyzed. (F) FISH of cells expressing wild-type FUN12 (Wt) or with FUN12 disruption (fun12) with ASH1-specific probes (ASH1 mRNA). Representative cells showing RNA localized at the bud tip (row 1), diffused in the bud (row 2), built-up in the neck (row 3), and scattered throughout mother and bud (row 4). Bar, 5 μm. (G) Percentage of cells showing localized and delocalized ASH1 mRNA. Localization of ASH1 mRNA was determined by FISH and cells were scored according to the pattern shown in F. (n) Total cells counted.
Although Fun12p is a general translation initiation factor, it is not essential for cell viability (Choi et al. 1998). To test if Fun12p was required for ASH1 translation, we disrupted FUN12 in a strain expressing Ash1p-Myc (fun12). The protein concentrations of Ash1, Pgk1, and Nop1 were reduced by deletion of FUN12. However, the reduction of Ash1p was considerably more significant than Pgk1p and Nop1p whose mRNAs do not contain consensus Puf6p-binding sites (Fig. 2C). We found the expression of Ash1p decreased by >80% relative to Pgk1p or Nop1p after the disruption of FUN12 (Fig. 2D). ASH1 mRNA levels were reduced by 50% relative to ACT1 in the fun12 strain (Fig. 2E), suggesting that both transcription and translation of ASH1 mRNA were reduced by FUN12 disruption. A GST-Fun12 fusion recombinant protein containing amino acid residues 396–1002 was functional in vivo, as it was able to fully complement the slow-growth phenotype of the fun12Δ strain and restore protein synthesis in vitro (Choi et al. 1998, 2000). Truncated Fun12p complemented the expression of Ash1p in a fun12Δ strain when introduced on a plasmid (Supplemental Fig. 2B). Therefore, this truncated Fun12p could form the initiation complex on ASH1 mRNAs, as does the full-length Fun12p in vivo. FUN12 overexpression increased the concentration of Ash1p by twofold in comparison with Pgk1p but had only a marginal difference at the mRNA level (Supplemental Fig. 3), consistent with the requirement for Fun12p in ASH1 mRNA translation.
Although fun12 cells grow slower than wild type (Choi et al. 1998), the cell morphology of fun12 has been shown to be normal (Narayanaswamy et al. 2006) and phalloidin staining displayed normal actin organization (data not shown). We then analyzed the effect of FUN12 disruption on ASH1 mRNA localization. ASH1 mRNA was delocalized in the fun12 mutant (Fig. 2F). In the wild-type strain, 80% of ASH1 mRNA was localized at the distal cortex of the bud. However, in the fun12 mutant, ASH1 mRNA was localized diffusely within the bud (62%), in the neck (16%), or in the mother and bud (12%) (Fig. 2G). This pattern of ASH1 mRNA in fun12 cells was different from the she mutants where the ASH1 mRNA was mostly diffused throughout mother and bud (Long et al. 1997). To test whether Fun12p was also required for the localization of other mRNAs, we examined the localization of IST2 mRNA (Takizawa et al. 2000), which contains putative Puf6p-binding sites at the 3′UTR (Gu et al. 2004). IST2 mRNA was found diffusely distributed within the bud in both fun12 and puf6 mutants (Supplemental Fig. 4), similar to the pattern observed for ASH1 mRNA.
Both the N-terminal region and PUF domain of Puf6p are required for translational repression
Puf6p contains a PUF domain and a distinct N-terminal region with low homology compared with other PUF proteins (Supplemental Fig. 5). We generated two fragments of Puf6 to dissect their functional roles (Fig. 3A): N120 (N-terminal region outside of the PUF domain), and C536 (PUF domain plus C terminus). Since the PUF domain (amino acids 121–565) was insoluble when expressed in Escherichia coli, we designed a fragment (C536) containing both the PUF domain and the C terminus of Puf6p (amino acids 121–656). The recombinant proteins of full-length, N120, and C536 with N-terminal His6 tag were purified from E. coli (Fig. 3B). N120 showed a higher molecular weight (∼33 kDa) than expected (18.7 kDa) by SDS-PAGE. This might be due to insufficient binding of SDS to the Asp/Glu-rich region in the N-terminal region of Puf6p (Gu et al. 2004).
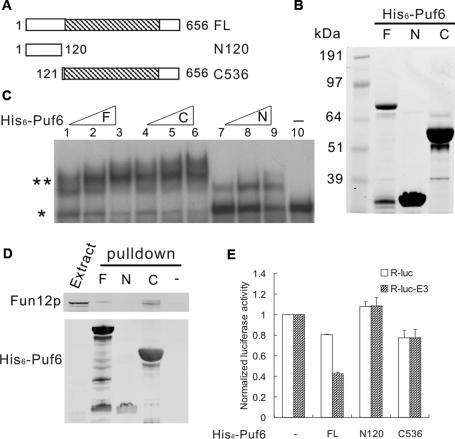
Figure 3.
PUF domain is necessary but not sufficient for translational repression. (A) Schematic describing Puf6p fragments in constructs for recombinant protein expression. Numbers indicate positions of amino acid residue from start methionine of Puf6p. The hatched region shows the PUF domain. (B) Full-length (F), N120 (N), and C536 (C) recombinant proteins of 5 μg each were separated by 4%–12% SDS-PAGE and stained by Coomassie blue. (C) Gel mobility shift assay for miniORF-E3 RNA with increasing concentrations (22 pmol, 44 pmol, and 66 pmol) of full-length (F), C536 (C), N120 (N) recombinant proteins or no protein (shown in lane 10). (*) Free probe; (**) bound probe. (D) Pull-down assays for yeast extract containing Fun12p-Tap with full-length (F), N120 (N), and C536 (C) recombinant His6-Puf6 proteins. Twenty percent of total input was loaded. (−) Pull-down assay without His6-Puf6. (E) Luciferase assay for R-luc and R-luc-E3 in vitro translated in yeast extract and normalized to reaction without His6-Puf6 (−) for each reporter. RNA (17 nM) was preincubated with full-length (FL), N120, and C536 recombinant His6-Puf6 proteins at a protein-to-RNA molar ratio of 80.
We tested RNA-binding activity of these three Puf6 recombinant proteins to E3 RNA using a polyacrylamide gel electrophoretic assay. Both C536 and full-length Puf6 bound to the E3 RNA and resulted in a band shift (Fig. 3C), consistent with the finding that PUF domain is an RNA-binding motif (Zamore et al. 1997). N120 showed a weaker shift with the RNA, although it does not contain a known RNA-binding motif.
We next examined the interaction of the three forms of recombinant Puf6 with Fun12p by pull-down assays. The recombinant Puf6 were incubated with yeast extract containing Tap-tagged Fun12p and purified on nickel beads. We found Fun12p-Tap was retained by C536 and full-length Puf6, but not by N120 (Fig. 3D), suggesting that the PUF domain is required for the interaction with Fun12p. This result also confirms the interaction of Puf6p and Fun12p from the co-IP studies.
Since C536 interacts with both Fun12p and RNA, it could be sufficient to repress translation of E3 RNA. To test this, we performed in vitro luciferase translation assays using yeast extract and different forms of Puf6. As shown in Figure 3E, in comparison with R-luc RNA, full-length His6-Puf6 repressed translation of R-luc-E3 RNA specifically by half. This repression was comparable with the results using the PhosphorImager (Fig. 1B), confirming luciferase assays as a quantitative measure of in vitro translated protein. In contrast, C536 repressed both R-luc-E3 and R-luc RNA equivalently by 20% in comparison with the reaction without recombinant proteins (Fig. 3E), indicating that the PUF domain alone was insufficient for E3-specific translational repression. Repression by N120 was not observed for either R-luc-E3 or R-luc RNA (Fig. 3E). These results suggest that both the N-terminal region and PUF domain are required for E3-specific translational repression.
Puf6p is phosphorylated by protein kinase CK2
One role for N120 could be as a regulatory domain for Puf6p-mediated repression. Cka2p, a catalytic subunit of protein kinase CK2, was also detected in the She2p-Tap affinity purification together with Puf6p (Gu et al. 2004; data not shown). Global protein interaction studies showed that Puf6p associated with each of the two catalytic subunits of CK2 (Ho et al. 2002), suggesting that Puf6p may be a substrate for CK2. Relevant to this, we found six potential CK2 phosphorylation sites in N120 region but only one in the C536 region.
We first tested whether Puf6p was phosphorylated in vivo using a sensitive noncovalent fluorescent dye staining technology for the detection of phosphoserine-, phosphothreonine-, and phosphotyrosine-containing proteins displayed on SDS-PAGE (Pro-Q diamond phosphoprotein gel staining, Molecular Probes). The Puf6p-Tap purified from yeast extracts was found to be phosphorylated (Fig. 4A). The phosphorylation of Puf6p was specific as the untagged cell did not show a phosphoprotein band with the similar gel mobility (Supplemental Fig. 6). The phosphorylation of endogenous Puf6p was further confirmed by treatment with λ phosphatase (λ ppase), which can remove phosphates from both Ser/Thr and Tyr. After λ ppase treatment, a faster-migrating band for Puf6p-Tap was detected (Fig. 4B), suggesting phosphate groups were removed from Puf6p. The λ ppase treatment also caused a faster migration of Puf6p-HA purified from yeast, confirming that the phosphorylation is on Puf6p and not the tag (Fig. 4B). Only a single band of Puf6p was detected by Western blot before the treatment of λ ppase, suggesting that the majority of endogenous Puf6p may be phosphorylated.
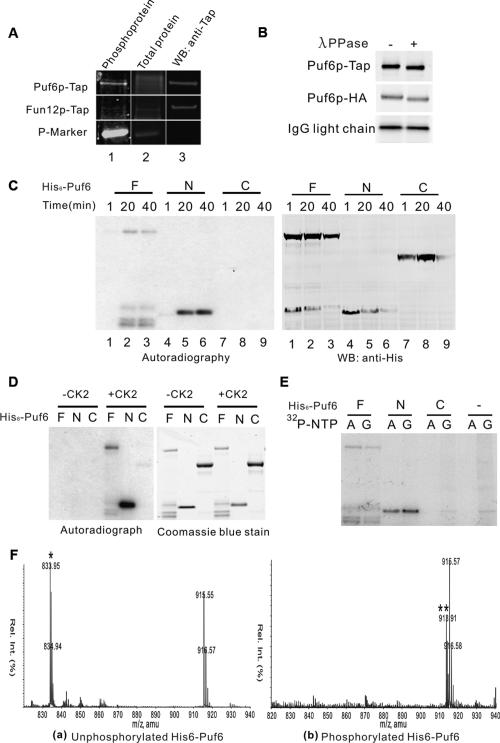
Figure 4.
Puf6p is phosphorylated by CK2 on the N-terminal region. (A) Yeast Puf6p is phosphorylated. Proteins in yeast extract from cells expressing Puf6p-Tap or Fun12p-Tap were affinity-purified on matrix-bound IgG. The IgG bead eluates were analyzed by Pro-Q Diamond gel staining for phosphoprotein (column 1), then SYPRO Ruby gel staining for total protein (column 2), and then processed for immnostaining with anti-Tap antibody (column 3). Fun12p-Tap was used as a negative control for phosphoprotein stain. (P-Marker) Phosphoprotein marker. (B) Proteins in yeast extract from cells expressing Puf6p-Tap or Puf6p-HA were affinity-purified and subjected to λ ppase assay. Eluates were analyzed by Western blotting with anti-HA and anti-Tap antibodies. Puf6p-tap and Puf6p-HA were precipitated by IgG and anti-HA antibody-coupled beads, respectively. IgG light chain was used as an internal control for the gel mobility shift. (C) In vitro phosphorylation assay with yeast extract and [γ-32P]-ATP. Proteins were sampled as indicated during incubation and analyzed by autoradiograph (left) and Western blotting (right). (F) full-length recombinant proteins; (N) N120 recombinant proteins; (C) C536 recombinant proteins. (D) In vitro phosphorylation assay with CK2 from sea star and [γ-32P]-ATP. Proteins were analyzed by autoradiograph (left) and Coomassie blue staining (right), respectively. (E) In vitro phosphorylation assay with yeast extract and [γ-32P]-ATP or [γ-32P]-GTP as above. Proteins were analyzed by autoradiograph. (F) Mass spectra of the tryptic peptides from unphosphorylated (panel a) and phosphorylated His6-Puf6 (panel b). In panel a, the species observed at 833.9 (m/z) corresponds to the doubly charged ion of the peptide ISIDSSDEESELSKK (1665.74 Da). The ion that has a mass of 80 higher than ISIDSSDEESELSKK at 913.9 (**) is observed only from the trypsin digestion of phosphorylated His6-Puf6. The increase of 80 on the ion at 913.91 corresponds to a mass of 160 as it is doubly charged, which is expected for addition of two phosphate groups (80 Da for one phosphate group) on the ion.
To test which region of Puf6p was phosphorylated, we performed an in vitro phosphorylation assay with recombinant Puf6 proteins. Full-length Puf6 and N120, but not C536, were phosphorylated when incubated with yeast extracts (Fig. 4C). A similar pattern was observed in the phosphorylation assay using CK2 purified from sea star (Fig. 4D), suggesting that phosphorylation of His6-Puf6 in yeast extract is likely due to endogenous CK2. A special feature of CK2 is that it can use both ATP and GTP as phosphate donors (Gatica et al. 1993). We found that yeast extract phosphorylated full-length Puf6 and N120 but not C536 in the presence of [γ-32P]-GTP or [γ-32P]-ATP (Fig. 4E). The phosphorylation of Puf6p by yeast CK2 was further confirmed by the finding that recombinant Puf6 was efficiently phosphorylated by yeast CK2 purified from cells expressing Cka2p-Tap (Supplemental Fig. 7).
To identify the phosphorylation sites, the recombinant full-length His6-Puf6 was phosphorylated in yeast extract and subjected to mass spectrometry analysis. Two phosphorylation sites, Ser34 and Ser35, were identified (Fig. 4F). This was consistent with the result from a recent proteomic study that two phosphorylated serines (Ser35 and either Ser31 or Ser34) were identified on endogenous Puf6p (Chi et al. 2007). Ser34 and Ser35 (within 33DSSDEE38) are canonical CK2 sites: S/T-XX-D/E (X for any nonbasic amino acid) (Meggio and Pinna 2003), and Ser31 could be primed to become a CK2 site by phosphorylation on Ser34 (Roach 1991). The agreement of the mass spectrometry with the results from in vitro phosphorylation assays strongly suggests that Puf6p is phopshorylated by CK2 on the N-terminal region in vivo, and yeast extract competent for translation also recapitulates this phosphorylation on recombinant Puf6.
CK2 phosphorylation of Puf6p relieves translational repression
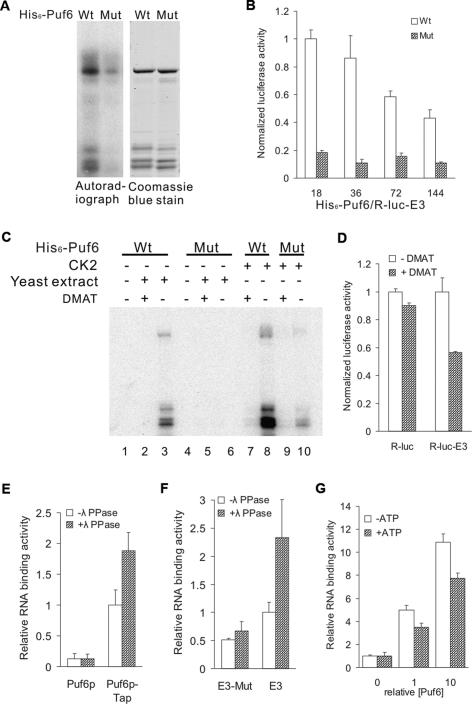
To test whether phosphorylation of Puf6p affects its function in regulating translation, we generated Ser-to-Ala point mutants of the potential phosphorylation sites (Ser31Ala, Ser34Ala, and Ser35Ala). Phosphorylation of Puf6 with all three point mutations by CK2 was reduced by 90% (Fig. 5A). We preincubated the R-luc-E3 RNA with either wild-type or mutant Puf6 and measured translation of the reporter by luciferase activity. Strikingly, in comparison with wild-type Puf6, the mutant His6-Puf6 repressed translation much more efficiently (Fig. 5B). At protein-to-RNA ratios of 18 and 36, the luciferase translated in the presence of the mutant Puf6p was <20% of the wild-type His6-Puf6, suggesting that phosphorylation of Puf6 down-regulated its repressing activity.
Figure 5.
CK2 phosphorylation relieves translational repression by Puf6p. (A) In vitro phosphorylation assay with rat CK2 and [γ-32P]-ATP. Wild-type (Wt) and Ser31,34,35Ala mutant (Mut) His6-Puf6 were analyzed by autoradiograph and Coomassie blue staining. (B) Luciferase assay for R-luc-E3 in vitro translated in yeast extract. RNAs were preincubated with wild-type (Wt) or Ser31,34,35 → Ala mutant (Mut) His6-Puf6. Luciferase activities were normalized to reaction with wild-type His6-Puf6 at a protein-to-RNA molar ratio of 18. Protein-to-RNA molar ratio varied with [RNA] = 17 nM (X-axis). (C) In vitro phosphorylation assay of Puf6 with rat CK2 or yeast extract and [γ-32P]-ATP with or without DMAT (25 μM). Wild-type (Wt) and Ser31,34,35Ala mutant (Mut) His6-Puf6 were analyzed by autoradiograph. (D) Luciferase assay for R-luc-E3 and R-luc in vitro translated in yeast extract. RNA was preincubated with wild-type His6-Puf6 at protein-to-RNA molar ratio of 36 with [RNA] = 17 nM. Luciferase activity was normalized to reaction without DMAT for each reporter. (E–G) Phosphorylation of Puf6 reduces RNA binding. (E) RNA-binding assay with 32P-labeled miniORF-E3 RNA and Puf6p-Tap affinity-purified from yeast extract. Extracts containing Puf6p-Tap or untagged Puf6p were incubated with IgG-coated beads and immobilized proteins were treated or not treated with λ ppase before the RNA-binding assay. RNA-binding activity was normalized to the Puf6p-Tap extract without λ ppase treatment. (F) RNA-binding assay with 32P-labeled miniORF-E3 RNA and Puf6p-Tap affinity-purified from yeast extract as described above. The two UUGU sequences essential for Puf6p binding were deleted in E3 (miniORF-E3-M). RNA-binding activity was normalized to the Puf6p-Tap extract without λ ppase treatment. (G) RNA-binding assay with 32P-labeled miniORF-E3 RNA and recombinant His-Puf6 immobilized on nickel beads. Recombinant His-Puf6 was phosphorylated with rat liver CK2 (+ATP) or not (−ATP) before the RNA-binding assay. RNA-binding activities were normalized to binding assays without recombinant Puf6.
To test whether CK2 affects the repression by Puf6p in a similar manner, we performed an in vitro translation assay with wild-type Puf6 using yeast extract treated with DMAT (2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole), an inhibitor of CK2 (Pagano et al. 2004). Phosphorylation of wild-type Puf6, either by yeast extracts or purified CK2, was significantly reduced in the presence of DMAT (Fig. 5C, cf. lanes 2 and 3, and lanes 7 and 8). As a result, the luciferase translated from R-luc-E3 RNA was reduced by 44% in the presence of DMAT (Fig. 5D). In comparison, the translation of R-luc RNA without the E3 Puf6p-binding element was affected only marginally. These results suggest that CK2-mediated phosphorylation of Puf6 decreases its repressing activity.
One of the mechanisms by which CK2 phosphorylation could affect Puf6p activity may be through changing the affinity of Puf6p for RNA. To test this possibility, we performed an RNA-binding assay using endogenous Puf6p treated with λ ppase. Puf6p-tap was affinity-purified on matrix-bound IgG from yeast extracts and treated with λ ppase. We then incubated the protein with 32P-labeled E3 RNA. The RNA bound to the IgG beads was eluted and quantified by radioactivity. We found that the E3 RNA was specifically retained by Puf6p-Tap and the retention increased after the treatment with λ ppase (Fig. 5E). The binding specificity was tested by a mutant E3 RNA that contained mutations in the elements essential for Puf6p binding (Gu et al. 2004). In comparison with wild-type E3, the mutant E3 RNA was retained less efficiently by Puf6p-Tap (Fig. 5F). In addition, binding of the mutant E3 was not changed after λ ppase treatment (Fig. 5F), suggesting that the effect of phosphorylation on RNA binding was specific for Puf6p. We also found that E3 RNA bound less efficiently to Puf6 phosphorylated by purified CK2 than unphosphorylated Puf6 (Fig. 5G). These results suggest that phosphorylation of Puf6p by CK2 can reduce RNA binding and thus affect the repressing activity of Puf6p, similar to the role of phosphorylation in regulating Khd1p, another translational repressor for ASH1 mRNA (Paquin et al. 2007).
CK2 phosphorylation of Puf6p is required for ASH1 mRNA localization and translation
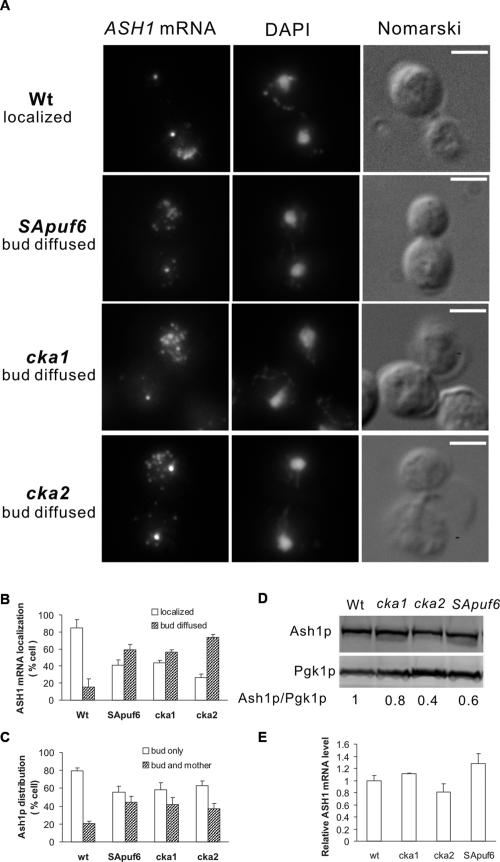
The abrogation of Puf6p translational repression by CK2 could release the translational control of ASH1 mRNA by Puf6p. Since translation is required for proper localization of ASH1 mRNA (Chartrand et al. 2002; Irie et al. 2002), a defect in translational control by Puf6p may affect ASH1 mRNA localization. To test this, a puf6 allele with the identified CK2 phosphorylation sites (Ser31, Ser34, and Ser35) mutated to Ala was integrated into the endogenous PUF6 locus. This strain was designated SApuf6. Cell growth and morphology appeared normal. The amounts and nuclear localization of the mutant SApuf6p-GFP were similar to wild-type Puf6p-GFP (data not shown). The mutant SApuf6p still interacted with Fun12p as it coimmunoprecipitated with Tap-tagged Fun12p (data not shown). We found that 60% ASH1 mRNA was diffusely distributed in the bud of this SApuf6 strain compared with 15% in the wild type (Fig. 6A,B). The effect of CK2-mediated Puf6p phosphorylation on ASH1 mRNA localization was further examined in cells with deletions of CKA1 or CKA2, two genes encoding catalytic subunits of CK2. Although both catalytic subunits were synthetic lethal (Padmanabha et al. 1990), loss of either catalytic subunit alone was tolerated and cells appeared phenotypically indistinguishable from wild type (Chen-Wu et al. 1988; Padmanabha et al. 1990). Actin structure appeared normal by phalloidin staining in those mutants (Supplemental Fig. 8). Strikingly, both cka1 and cka2 cells showed abnormalities in ASH1 mRNA localization (Fig. 6A). In the cka1 and cka2 strains, 56% and 72% of ASH1 mRNA were diffusely localized within the bud, respectively (Fig. 6B). The similarity of the effect in ASH1 mRNA localization observed in SApuf6, cka1, and cka2 supports the conclusion that CK2 phosphorylation of Puf6p was required for proper localization of ASH1 mRNA at the bud cortex. As expected, the Ash1p distribution was also affected in SApuf6, cka1, and cka2 mutants with an increase in cells, with Ash1p symmetrically partitioned in daughter and mother cells compared with wild type (Fig. 6C).
Figure 6.
CK2 phosphorylation is required for ASH1 mRNA localization and translation. (A) FISH of cells expressing wild-type PUF6, CKA1, and CKA2 (Wt), or mutant puf6 with Ser31,34,35 → Ala mutation (SApuf6), or with disruption of CKA1 (cka1) or CKA2 (cka2) using ASH1-specific probes (red). Representative cells show ASH1 mRNA localized at the bud tip (row 1) or diffuse in the bud (rows 2–4). Bar, 5 μm. (B) Percentages of cells showing localized and diffused ASH1 mRNA in wild-type (Wt), Ser31,34,35 → Ala puf6 mutant (SApuf6), cka1, and cka2 strains. Localization of ASH1 mRNA was determined by FISH and cells were scored according to the pattern shown in A. Error bars indicate SD for three experiments, each with n = 100 cells. (C) Percentages of cells showing asymmetric and symmetric distribution of Ash1p. Ash1p localization was determined by immunofluorescence. Error bars indicate SD for three experiments, each with n = 100 cells. (D) Extracts from cells expressing wild-type PUF6 (Wt), cka1, cka2, or mutant puf6 with Ser31,34,35 → Ala mutation (SApuf6) were analyzed by Western blotting. Nine repeats of the c-myc peptide sequence were inserted at the C terminus of Ash1p in each strain. Numbers indicate Ash1p levels relative to Pgk1p. (E) ASH1 mRNA levels quantified by real-time PCR and normalized to PGK1 mRNA.
To assess whether the phosphorylation of Puf6p might affect the translation of endogenous ASH1, we examined the protein levels of Ash1p in wild-type, cka1, cka2, and SApuf6 mutant strains. Ash1p levels decreased by 40% ± 10% (P < 0.05) in the SApuf6 mutant, 20% in cka1, and 60% in cka2 mutants compared with the wild-type strain (Fig. 6D). The mRNA levels of ASH1 increased by 20% in the SApuf6 mutant (Fig. 6E), suggesting that the decrease in Ash1p was mainly due to translational repression in SApuf6 mutant strain. The phosphorylation status of endogenous Puf6p in cka1, cka2, and SApuf6 mutants was examined by λ ppase treatment and phosphoprotein staining. We found the endogenous Puf6p still had some phosphorylation in cka1, cka2, and SApuf6 mutants (Supplemental Fig. 9), indicating there might be other potential kinases or sites involved in phosphorylation of Puf6p.
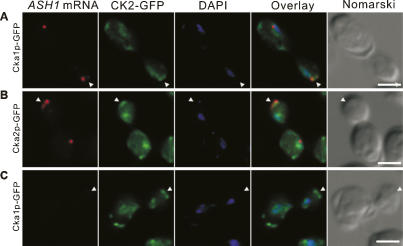
The release of Puf6p requires coordination with the transport of ASH1 mRNA. Therefore, it is reasonable to hypothesize that CK2 phosphorylation of Puf6p occurs at the bud cortex where ASH1 mRNA is localized. To test this, we performed fluorescent in situ hybridization (FISH) in cells expressing GFP-tagged Cka1p or Cka2p. CK2 has been reported as primarily nuclear in yeast (Poole et al. 2005). Our results showed that CK2 also accumulated at the bud cortex and colocalized with the ASH1 mRNA (Fig. 7A,B). The cortical CK2 was detectible in cells with small buds that had not started expression of ASH1 mRNA (Fig. 7C), suggesting that partition of the cortical CK2 occurred prior to ASH1 mRNA localization. Our findings suggest that Puf6p could be phosphorylated at the bud cortex and released from localized ASH1 mRNA, resulting in the localized synthesis of Ash1p.
Figure 7.
ASH1 mRNA colocalizes with CK2 catalytic subunits at the bud cortex. ASH1-FISH (red) on cells expressing Cka1p-GFP or Cka2p-GFP (green). Overlay showing ASH1 mRNA, GFP from CK2 catalytic subunits, and DAPI. Representative cells were in late anaphase with ASH1 expression (A,B) and cells in earlier cell cycle stage without ASH1 expression (C). Bar, 5 μm. Arrowheads indicate bud tip.
Discussion
Puf6p suppresses ASH1 mRNA translation via Fun12p/eIF5B
Puf6p belongs to a highly conserved family of RNA-binding proteins that are involved in regulating mRNA translation and stability (Spassov and Jurecic 2003). Binding of Puf6p to the 3′UTR of ASH1 mRNA could potentially affect events occurring at the 5′ of the transcript by interacting with a general translation factor(s) (Wickens et al. 2002). eIF5B (Fun12p in S. cerevisiae) is such a general translation factor assisting 60S ribosomal subunit joining in the translation initiation (Pestova et al. 2000). We showed that Puf6p interacts with eIF5B/Fun12p and this interaction may lead to the translational repression of ASH1 mRNA. First, the binding of Puf6p interferes with 80S ribosomal complex assembly on the ASH1 mRNA, which could result from the interference with the function of Fun12p in translation initiation. Second, Ash1p levels decrease in cells with FUN12 disruption and increase when FUN12 is overexpressed, suggesting that Fun12p is specifically required for ASH1 mRNA translation. Although identified as a general translational factor (Choi et al. 1998; Lee et al. 2002), eIF5B/Fun12p has been shown to regulate translation for many specific transcripts. Interaction of eIF5B and VASA is essential for translational activation of gurken mRNA (Carrera et al. 2000; Johnstone and Lasko 2004), one of the localized transcripts important for the embryonic development of Drosophila. Interaction of human eIF5B and HIV-1 matrix was thought to generate a pool of ribosome-free mRNA for virion packaging by repressing translation initiation (Wilson et al. 1999). In S. cerevisae, the poly(A)-binding protein (PABP)-mediated translational regulation of poly(A) mRNA is eIF5B/eIF5-dependent (Searfoss et al. 2001). In addition, the Fun12p–Puf6p association was abrogated when the N-terminal region of Fun12p was truncated (data not shown). Truncated Fun12p rescued the expression of Ash1p but not ASH1 mRNA localization (data not shown), supporting the fact that the Fun12p–Puf6p interaction was critical for proper regulation of ASH1 mRNA in vivo.
The C-terminal region and PUF domain (C536) can interact with Fun12p and bind RNA. However, this fragment of Puf6p is not sufficient to repress translation, which highlights the essential role of the N-terminal region of Puf6p. This is in contrast to Drosophila Pumilio (a PUF family member), for which expression of the RNA-binding domain is sufficient to rescue abdominal segmentation defects in pum mutant embryos (Wharton et al. 1998). Since the interaction between Puf6p and Fun12p is dependent on RNA, the finding that the N-terminal region did not pull down Fun12p from yeast extracts does not rule out the possibility that it could interact with Fun12p in vivo. However, it is possible that the N120 region of Puf6p could potentially interfere with the function of Fun12p even if it does not directly interact with Fun12p; e.g., through interaction with other factors that would otherwise interact with Fun12p. One candidate is eIF1A, which has been shown to interact directly with eIF5B and this interaction is required for translation initiation (Choi et al. 2000). If N120 competed with Fun12p to interact with eIF1A, it could also interdict the function of Fun12p in translation initiation. Therefore, we speculate that the N-terminal region of Puf6p might directly interact with Fun12p or compete with Fun12p for other factors required for the function of Fun12p in translation initiation.
Phosphorylation by CK2 releases translational control by Puf6p
The N-terminal region of Puf6p is critical for translational repression and contains the identified CK2 phosphorylation sites. CK2 is a ubiquitous Ser/Thr protein kinase that acts as a global regulator of cellular function (Litchfield 2003; Canton and Litchfield 2006). Mutation of the identified CK2 phosphorylation sites in Puf6p significantly increases the translational repression that was corroborated by the CK2 inhibitor DMAT. Consistent with the in vitro studies, Ash1p levels decrease in SApuf6 strain, supporting the conclusion that CK2 phosphorylation is necessary to release translational repression of ASH1 mRNA in cells. In comparison with the in vitro data, the difference in Ash1p levels in vivo was modest, which might be due to the nuclear localization signal (NLS) on Puf6p. The NLS is still functional in the SApuf6p because it shows a nuclear distribution similar to the wild-type Puf6p. This would ameliorate the repression of the mutant Puf6p in the cell as the NLS could force Puf6p to be segregated from the mRNA. Taken together, these results suggest that N120 plays a role in both translational repression and its release mediated by Puf6p, which is consistent with the finding that truncation of the N120 fragment only caused a marginal change in Ash1p levels in vivo (Supplemental Fig. 10). Other potential phosphorylation sites are present in Puf6p in addition to the identified CK2 phosphorylation sites Ser31, Ser34, and Ser35, but the SApuf6 mutant showed a pronounced effect on the expression of Ash1p and ASH1 mRNA localization, suggesting that these identified CK2 phosphorylation sites are critical in regulating Puf6p repression activity.
One possible mechanism for phosphorylation to release translational control is to reduce RNA binding of the repressor. Src phosphorylation reduced the binding of β-actin mRNA to its translational repressor ZBP1 (Huttelmaier et al. 2005). Recently, Paquin et al. (2007) showed that phosphorylation by CK1 reduced RNA binding to Khd1p and released its translational control on ASH1 mRNA. In this study, we found that the phosphorylation of Puf6p significantly reduced RNA binding, and these results indicate that CK2 phosphorylation might induce the dissociation of Puf6p from ASH1 mRNA or Fun12p and thereby release the translational repression. Interestingly, phosphorylation has also been implicated in the translational control by VASA, an eIF5B-interacting translational regulator in Drosophila. VASA activates the translation of several maternal mRNAs, and phosphorylation of VASA has been linked to a down-regulation of its activity in translation of gurken mRNA (Ghabrial and Schupbach 1999). It is possible that phosphorylation of VASA would also reduce its RNA binding.
CK2 regulates cell cycle and cell polarity in S. cerevisiae (Hanna et al. 1995; Rethinaswamy et al. 1998). ASH1 mRNA localization is cell cycle-regulated and actin cytoskeleton-dependent (Bobola et al. 1996; Long et al. 1997). The phosphorylation of Puf6p by CK2 could be a temporally and spatially regulated event that may be restricted to the bud cortex where ASH1 mRNA localizes. Compatible with this hypothesis, CK2 has been identified to associate with the Arp2/3 complex (Schaerer-Brodbeck and Riezman 2003) that colocalizes with filamentous actin in highly dynamic regions of the cell cortex (Moreau et al. 1996; Winter et al. 1997). Our study showed that the two catalytic subunits of CK2 were enriched at the bud tip, supporting the hypothesis that Puf6p is phosphorylated by CK2 when it reaches the bud cortex. Interestingly, the CK2 catalytic subunits were found localized to the rough ER in mammalian cells (Faust et al. 2001) and cortical ER is also localized at the bud tip in yeast cells (Preuss et al. 1991). Our findings that the catalytic subunits of CK2 localized to the bud cortex suggest that yeast CK2 catalytic subunits might also localize to the cortical ER in yeast, which has membrane-associated ribosomes.
Translation is required for anchoring ASH1 mRNA to the bud cortex
Translation is important for proper localization of ASH1 mRNA. Cells overexpressing Khd1p, a translational repressor, localize ASH1 mRNA less efficiently (Irie et al. 2002). Our studies find that loss of Fun12p reduced Ash1p levels and abrogated ASH1 mRNA localization. The delocalized ASH1 mRNA had a diffusion pattern different from the she mutants (Long et al. 1997) but is a phenocopy of the atg-mutant ASH1, which is found diffusely within the bud (Irie et al. 2002). This suggests that delocalization of ASH1 mRNA in the fun12 strain may result from a deficient translation but is not a secondary effect of a transport defect. The diffusion pattern of ASH1 in fun12 cells is similar to what has been found in the cells with gene disruptions of Bud6p/Aip3p or Bni1p/She5p (Beach et al. 1999). In those mutants, GFP-labeled ASH1 transcripts migrated to the bud but failed to be immobilized at the bud tip (Beach et al. 1999). Therefore, our results support the model that translation is required for ASH1 mRNA anchoring. We found that SApuf6 cells showed a similar but milder phenotype of both RNA delocalization and Ash1p levels compared with fun12 cells. This correlation between RNA delocalization and Ash1p levels confirms that ASH1 mRNA localization and translation is a well-coordinated process and suggests that the release of Puf6p for translation is important for the proper anchoring of ASH1 mRNA at the bud tips. It could also be possible that the polysomes or the nascent chains of Ash1p immobilize ASH1 mRNA at the bud cortex. Recently, it has been suggested that translation of ASH1 occurs via specific ribosomes (Komili et al. 2007) and, if so, may help to explain the specific action of Fun12p for its translation.
In conclusion, we identified a possible mechanism of the translational repression by Puf6p and propose that the repression may be released by CK2 phosphorylation. Our work advances the understanding of the relationship and coordination between RNA localization and translation. Premature translation due to the lack of a translational repressor can result in ASH1 mRNA delocalization, which is probably caused by interference with the transport machinery. Conversely, derepression is also required for stringent ASH1 mRNA localization, which might reflect a unique role for translation in anchoring ASH1 mRNA at the bud tip. Future work will detail the temporal and structural events of the protein–protein interactions and protein–RNA interactions that mediate this highly complex regulatory process.
Materials and methods
Yeast strains and growth media
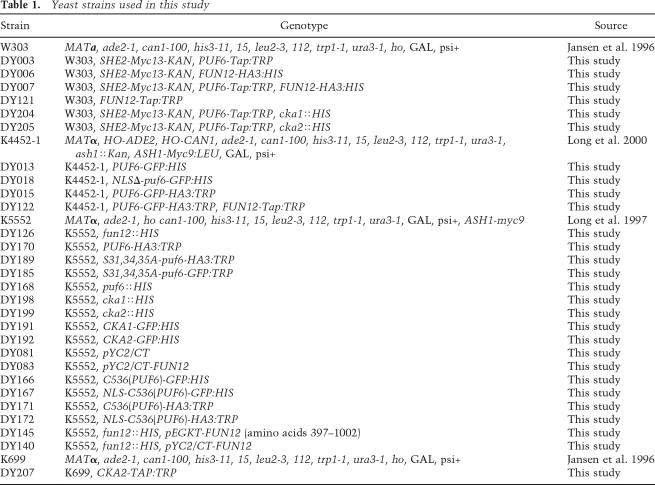
Yeast cells were grown either in rich media or in the synthetic media lacking the nutrients indicated. Yeast strains used are listed in Table 1. Transformation was performed as described (Gietz and Woods 2002). FUN12, PUF6, CKA1, and CKA2 genes were disrupted using an HIS disruption cassette amplified by PCR from plasmid pFA6a-His3MX6 (Wach et al. 1997). Strains expressing Fun12-HA, Puf6-HA, Cka1-GFP, Cka2-GFP, or Fun12-Tap were obtained through insertion of an HA-HIS cassette amplified from pFA6a-3HA-His, a GFP-HIS cassette amplified from pFA6a-GFP(S65T)-His (Longtine et al. 1998), or a Tap-TRP cassette amplified from pBS1479 (Rigaut et al. 1999). The strain expressing Puf6-GFP-HA was obtained by two sequential insertions of cassettes amplified from pFA6a-GFP(S65T)-His and pFA6a-3HA-TRP (Longtine et al. 1998). The strain expressing Puf6S31,34,35A was obtained through insertion of a Puf6S31,34,35A-3HA-TRP cassette in the pufΔ strain. This cassette was obtained by conjugating a 3xHA-TRP cassette from pFA6a-3HA-TRP (Longtine et al. 1998) with the Puf6S31,34,35A construct derived from plasmid pET30a-Puf6S31,34,35A through PCR. The HA tag was fused in-frame to the C terminus of the Puf6S31,34,35A construct. The transformed strains were selected in appropriate synthetic medium plus dextrose (Difco). Insertion of each cassette was verified by genomic PCR.
Table 1.
Yeast strains used in this study
Plasmid construction
Plasmids used are summarized in Supplemental Table 1. The coding region of renilla luciferase was amplified by PCR from pGL3 (Promega) with one primer designed with the SP6 promoter (ATTTAGGTGACACTATAGAATACAA) sequence at the 5′ end and the other with 30 dTs at the 5′ end. The PCR product was cloned into the pPCR4-TOPO vector (Invitrogen), generating pDP050. The E3 element of ASH1 (121 nt of the 3′UTR and 15 nt of the coding sequence) was amplified by PCR from yeast genomic DNA with primers designed to generate XhoI and SmaI cloning sites. The E3 was subcloned into plasmid pDP050 with XhoI/SmaI. The two UUGU elements for Puf6p binding (Gu et al. 2004) were deleted in E3 with a PCR-directed mutagenesis method (Sambrook et al. 2001), and wild-type E3 in pR-luc-E3 was replaced by mutant E3 using SmaI/XhoI. pMiniORF-E3 and pMiniORF-E3-M were obtained through SfuI/XhoI digestion of pR-luc-E3 and pR-luc-E3-M followed by ligation of the backbone. The resulting plasmid has a coding region for the first three amino acids of renilla luciferase followed by the E3 of ASH1. The coding region of PUF6 was amplified by PCR from yeast genomic DNA with primers designed to generate BglII and SalI sites at the two ends and subcloned to pET30a (Qiagen) with BamHI/XhoI. The coding sequence for amino acids 1–120 and 120–656 of Puf6 were amplified by PCR and subcloned into the pET30a expression vector with EcoRV/HindIII. The codons for Ser31, Ser34, and Ser35 of Puf6 were mutated to Ala (gCA, gCa, gCT) by PCR-directed mutagenesis in pET30a-puf6S31,34,35A Puf6. All of the constructs were confirmed by DNA sequencing.
Expression and purification of recombinant His6-Puf6
E. coli strain BL21(DE3) (Novagen) was transformed with plasmids pET30a-Puf6, pET30a-N120, pET30a-C536, and pET30a-Puf6S31,34,35A and cultured in Luria-Bertani broth with 100 mg/L ampicillin at 37°C. Cultures were grown to an optical density of 0.8, measured at 600 nm (OD600), and induced with 0.5 mM isopropyl thio-β-D-galactoside (IPTG) for 4 h at 37°C. Cells were harvested and lysed by sonication in lysis buffer (10 mM Na-KHPO4 at pH 7.4, 1 M NaCl, 10 mM β-mercaptoethanol) plus one tablet of complete EDTA-free protease inhibitor cocktail (Roche). The soluble fraction after centrifugation was incubated with HIS-select HF Nickel affinity gel (Sigma) and eluted with 200 mM imidazole in lysis buffer. The purified proteins were then dialyzed against buffer (10 mM Na-KHPO4 at pH 7.4, 137 mM NaCl) and concentrated by ultrafree concentrators (Millipore).
Gel mobility shift assay
A 32P-labeled RNA probe was generated by SP6 polymerase-directed in vitro transcription from the SmaI linearized plasmid pMiniORF-E3. The RNA transcribed from the construct was purified as described (Gu et al. 2004). The RNA probe (10,000 counts per minute [cpm]) was incubated with recombinant His6-Puf6 for 30 min in a 20-μL binding solution (20 mM Tris at pH 7.4, 50 mM KCl, 3 mM MgCl2, 2 mM dithiothreitol, 5% Glycerol) at room temperature. Nonspecific RNA–protein interaction was minimized by incubation with 5 mg/mL heparin for 10 min. RNA–protein complexes were separated in a 4% native gel and analyzed by autoradiography.
In vitro translation and luciferase assays
Yeast extracts were prepared as described (Iizuka et al. 1994) with the following modifications. Yeast cells were harvested in late log phase, washed twice in buffer A [20 mM HEPES-KOH at pH 7.4, 100 mM KOAc, 2 mM Mg(Oac)2, 2 mM dithiothreitol], resuspended in buffer A supplemented with protease inhibitor cocktail tablets (Roche) and snap-frozen in liquid N2. The frozen cells were ground in a mixer mill MM 301 (Retsch). The grinding jars were shaken vigorously in a horizontal motion at a frequency of 30 per second for seven 3-min cycles of breakage with 3-min chilling in liquid N2 in between cycles. The ground powder was thawed at 4°C and a 30,000g supernatant was prepared. Endogenous amino acid pools and low-molecular-weight inhibitors were removed by PD-10 column (Amersham). Capped and polyadenylated transcripts for the in vitro translation assay were synthesized with Mmessage Mmachine SP6 kit (Ambion) from HindIII linearized plasmid pR-luc-E3 and pR-luc. RNA was purified by DNase I digestion, phenol/chloroform extraction, and ethanol precipitation. The integrity of the transcribed RNA was verified by electrophoresis in 1% agarose gels containing 1 μg/mL ethidium bromide. In vitro translation assays were performed as described (Iizuka et al. 1994) with the following modification. RNA reporters were heated for 10 min at 65°C and cooled for 10 min at room temperature. RNA reporters of 100 ng were preincubated with recombinant His6-Puf6 as indicated in the figure legends for 30 min at room temperature and then incubated with yeast extract in the translation buffer for 40 min at 18°C as described (Iizuka et al. 1994). For autoradiography, minus Met amino acid (Promega) was used with [35S]-Met (15 mCi/mL; Amersham). For the luciferase assay, the complete amino acid mixture (Promega) was supplied to the reaction. After incubation with extracts, the total mixture was used for the luciferase assay using a renilla substrate (Promega). All luciferease assays were performed three times and the average results are shown.
Sucrose gradient analysis
Cold and 32P-labeled transcripts were generated by SP6 polymerase-directed in vitro transcription from the SmaI linearized plasmid pMiniORF-E3. 32P-labeled RNA (16 ng) was preincubated with 1 μg of full-length His6-Puf6 or the indicated components in the figure legends for 30 min at room temperature. The in vitro translation reaction was then assembled as described above and loaded onto 12 mL of 7%–47% linear sucrose gradients and subjected to centrifugation at 40,000 rpm for 2.5 h in an SW41 rotor (Beckman). Fractions (0.5 mL each) were collected from top to bottom and the A254 profile of the fractions was determined with a PharmaciaLKB Uvicord SII detector (Pharmacia) equipped with an autodensi-flow (Labconco). The radioactivity of each fraction was measured by a scintillation counter. The experiments under the condition of Puf6p incubation were repeated in 5%–25% sucrose gradient in the SW51Ti and SW41 rotors, respectively.
Co-IP, pull-down, and Western blot analysis
Cells were lysed in IP buffer (50 mM Tris at pH 7.4, 10 mM MgCl2, 100 mM NaCl, 1 mM EDTA, 10% Glycerol supplied with RNaseOUT and Protease inhibitors). Cell extracts containing 1 mg of total protein were used for co-IP and pull-down assays. For co-IPs, the yeast extract was incubated with IgG Sepharose 6 Fast Flow beads (Amersham) equilibrated in IP buffer in a volume of 200 μL for 2 h at 4°C. Beads were washed with 500 μL of buffer B (10 mM Tris at pH 8.0, 150 mM NaCl, 0.1% NP40), resuspended in SDS sample buffer, boiled, and analyzed by Western blotting. For pull-down assays, yeast extract was incubated with 946 pmol of each recombinant protein for 1 h at 4°C and pulled down by HIS-select HF Nickel affinity gel (Sigma). Beads were washed with 500 μL of buffer B (10 mM Tris at pH 8.0, 150 mM NaCl, 0.1% NP40), resuspended in SDS sample buffer, boiled, and analyzed by Western blotting. Western blot analyses were performed according to standard procedure. IR dye-conjugated secondary antibodies (Rockland) were used at 1:10,000 dilution and detected with LI-COR Odyssey Infrared Imaging (Li-cor, Inc.). Anti-HA (Roche, 1583816), anti-c-Myc (Molecular Probes, A-21280), anti-Pgk1 (Molecular Probes, A-6457), and anti-Tap (Open Biosystems, CAB1001) antibodies were used for detection of proteins expressed in yeast cells. Recombinant Puf6 purified from E. coli was detected by an anti-His antibody (Invitrogen, R930-25). Anti-Rpl3p and anti-Nop1 antibodies were kind gifts from Dr. Jonathan Warner and Dr. Tom Meier (Albert Einstein College of Medicine), respectively.
Real-time PCR
Yeast RNA was prepared using an RNeasy kit according to the manufacturer’s instructions (Qiagen). Real-time PCR was performed with ASH1-, ACT1-, and PGK1-specific primers using FastStart SYBR Green Master Mix and the LightCycler instrument (Roche) as instructed by the manufacturers.
Phosphoprotein staining, λ ppase, and RNA-binding assays
Cell extract containing Puf6p-Tap was incubated with IgG-coated Sepharose 6 Fast Flow beads (Amersham Biosciences) equilibrated in buffer B (10 mM Tris at pH 8.0, 150 mM NaCl, 0.1% NP40) for 2 h at 4°C. The extract was purified by anti-HA antibody-coupled protein G beads. Beads were washed, resuspended in SDS sample buffer, boiled, and separated on 4%–12% SDS-PAGE gels (Invitrogen). The gel was stained with Pro-Q Diamond Phosphoprotein Gel Stain and SYPRO Ruby (Molecular Probes) as instructed by the manufacturer. After Ruby staining, the gel was analyzed by standard Western blotting. PeppermintStick Phosphoprotein Molecular Weight Standard is from Molecular Probes. For the λ ppase assay, proteins affinity-purified on IgG-coupled beads were treated with 3200 U of λ ppase (New England Biolabs) for 1 h at 30°C and the eluates were analyzed by Western blotting as described above. For RNA-binding assays, the 32P-labeled RNA probe was generated by SP6 polymerase-directed in vitro transcription from the SmaI linearized plasmids pMiniORF-E3 and pMiniORF-E3-M, as described above. Labeled RNA was incubated with affinity-purified proteins from yeast extract bound to IgG-coupled beads in RNA-binding solution (20 mM Tris at pH 7.4, 50 mM KCl, 3 mM MgCl2, 2 mM dithiothreitol, 5% Glycerol) for 30 min at room temperature. Protein–RNA complexes were eluted with preheated SDS sample buffer and radioactivity from the liquid phase was measured using a scintillation counter.
In vitro phosphorylation assay
Recombinant Puf6 was incubated with yeast extract as described in the in vitro translation assay except for the following changes. RNA reporter and the amino acid mix were omitted from the reaction, and the ATP concentration was lowered to 0.1 mM. Where indicated, [γ-32P]-ATP or [γ-32P]-GTP (4 μCi per sample; Amersham) was used. Reactions were terminated by addition of 500 μL of binding buffer (10 mM Na-KHPO4 at pH 7.4, 1 M NaCl, 10 mM β-mercaptoethanol). Recombinant Puf6 used in the assay was precipitated with HIS-select HF Nickel affinity gel (Sigma) and analyzed by autoradiograph and Western blotting. Two sources of CK2 were used as indicated in the legends. Pure CK2 from sea star Pisaster ochraceus (Upstate Biotechnology) was incubated with recombinant protein for 1 h at 37°C as described (Meier 1996). Pure CK2 from rat liver (Promega) was incubated with recombinant protein in buffer (25 mM Tris at pH 7.4, 200 mM NaCl, 10 mM MgCl2, 0.1 mM ATP) for 30 min at 37°C. Where indicated, 25 μM DMAT (EMD) was added to the yeast extract 30 min before the phosphorylation assay or supplied to the pure CK2 reaction. For mass spectrometry, 46 μg of full-length His6-Puf6 were incubated with yeast extract for 2.5 h at 18°C. His6-Puf6 was pulled down with HIS-select HF Nickel affinity gel (Sigma) and eluted with preheated SDS sample buffer. Eluted proteins and pure His6-Puf6 were separated on 4%–12% SDS-PAGE gels and stained with Coomassie blue. Gel bands containing phosphorylated and unphosphorylated His6-Puf6 were excised and subjected to MALDI-TOF at Rockefeller University.
FISH and immunofluorescence
Yeast cells were grown to early or mid-log phase and processed for in situ hybridization and immunofluorescence staining as described in Chartrand et al. (2000). Briefly, yeast spheroplasts were hybridized with a pool of Cy3-conjugated ASH1 DNA oligonucleotide probes (Long et al. 1997). Cells were imaged on a BX61 upright wide-field, epifluorescence microscope. An optically sectioned three-dimensional image stack of each field was acquired with a slice spacing of 200 nm in the Z-axis with a CoolSnap HQ CCD camera (Photometrics) operated by IPLab software (BD Biosciences). Each image stack was combined using a maximum intensity projection algorithm. Image stacks from the GFP signal were deconvolved using a classic maximum likelihood estimation algorithm in Huygens Professional (Scientific Volume Imaging) to increase the sensitivity of detection. The method of quantitative measurement on the localization of ASH1 mRNA and Ash1p has been described previously (Chartrand et al. 2002). ASH1 mRNA was classified as localized when it was predominantly in the bud tip showing a crescent localization pattern. ASH1 mRNA was classified as delocalized when it was diffusely distributed in the bud or in both mother and daughter cells.
Statistical analysis
The results are shown as means ± S.D. Statistical analysis was performed by the Student’s t-test using SigmaPlot 9.0. Significance was accepted at P values of <0.05.
Acknowledgments
We thank Shubhendu Ghosh (Allan Jacobson Laboratory, University of Massachussetts at Worcester) for the help in the development of in vitro translation assay with yeast extract. We thank the input from the yeast group of the laboratory, Daniel Zenklusen, Tatjana Trcek, and Erin Powrie. We also thank Shailesh Shenoy for help in microscopy and image processing. This work was supported by NIH GM57071 to R.H.S.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1611308.
References
- Beach D.L., Salmon E.D., Bloom K. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- Bobola N., Jansen R.P., Shin T.H., Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- Bohl F., Kruse C., Frank A., Ferring D., Jansen R.P. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton D.A., Litchfield D.W. The shape of things to come: An emerging role for protein kinase CK2 in the regulation of cell morphology and the cytoskeleton. Cell. Signal. 2006;18:267–275. doi: 10.1016/j.cellsig.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Carrera P., Johnstone O., Nakamura A., Casanova J., Jackle H., Lasko P. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol. Cell. 2000;5:181–187. doi: 10.1016/s1097-2765(00)80414-1. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Meng X.H., Singer R.H., Long R.M. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr. Biol. 1999;9:333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Bertrand E., Singer R.H., Long R.M. Sensitive and high-resolution detection of RNA in situ. Methods Enzymol. 2000;318:493–506. doi: 10.1016/s0076-6879(00)18072-3. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Singer R.H., Long R.M. RNP localization and transport in yeast. Annu. Rev. Cell Dev. Biol. 2001;17:297–310. doi: 10.1146/annurev.cellbio.17.1.297. [DOI] [PubMed] [Google Scholar]
- Chartrand P., Meng X.H., Huttelmaier S., Donato D., Singer R.H. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol. Cell. 2002;10:1319–1330. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- Chen-Wu J.L., Padmanabha R., Glover C.V. Isolation, sequencing, and disruption of the CKA1 gene encoding the α subunit of yeast casein kinase II. Mol. Cell. Biol. 1988;8:4981–4990. doi: 10.1128/mcb.8.11.4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi A., Huttenhower C., Geer L.Y., Coon J.J., Syka J.E., Bai D.L., Shabanowitz J., Burke D.J., Troyanskaya O.G., Hunt D.F. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S.K., Lee J.H., Zoll W.L., Merrick W.C., Dever T.E. Promotion of Met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science. 1998;280:1757–1760. doi: 10.1126/science.280.5370.1757. [DOI] [PubMed] [Google Scholar]
- Choi S.K., Olsen D.S., Roll-Mecak A., Martung A., Remo K.L., Burley S.K., Hinnebusch A.G., Dever T.E. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol. Cell. Biol. 2000;20:7183–7191. doi: 10.1128/mcb.20.19.7183-7191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S.R., Kemmeren P., Zhao X.C., Greenblatt J.F., Spencer F., Holstege F.C., Weissman J.S., Krogan N.J. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell. Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- Corral-Debrinski M. mRNA specific subcellular localization represents a crucial step for fine-tuning of gene expression in mammalian cells. Biochim. Biophys. Acta. 2007;1773:473–475. doi: 10.1016/j.bbamcr.2006.06.008. [DOI] [PubMed] [Google Scholar]
- Darzacq X., Powrie E., Gu W., Singer R.H., Zenklusen D. RNA asymmetric distribution and daughter/mother differentiation in yeast. Curr. Opin. Microbiol. 2003;6:614–620. doi: 10.1016/j.mib.2003.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever T.E. Gene-specific regulation by general translation factors. Cell. 2002;108:545–556. doi: 10.1016/s0092-8674(02)00642-6. [DOI] [PubMed] [Google Scholar]
- Du T.G., Schmid M., Jansen R.P. Why cells move messages: The biological functions of mRNA localization. Semin. Cell Dev. Biol. 2007;18:171–177. doi: 10.1016/j.semcdb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Faust M., Jung M., Gunther J., Zimmermann R., Montenarh M. Localization of individual subunits of protein kinase CK2 to the endoplasmic reticulum and to the Golgi apparatus. Mol. Cell. Biochem. 2001;227:73–80. [PubMed] [Google Scholar]
- Gatica M., Hinrichs M.V., Jedlicki A., Allende C.C., Allende J.E. Effect of metal ions on the activity of casein kinase II from Xenopus laevis. FEBS Lett. 1993;315:173–177. doi: 10.1016/0014-5793(93)81157-u. [DOI] [PubMed] [Google Scholar]
- Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Ghabrial A., Schupbach T. Activation of a meiotic checkpoint regulates translation of Gurken during Drosophila oogenesis. Nat. Cell Biol. 1999;1:354–357. doi: 10.1038/14046. [DOI] [PubMed] [Google Scholar]
- Gietz R.D., Woods R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Gonzalez I., Buonomo S.B., Nasmyth K., von Ahsen U. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr. Biol. 1999;9:337–340. doi: 10.1016/s0960-9822(99)80145-6. [DOI] [PubMed] [Google Scholar]
- Gu W., Deng Y., Zenklusen D., Singer R.H. A new yeast PUF family protein, Puf6p, represses ASH1 mRNA translation and is required for its localization. Genes & Dev. 2004;18:1452–1465. doi: 10.1101/gad.1189004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna D.E., Rethinaswamy A., Glover C.V. Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:25905–25914. doi: 10.1074/jbc.270.43.25905. [DOI] [PubMed] [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore L., Adams S.L., Millar A., Taylor P., Bennett K., Boutilier K., et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Huttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G.J., Condeelis J., Singer R.H. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- Iizuka N., Najita L., Franzusoff A., Sarnow P. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:7322–7330. doi: 10.1128/mcb.14.11.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie K., Tadauchi T., Takizawa P.A., Vale R.D., Matsumoto K., Herskowitz I. The Khd1 protein, which has three KH RNA-binding motifs, is required for proper localization of ASH1 mRNA in yeast. EMBO J. 2002;21:1158–1167. doi: 10.1093/emboj/21.5.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R.P., Dowzer C., Michaelis C., Galova M., Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- Johnstone O., Lasko P. Interaction with eIF5B is essential for Vasa function during development. Development. 2004;131:4167–4178. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- Komili S., Farny N.G., Roth F.P., Silver P.A. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:450–451. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon S., Schnapp B.J. RNA localization: SHEdding light on the RNA-motor linkage. Curr. Biol. 2001;11:R166–R168. doi: 10.1016/S0960-9822(01)00084-7. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Pestova T.V., Shin B.S., Cao C., Choi S.K., Dever T.E. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc. Natl. Acad. Sci. 2002;99:16689–16694. doi: 10.1073/pnas.262569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litchfield D.W. Protein kinase CK2: Structure, regulation and role in cellular decisions of life and death. Biochem. J. 2003;369:1–15. doi: 10.1042/BJ20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R.M., Singer R.H., Meng X., Gonzalez I., Nasmyth K., Jansen R.P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- Long R.M., Gu W., Lorimer E., Singer R.H., Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p–She3p complex to ASH1 mRNA. EMBO J. 2000;19:6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Meggio F., Pinna L.A. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- Meier U.T. Comparison of the rat nucleolar protein nopp140 with its yeast homolog SRP40. Differential phosphorylation in vertebrates and yeast. J. Biol. Chem. 1996;271:19376–19384. [PubMed] [Google Scholar]
- Moreau V., Madania A., Martin R.P., Winson B. The Saccharomyces cerevisiae actin-related protein Arp2 is involved in the actin cytoskeleton. J. Cell Biol. 1996;134:117–132. doi: 10.1083/jcb.134.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanaswamy R., Niu W., Scouras A.D., Hart G.T., Davies J., Ellington A.D., Iyer V.R., Marcotte E.M. Systematic profiling of cellular phenotypes with spotted cell microarrays reveals mating-pheromone response genes. Genome Biol. 2006;7:R6. doi: 10.1186/gb-2006-7-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabha R., Chen-Wu J.L., Hanna D.E., Glover C.V. Isolation, sequencing, and disruption of the yeast CKA2 gene: Casein kinase II is essential for viability in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990;10:4089–4099. doi: 10.1128/mcb.10.8.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano M.A., Meggio F., Ruzzene M., Andrzejewska M., Kazimierczuk Z., Pinna L.A. 2-Dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole: A novel powerful and selective inhibitor of protein kinase CK2. Biochem. Biophys. Res. Commun. 2004;321:1040–1044. doi: 10.1016/j.bbrc.2004.07.067. [DOI] [PubMed] [Google Scholar]
- Paquin N., Menade M., Poirier G., Donato D., Drouet E., Chartrand P. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol. Cell. 2007;26:795–809. doi: 10.1016/j.molcel.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Pestova T.V., Lomakin I.B., Lee J.H., Choi S.K., Dever T.E., Hellen C.U.T. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- Poole A., Poore T., Bandhakavi S., McCann R.O., Hanna D.E., Glover C.V. A global view of CK2 function and regulation. Mol. Cell. Biochem. 2005;274:163–170. doi: 10.1007/s11010-005-2945-z. [DOI] [PubMed] [Google Scholar]
- Preuss D., Mulholland J., Kaiser C.A., Orlean P., Albright C., Rose M.D., Robbins P.W., Botstein D. Structure of the yeast endoplasmic reticulum: Localization of ER proteins using immunofluorescence and immunoelectron microscopy. Yeast. 1991;7:891–911. doi: 10.1002/yea.320070902. [DOI] [PubMed] [Google Scholar]
- Rethinaswamy A., Birnbaum M.J., Glover C.V. Temperature-sensitive mutations of the CKA1 gene reveal a role for casein kinase II in maintenance of cell polarity in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:5869–5877. doi: 10.1074/jbc.273.10.5869. [DOI] [PubMed] [Google Scholar]
- Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Roach P.J. Multisite and hierarchal protein phosphorylation. J. Biol. Chem. 1991;266:14139–14142. [PubMed] [Google Scholar]
- Sambrook J., Fritsch E., Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Schaerer-Brodbeck C., Riezman H. Genetic and biochemical interactions between the Arp2/3 complex, Cmd1p, casein kinase II, and Tub4p in yeast. FEM. Yeast Res. 2003;4:37–49. doi: 10.1016/S1567-1356(03)00110-7. [DOI] [PubMed] [Google Scholar]
- Searfoss A., Dever T.E., Wickner R. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: Relations of Pab1p, eukaryotic translation initiation factor 5b (Fun12p), and Ski2p-Slh1p. Mol. Cell. Biol. 2001;21:4900–4908. doi: 10.1128/MCB.21.15.4900-4908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov D.S., Jurecic R. The PUF family of RNA-binding proteins: Does evolutionarily conserved structure equal conserved function? IUBMB Life. 2003;55:359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: The intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Takizawa P.A., Vale R.D. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc. Natl. Acad. Sci. 2000;97:5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa P.A., Sil A., Swedlow J.R., Herskowitz I., Vale R.D. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- Takizawa P.A., DeRisi J.L., Wilhelm J.E., Vale R.D. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- Trachsel H., Erni B., Schreier M.H., Staehelin T. Initiation of mammalian protein synthesis. II. The assembly of the initiation complex with purified initiation factors. J. Mol. Biol. 1977;116:755–767. doi: 10.1016/0022-2836(77)90269-8. [DOI] [PubMed] [Google Scholar]
- Wach A., Brachat A., Alberti-Segui C., Rebischung C., Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wharton R.P., Sonoda J., Lee T., Patterson M., Murata Y. The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell. 1998;1:863–872. doi: 10.1016/s1097-2765(00)80085-4. [DOI] [PubMed] [Google Scholar]
- Wickens M., Bernstein D.S., Kimble J., Parker R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Wilson S.A., Sieiro-Vazquez C., Edwards N.J., Iourin O., Byles E.D., Kotsopoulou E., Adamson C.S., Kingsman S.M., Kingsman A.J., Martin-Rendon E. Cloning and characterization of hIF2, a human homologue of bacterial translation initiation factor 2, and its interaction with HIV-1 matrix. Biochem. J. 1999;342:97–103. [PMC free article] [PubMed] [Google Scholar]
- Winter D., Podtelejnikov A.V., Mann M., Li R. The complex containing actin-related proteins Arp2 and Arp3 is required for the motility and integrity of yeast actin patches. Curr. Biol. 1997;7:519–529. doi: 10.1016/s0960-9822(06)00223-5. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Williamson J.R., Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]