Abstract
During transcription termination by RNA polymerase II on protein-coding genes, the nuclear 5′ exonuclease Rat1/Xrn2 degrades the nascent transcript downstream from the polyadenylation site and “torpedoes” the polymerase. We report that the activity of Rat1 is also required for efficient termination by RNA polymerase I (Pol I) on the rDNA. In strains lacking catalytically active Rat1 or its cofactor Rai1, Pol I reads through the major, “Reb1-dependent” terminator (T1) but stops downstream at the “fail-safe” terminator (T2) and replication fork barrier (RFB). The absence of both Rat1 and the RFB-binding protein Fob1 increased Pol I read-through of T2 and the RFB. We propose that cotranscriptional cleavage of the pre-rRNA by the endonuclease Rnt1 generates a loading site for the Rat1/Rai1 complex, which then degrades the nascent transcript. When Rat1 catches Pol I, which is predicted to be paused at T1, transcription is terminated.
[Keywords: Ribosome synthesis, rRNA synthesis, RNA polymerase I, exonuclease, transcription termination]
Under good growth conditions each Saccharomyces cerevisiae cell will produce ∼2000 mature ribosomes per minute, requiring the synthesis of some 14 Mb of pre-rRNA and 160,000 ribosomal proteins. The rDNA genes in S. cerevisiae form an array of 150–200 repeats located on chromosome XII, around one-half of which are actively transcribed. The high rate of ribosome synthesis means that these rDNA repeats must each give rise to >20 pre-rRNA transcripts per minute. Each rDNA repeat contains the 35S gene, transcribed by RNA polymerase I (Pol I), and the intergenic spacer regions (IGS1 and IGS2). The 5S gene, which is independently transcribed by RNA Polymerase III (Pol III), separates IGS1 from IGS2 (see Fig. 1A). IGS1 contains the replication fork barrier (RFB) region extending from +282 to +418 from the 25S sequence (see Fig. 2A). Fob1 protein binds to the RFB. These establish a DNA–protein structure that blocks DNA replication forks from moving in the opposite direction to the Pol I transcription machinery, preventing their collision (Takeuchi et al. 2003; Huang et al. 2006).
Figure 1.
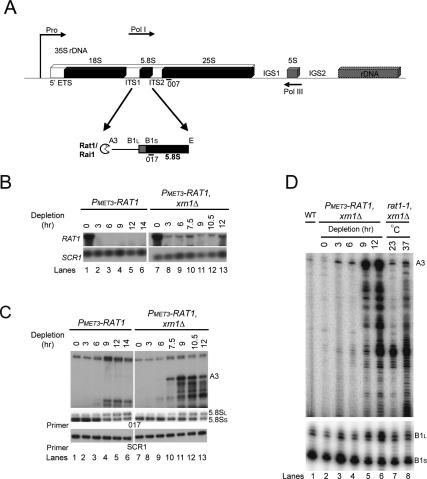
Rat1 depletion leads to defects in 5.8S rRNA processing. (A) Schematic of an rDNA repeat in S. cerevisiae showing the 5′ processing of 5.8S rRNA. The rDNA repeat includes the 35S pre-RNA gene, which is transcribed by Pol I and processed to mature 18S, 5.8S, and 25S rRNAs. (Pro) Pol I promoter. The 35S genes are separated by IGS1 and IGS2 and the 5S rRNA gene, which is transcribed by Pol III in the opposite direction (indicated by arrows). The pre-rRNA is cleaved at site A3 by the endonuclease RNase MRP (Lygerou et al. 1996) and then processed by Rat1/Rai1 to site B1S, the 5′ end of the 5.8SS rRNA. A distinct minor pathway, which is independent of Rat1 activity, generates the 5′ end of the longer 5.8SL rRNA at site B1L. Processing in ITS2 generates the 3′ end of both 5.8S species at site E. (B) Rat1 mRNA depletion. Total RNA was extracted from PMET3-RAT1 and PMET3-RAT1, xrn1Δ strains grown at 30°C in the absence of methionine (0-h samples) or in the presence of 5 mM methionine for the times indicated and resolved on a 1.2% agarose/glyoxal gel. RAT1 mRNA was detected with a random primed probe (see Materials and Methods; Supplemental Table S2). (Bottom panel) As a control, the abundant cytoplasmic SCR1 RNA was detected with oligonucleotide SCR1 (see Supplemental Table S2). (C) Northern analysis of 5.8S rRNA maturation. RNAs extracted as above were resolved on an 8% polyacrylamide/urea gel. Mature 5.8SS and 5.8SL rRNAs and extended pre-5.8S species (A3) were detected with probe 017 (see Supplemental Table S2). The top panel (A3) shows a longer exposure than the middle panel (5.8SS and 5.8SL). (Bottom panel) As a control, the abundant cytoplasmic SCR1 RNA was detected with oligonucleotide SCR1. (D) Primer extension analysis of 5.8S rRNA maturation. RNA was extracted from wild-type and PMET3-RAT1, xrn1Δ strains as above and from rat1-1, xrn1Δ strain grown at 25°C and following transfer for 2 h to 37°C. Primer extension products using oligonucleotide 017 were resolved on a 6% polyacrylamide/urea gel. The primer extension stops at B1L and B1S represent the 5′ ends of the mature 5.8SS and 5.8SL rRNAs, respectively. The bottom panel shows a shorter exposure than the top panel.
Figure 2.
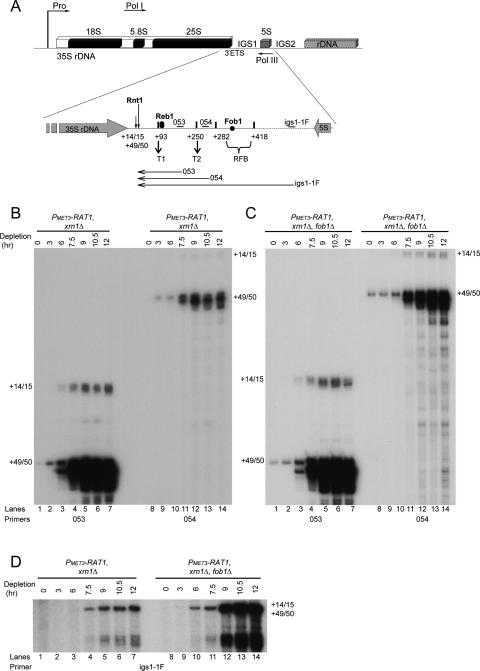
The 5′ ends of IGS1 transcripts map to the Rnt1 cleavage sites at +14/15 and +49/50 in the 3′-ETS. (A) Schematic of an rDNA repeat in S. cerevisiae (as in Fig. 1A). Rnt1 cleaves the 35S pre-rRNA in the 3′-ETS in IGS1 at +14/15 and +49/50 relative to the 3′ end of the 25S rRNA sequence. Pol I transcription predominantly terminates at the “Reb1-dependent” terminator (T1) located at approximately +93 nt. Reb1 binds a few nucleotides downstream from +93. A “fail-safe” terminator (T2) is located at approximately +250, followed by the RFB, which binds Fob1 and extends from +282 to +418. Oligonucleotides used (see also Supplemental Table S2) are indicated by small horizontal bars above the rDNA. Probe 053 is located between T1 and T2 at position +180, probe 054 is located between T2 and the RFB at +264, and probe igs1-1F is located between the RFB and the 5S rRNA gene at +832. Horizontal arrows below the rDNA indicate the RNA species detected by primer extensions. (B–D) RNAs extracted from strains PMET3-RAT1, xrn1Δ and PMET3-RAT1, xrn1Δ, fob1Δ as described in Figure 1B were processed for primer extension using oligonucleotides 053, 054, or igs1-1F. Products were resolved on 6% polyacrylamide/urea gels. Primer extension stops at +14/15 and +49/50 are labeled.
Analyses of Pol I transcription performed in vitro and in vivo in yeast showed that ∼90% of transcripts terminate at the “Reb1-dependent” or “primary” terminator (herein referred to as T1) (see Fig. 2A). This is located at a position ∼93 nucleotides (nt) downstream from the 3′ end of the 25S sequence in the 3′-external transcribed spacer (3′-ETS) (Lang and Reeder 1993; Reeder and Lang 1994, 1997; Prescott et al. 2004). Approximately 10% of Pol I molecules read through T1 and stop at the downstream “fail-safe” terminator (herein referred to as T2) (see Fig. 2A). T2 was initially reported to lie at +210 from the 25S sequence (van der Sande et al. 1989), but subsequent analyses mapped it close to around position +250 (Reeder et al. 1999).
At T1, termination occurs 12–20 nt upstream of the binding site for the Reb1 protein, within a T-rich element that encodes the last 10–12 nt of the terminated transcript (Reeder and Lang 1997). In the mouse, the Reb1 homolog TTF1 cooperates with a transcript release factor called PTRF in conjunction with the T-rich DNA sequence to induce transcription termination and dissociate Pol I and the transcript from the template DNA (Jansa and Grummt 1999). No equivalent transcript release factor has been identified in yeast, but in vitro and in vivo data suggested the existence of such factors (Reeder and Lang 1997; Tschochne and Milkereit 1997; Jansa and Grummt 1999). At T2 in yeast, termination also occurs at or close to an extended, highly T-rich element (Reeder et al. 1999), but the specific protein factors involved have not yet been identified.
Deletion of the RPA12 gene, which encodes a small subunit of Pol I, leads to increased read-through of T1 (Prescott et al. 2004). Rpa12 is homologous with the small RNA polymerase II (Pol II) subunit Rpb9, which is similarly implicated in transcription termination (Awrey et al. 1997). Inactivation of the chromatin remodeling ATPases Chd1, Isw1, and Isw2 also caused defects in transcription termination by both Pol I and Pol II (Alen et al. 2002; Jones et al. 2007). These observations pointed to similarities in the termination of transcription by Pol I and Pol II. Efficient transcription termination on mRNA genes requires the catalytic activity of the highly processive nuclear 5′–3′ exonuclease Rat1 (Xrn2 in humans) (Kim et al. 2004; West et al. 2004; Luo et al. 2006; Kaneko et al. 2007). Cotranscriptional cleavage by the polyadenylation machinery at the 3′ end of the pre-mRNA allows Rat1 to attack the free 5′ end of the downstream cleavage product. When Rat1 catches up with Pol II the transcription terminates. Rat1 is thus said to “torpedo” the polymerase.
Xrn1, the second, mainly cytoplasmic, 5′–3′ exonuclease in yeast, can degrade many 5′-extended RNAs that would otherwise accumulate in the absence of Rat1, including 5′-extended pre-5.8S and 25S pre-rRNAs (Henry et al. 1994; Geerlings et al. 2000). Xrn1 is detected exclusively in the cytoplasm of wild-type cells, and artificial relocation of Xrn1 to the nucleus, by fusion with a strong nuclear localization signal, suppresses the lethality of the rat1-1 mutation (Johnson 1997; Luo et al. 2006). This implies that there normally can be little if any nuclear Xrn1, even in the absence of fully functional Rat1. Moreover, Xrn1 cannot replace Rat1 for its role in mRNA transcription termination in the rat1-1 mutant even when targeted to the nucleus (Luo et al. 2006).
During Pol I transcription, the RNase III-like endonuclease Rnt1 cleaves the nascent pre-rRNA across a stem–loop structure within the 3′-ETS, at positions +14/15 and +49/50 relative to the 3′ end of the 25S rRNA sequence (see Fig. 2A; Kufel et al. 1999). Rnt1 was detected at the site of rRNA transcription (Henras et al. 2004), and can interact physically with Pol I subunits including Rpa12 (Catala et al. 2008). Moreover, loss of Rnt1 activity influences rDNA transcription and chromatin structure (Catala et al. 2008), consistent with cotranscriptional activity. A rnt1Δ strain showed increased read-through of T1 with most Pol I stopping at T2 (Reeder et al. 1999; Prescott et al. 2004), and read-through transcripts extending to T2 were also detected in rnt1-ts strains (Catala et al. 2008), indicating that cotranscriptional cleavage is important for efficient termination at T1. Rnt1 cleavage generates products with 5′-monophosphate termini, the preferred substrates for Rat1 (Stevens and Poole 1995), which was reported previously to participate in the degradation of the 3′-cleavage product (Kufel et al. 1999), raising the interesting possibility that Rat1 might also “torpedo” Pol I during transcription termination.
Here we show that Rat1 plays a role in Pol I transcription termination. When Rat1 is absent from the cells, Pol I fails to efficiently terminate at T1 but mainly stops at T2 and at the RFB. In the absence of both Rat1 and Fob1, read-through of T2 and the RFB was substantially increased. We further show that the catalytic activity of Rat1 is essential for Pol I transcription termination to occur, suggesting that Rat1 “torpedoes” the polymerase on rDNA genes during termination, as is the case for Pol II on mRNA-coding genes.
Results
Rat1 and Rai1, but not Xrn1, contribute to productive 5.8S rRNA maturation
The rat1-1 mutation is temperature-sensitive lethal and leads to the accumulation of 5′-extended forms of the 5.8S rRNA at nonpermissive temperatures (Amberg et al. 1992). However, very little of the extended RNA was detected in the rat1-1 single mutant (Henry et al. 1994). This apparently poor penetrance of the rat1-1 mutation led us to assess whether a stronger phenotype could be generated by placing the RAT1 gene under the control of a conditional MET3 promoter. Yeast cells carrying PMET3-RAT1 were pregrown in SD medium lacking methionine then shifted to SD + 5 mM methionine to inhibit RAT1 expression. The levels of RAT1 mRNA were substantially reduced 3 h after methionine addition (Fig. 1B, lanes 1–6).
Maturation by Rat1 is required to form the 5′ end of the major, short form of 5.8S rRNA (5.8SS) but is not required for formation of the longer 5.8SL rRNA (Henry et al. 1994). In the absence of functional Rat1, synthesis of 5.8SL is therefore favored. Northern analysis (Fig. 1C, lanes 1–6) using a probe that hybridizes to the 5.8S rRNA (probe 017) (Fig. 1A) showed that the ratio of 5.8SS to 5.8SL was reduced in the PMET3-RAT1 strain, commencing 9 h after methionine addition, indicating efficient Rat1 depletion. The 5′-extended forms of the 5.8S rRNA were not clearly accumulated in the PMET3-RAT1 single mutant strain, whereas species extended to the A3 cleavage site accumulated strongly in strains also lacking the 5′ exonuclease Xrn1 (Fig. 1C, top panels, cf. lanes 1–6 and 7–13). Xrn1 is exclusively detected in the cytoplasm (Johnson 1997) and, consistent with this, the presence or absence of Xrn1 did not affect the ratio between the 5.8SS and 5.8SL species (Fig. 1C, middle panels, cf. lanes 1–6 and 7–13).
Rat1 is associated with a cofactor, Rai1 (Xue et al. 2000), that was reported to enhance the activity of Rat1 in 5.8S rRNA processing and in transcription termination by Pol II (Xue et al. 2000; Kim et al. 2004). The absence of Rai1 modestly enhanced accumulation of 5′-extended 5.8S rRNA prior to depletion of Rat1 (0-h sample) in the PMET3-RAT1, xrn1Δ background (Supplemental Fig. S1A, lane 3). However, the rai1Δ single mutant strongly enhanced the 5.8SL to 5.8SS ratio, without clear accumulation of 5′-extended species (Supplemental Fig. S1A, lane 2), and the absence of Rai1 enhanced the deficit in the 5.8SL to 5.8SS ratio seen on Rat1 depletion (Supplemental Fig. S1B, cf. lanes 1–5 and 8–12). These data are consistent with a “window of opportunity” for processing by Rat1 from A3 to B1S to generate 5.8SS (Fig. 1A). In the absence of Rai1, Rat1 may load more slowly or act less processively, favoring the alternative processing at B1L. A similar situation occurs in the absence of Rat1 where Xrn1 does not contribute to productive rRNA maturation at B1S, although it can degrade 5′ extensions present on the 5.8S precursors (Fig. 1C, cf. lanes 1–6 and 7–13).
Rat1 is also responsible for 5′ maturation of the 25S rRNA (Geerlings et al. 2000). Primer extension with a primer within the mature 25S rRNA (probe 007) (Fig. 1A) revealed strong accumulation of the 26S pre-rRNA, which extends to the C2 cleavage site within ITS2 (internal transcribed spacer 2), in the PMET3-RAT1, xrn1Δ strain following methionine addition (data not shown). Comparison of the PMET3-RAT1, xrn1Δ and rat1-1, xrn1Δ strains (Fig. 1D) revealed stronger accumulation of pre-5.8S and 5.8SL in the PMET3-RAT1, xrn1Δ strain after 12 h of depletion (Fig. 1D, cf. lanes 2–6 and 7,8). PMET3-RAT1 strains were therefore used for further analyses.
Transcripts from IGS1 are stabilized in the absence of Rat1 and Xrn1
If Rat1 plays a role in Pol I transcription termination then its depletion should lead to increased transcription extending beyond T1 into IGS1 (see Fig. 2A). To assess this, we initially performed primer extension analyses using primers complementary to sequences located downstream from the Rnt1 cleavage sites beyond the major T1 terminator. Probe 053 is located between T1 and T2, probe 054 is located between T2 and the RFB, and probe igs1-1F is located between the RFB and the 5S gene (Fig. 2A). Primer extension analyses were performed on RNA recovered from PMET3-RAT1 strains before and during Rat1 depletion. As the read-through transcripts are degraded by Xrn1 in the PMET3-RAT1 strain (see Supplemental Fig. S2), all of the following analyses were performed in xrn1Δ background. Probes 053 and 054 showed that extended transcripts accumulate progressively during Rat1 depletion, commencing 3 h after methionine addition (Fig. 2B). These have 5′ ends corresponding to the reported sites of Rnt1 cleavage sites at +14/15 or +49/50 (Kufel et al. 1999), with the predominant stop at +49/50 (Fig. 2B).
To assess whether the RFB also acts as a Pol I terminator, the gene encoding the RFB-binding protein Fob1 was deleted in the PMET3-RAT1, xrn1Δ double mutant strain (Fig. 2C,D). The absence of both Rat1 and Fob1 lead to an increase in the levels of all RNAs that extend through T2 (probe 054) (Fig. 2, cf. B and C, lanes 8–14) or both T2 and the RFB (probe igs1-1F) (Fig. 2D, cf. lanes 1–7 and 8–14). The species detected with probe igs1-1F also have a size consistent with 5′ ends at the Rnt1 cleavage sites, but the products are too long (783/818 nt) for these sites to be clearly resolved (Fig. 2D). Additional heterogeneous stops detected with probe igs1-1F (Fig. 2D, lanes 8–14) correspond to the approximate position of T2 and may represent premature primer extension stops due to the presence of the poly(U) tracts in this region.
These data indicate that during Rat1 depletion in the absence of Xrn1, RNAs accumulate that extend from the sites of cotranscriptional Rnt1 cleavage in IGS1 to beyond the major T1 terminator. The additional loss of Fob1 increased read-through of both T2 and the RFB, indicating that the RFB acts as an additional transcription terminator.
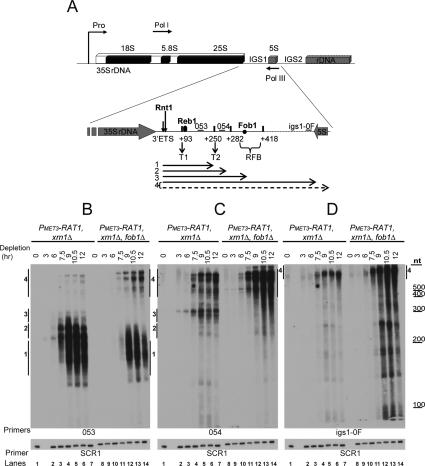
In order to analyze further the RNA species that accumulate during Rat1 depletion, we performed Northern analyses using primers 053, 054, and igs1-0F, which hybridizes at the 3′ end of IGS1 (Fig. 3A). Progressive accumulation of IGS transcripts was visible in the PMET3-RAT1, xrn1Δ double mutant strain commencing 3 h after methionine addition (Fig. 3B–D, lanes 1–7). Using primer 053, the major transcripts detected ranged from ∼160 to ∼300 nt (Fig. 3B, lanes 1–7). We labeled these species 1–3. Lower levels of longer transcripts were detected over ∼400 nt in length (species labeled 4). The 5′ ends of the transcripts predominantly lie at the major Rnt1 cleavage site at +49/50 (see Fig. 2), so the 3′ ends of the corresponding RNAs detected by Northern hybridization lie at positions 50 nt further into IGS1 relative to the 3′ end of the 25S sequence. The major RNA species accumulated following Rat1 depletion would therefore be consistent with termination around T2 (+250, species 1 and 2) and the RFB (+350, species 3) (see also Fig. 3A, horizontal arrows).
Figure 3.
RNAs transcribed from the IGS1 region accumulate when Rat1 and Xrn1 are absent. (A) Schematic of an rDNA repeat in S. cerevisiae (as in Fig. 2A). Oligonucleotides used (see also Supplemental Table S2) are indicated by small horizontal bars above the rDNA. Primers 053 and 054 were as in Figure 2A. Primer igs1-0F hybridizes to IGS1 immediately upstream of the 5S gene at +1054 relative to the 3′ end of the 25S rRNA sequence. Horizontal arrows below the rDNA indicate the RNA species (labeled 1–4) detected by Northern hybridizations. (B–D) Northern analyses. RNAs were extracted from the strains PMET3-RAT1, xrn1Δ, and PMET3-RAT1, xrn1Δ, fob1Δ as described for Figure 1B. Total RNA and an RNA ladder (RNA Century, Ambion) were resolved on an 8% polyacrylamide/urea gel. The same filter was probed sequentially with oligonucleotides 053 (B), 054 (C), igs1-0F (D), and SCR1.
In contrast to species 1 and 2, the longer transcripts (species 4) were detected also by primers 054 and igs1-0F (Fig. 3C,D, lanes 1–7), showing that these extend through T1, T2, and the RFB, as expected (see also Fig. 3A, horizontal arrows). Species 2, which from their gel mobility (over ∼200 nt) would be expected to extend beyond the position of oligo 054 downstream from T2, were detected with probe 053 but not probe 054 (Fig. 3, cf. B and C, lanes 1–7). Pol I can undergo iterative slippage at poly(dT) coding sequences, such as those located at T2, leading to addition of nontemplated 3′ poly(U) tracts in vitro (Jeong et al. 1996). This strongly suggests that transcripts 2 terminate around T2 but have been 3′-extended with poly(U).
We conclude that IGS1 transcripts accumulated in the absence of Rat1 and Xrn1 predominantly extend through T1 to the region that encompasses T2 and the RFB, with lower levels of species extending beyond the RFB.
In the PMET3-RAT1, xrn1Δ, fob1Δ triple mutant strain, the ∼300-nt bands predicted to arise from termination at the RFB were absent (species 3 in Fig. 3B,C, cf. lanes 1–7 and 8–14), and species corresponding to termination around T2 were greatly reduced in length and abundance (species 1 and 2 in Fig. 3B, cf. lanes 1–7 and 8–14). This was accompanied by a large accumulation of longer transcripts extending through the IGS1 region (species 4 in Fig. 3B–D, lanes 8–14). Analyses on agarose gels revealed that these long species have heterogeneous 3′ ends, mainly located between the 3′ end of IGS1 and the start of IGS2 (Supplemental Fig. S3). A very low level of transcripts extended through all of IGS1 and IGS2 to a position ∼300 nt upstream of the transcription initiation site of the downstream 35S gene (van der Sande et al. 1989).
These data show that the RFB acts as a terminator for Pol I molecules that read through both T1 and T2, with additional terminators located mainly between the 3′ end of IGS1 and the beginning of IGS2.
To assess the requirement for Rai1 in degradation of the IGS transcripts, we analyzed total RNAs from wild-type, rai1Δ, and PMET3-RAT1, xrn1Δ, rai11Δ strains. Only modest effects of rai1Δ were detected on the accumulation of the IGS1 transcripts (Supplemental Fig. S1C).
Rat1 associates with the 3′-ETS region of the rDNA
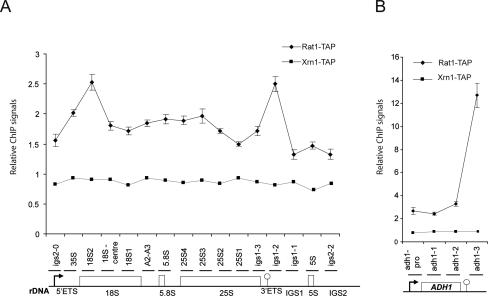
Previous chromatin immunoprecipitation (ChIP) analyses detected Rat1/Xrn2 downstream from the poly(A) site on mRNA-coding genes (Kim et al. 2004; Luo et al. 2006; Kaneko et al. 2007), consistent with its role in Pol II termination. To assess whether Rat1 functions directly in Pol I termination, we monitored the distribution of Rat1 over the rDNA by ChIP (Fig. 4A). Two peaks of Rat1 occupancy were localized to distinct regions, at the 5′ end of the 35S pre-rRNA-coding region and over the region of IGS1 that includes the T1, T2, and RFB terminators. In contrast, no association of Xrn1-TAP was detected under the same conditions, as expected from its reported cytoplasmic localization (Johnson 1997). As positive controls, two mRNA coding genes ADH1 (Fig. 4B) and CYC1 (data not shown) were also analyzed. ADH1 gave a strong Rat1-TAP ChIP signal, whereas CYC1 gene gave a lower signal, but higher than that seen for the rDNA IGS1 ChIP. These data are consistent with recruitment of Rat1 to transcripts derived from the 3′-ETS region of the 35S gene. The association with other regions of the rDNA may reflect the recruitment of Rat1 to other sites on the pre-rRNA. In contrast to Rat1, no ChIP signal above the background was detected for Rnt1-TAP (data not shown). The available data indicate that Rnt1 does bind to the pre-rRNA cotranscriptionally and is present at the site of transcription (Henras et al. 2004), but its transient association with the nascent transcript apparently is not captured by cross-linking in ChIP analyses.
Figure 4.
Rat1 but not Xrn1 can be cross-linked to the rDNA. ChIP analyses were performed for Rat1-TAP and Xrn1-TAP over the rDNA (A) and ADH1 (B). The promoter is depicted by an arrow. The “Reb1-dependent” terminator T1 (rDNA) and the poly(A) site (ADH1) are represented by lollipops. Horizontal bars above the rDNA or ADH1 denote the locations of products amplified by real-time PCR (see also Supplemental Table S3). QPCR values were corrected for background as described in the Materials and Methods. A value of 1 (Y-axis) indicates no signal above background. The mean of six independent experiments is shown with standard error for Rat1-TAP. The mean of two independent experiments is shown for Xrn1-TAP.
Pol I termination is impaired when Rat1 or the cofactor Rai1 is absent
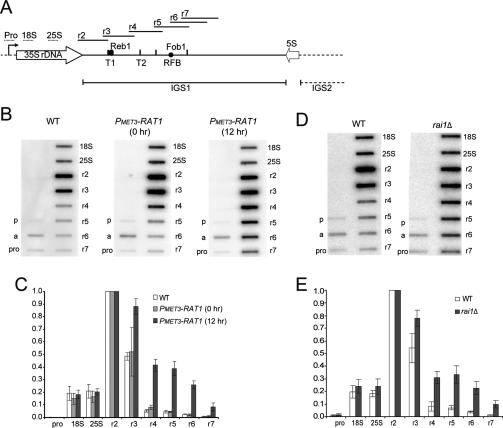
To determine whether Pol I termination is impaired in strains lacking Rat1, we performed transcription run-on analyses (TRO) using a previously described set of M13 phage probes (Prescott et al. 2004). These cover the IGS1 region from the 3′ end of 25S to 151 nt downstream from the RFB (probes r2–r7) (Fig. 5A). In wild-type cells, the TRO signal significantly declines immediately after T1 (probe r3), with background levels further downstream (probes r4–r7) (Fig. 5B–E). In PMET3-RAT1 cells grown under permissive conditions, the signals were similar to the wild type (0-h samples in Fig. 5B,C), whereas cells depleted for Rat1 (12-h samples in Fig. 5B,C) showed strongly increased TRO signals over the region extending from T1 to the RFB (probes r3–r5) with a smaller increase over the RFB region (r6 and r7). Cells lacking the Rat1 cofactor Rai1 also showed clearly increased read-through of T1 (Fig. 5D,E). In contrast, seven TRO replicates using the xrn1Δ strain failed to show any significant read-through of T1 (data not shown). Analyses of strains lacking Fob1 showed that there was no detectable read-through of T1 in the fob1Δ single mutant, but an increased TRO signal was observed over the RFB region (r6 and r7) in PMET3-RAT1, fob1Δ relative to PMET3-RAT1 after 12 h of depletion (data not shown).
Figure 5.
TRO shows read-through of the major Pol I terminator in strains lacking Rat1 or Rai1. (A) rDNA schematic showing the positions of M13 phage TRO probes depicted as horizontal bars above the rDNA sequence (see also Prescott et al. 2004). Labels are as in Figure 2A. (B,C) TRO analysis of wild-type and PMET3-RAT1 strains. (B) Representative TRO profiles are shown for the isogenic wild type grown in the presence of methionine (5 mM) and for PMET3-RAT1 cells grown in the absence (0 h) or presence of methionine for 12 h to allow depletion of Rat1. (pro) Promoter; (a) ACT1-positive control. (p) negative control. (C) Quantifications of TRO signals were corrected for background hybridization (probe p) and uracil content, and normalized to probe r2, which was arbitrarily set to 1. The mean of three independent experiments is shown with standard deviation. Lower signals over the regions encoding the 18S and 25S rRNAs were observed previously (see the Supplemental Material in Jones et al. 2007) and may reflect quenching of the signals by the highly abundant rRNAs. (D–E) TRO analysis of wild-type and rai1Δ strains (BY4741 background) grown in minimal medium at 30°C. D as B. E as C. Similar data (not shown) were observed with W303-1a background.
We conclude that the efficiency of Pol I transcription termination at the T1 terminator is reduced when Rat1 or the cofactor Rai1 is absent. The transcription read-through in the rai1Δ strain is in contrast to the lack of clear RNA stabilization (Supplemental Fig. S1C), confirming that the failure in RNA degradation is not causal in the termination defect. These data resemble the effects of Rat1, Rai1, and Xrn1 on 5′ processing of 5.8S (Fig. 1). Rat1 and Rai1 are required for efficient termination, whereas Xrn1 can degrade the IGS transcripts that would otherwise accumulate in the absence of Rat1 but does not participate in productive termination.
Pol I accumulates over the IGS1 region in the absence of Rat1
To confirm that the TRO data reflect transcription by RNA Pol I, we also monitored Pol I distribution over the IGS region in the presence or absence of Rat1 by ChIP using an epitope-tagged Pol I subunit (Rpa34-13MYC) (Fig. 6) or antibodies directed against Rpa190 (data not shown). Similar results were obtained for the two Pol I subunits.
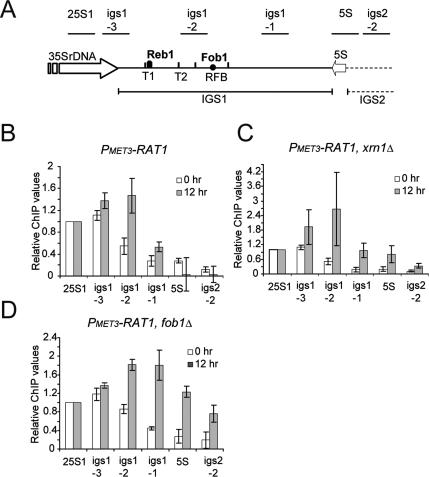
Figure 6.
Pol I occupancy over the IGS1 region is increased by depletion of Rat1. (A) rDNA schematic. Horizontal bars above the rDNA denote the locations of products amplified by real-time PCR (see also Supplemental Table S3). Labels are as in Figure 2A. (B–D) Profiles of Pol I cross-linking over the intergenic region of rDNA repeats. Aliquots from strains PMET3-RAT1 (B); PMET3-RAT1, xrn1Δ (C); and PMET3-RAT1, fob1Δ (D) were taken at 0 and 12 h of Rat1 depletion and processed for Pol I ChIP (Rpa34-13MYC). QPCR values were corrected for background as described in the Materials and Methods and normalized to 25S1, which was set arbitrarily to 1, in order to compensate for differences in immunoprecipitation efficiencies. The mean of three independent experiments is shown with standard error.
In wild-type cells (data not shown) or in PMET3-RAT1 cells grown under permissive conditions (0-h sample in Fig. 6B) a peak of Pol I association with the rDNA was seen with the igs1.3 primers, around the position of the T1 terminator (see Fig. 6A for locations of primers). This association decreased over the igs1.2 primers located over T2 and the RFB and reached background levels over the igs1.1 primers located further downstream in IGS1. Note that the close proximity of T2 and the RFB prevents clear discrimination between Pol I associated with these sites by ChIP. Following depletion of Rat1 (12-h sample in Fig. 6B) the peak of Pol I association was displaced from the igs1.3 primers (T1) to igs1.2 (T2/RFB region), consistent with transcription read-through of T1. The peaks of Pol I association were observed in the same positions using the PMET3-RAT1, xrn1Δ strain (Fig. 6C), although the intensity of the peaks at 12 h of depletion showed a large variation between different experiments. We conclude that the presence or absence of Xrn1 does not clearly affect Pol I transcription termination.
To assess the ability of the RFB to act as a transcription terminator, ChIP analyses were performed on a PMET3-RAT1, fob1Δ strain (Fig. 6D). Under permissive conditions for Rat1 expression, the absence of Fob1 did not affect Pol I occupancy across the IGS regions, consistent with TRO data (data not shown). However, following Rat1 depletion, the fob1Δ strain showed increased Pol I occupancy 3′ to the RFB (igs1.1, 5S and igs2.2 primers).
Together, these data demonstrate that Rat1 is required for normal transcription termination by RNA Pol I. Xrn1 does not contribute substantially to termination, but the RFB acts as a terminator when Rat1 is absent and Fob1 is present.
The catalytic activity of Rat1 is required for Pol I termination
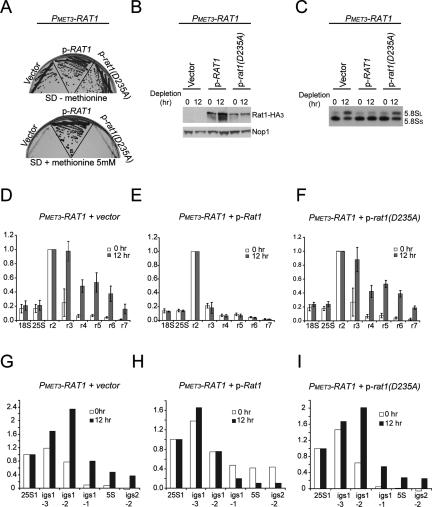
A Rat1 mutant that lacks catalytic activity cannot complement Pol II termination defects on mRNA-coding genes in rat1-1 strains at nonpermissive temperatures (Kim et al. 2004). To determine whether this is also the case for Pol I, the catalytically inactive mutant Rat1D235A (Kim et al. 2004) was expressed from a plasmid (kindly provided by Steven Buratowski, Harvard University) in the PMET3-RAT1 strain. As controls, the empty vector and a plasmid expressing wild-type Rat1 were also used. As expected, expression of the wild-type Rat1 allowed growth of the PMET3-RAT1 strain in the presence of 5 mM methionine, whereas no growth was seen in strains carrying the empty vector or expressing Rat1D235A (Fig. 7A). This shows that the catalytic activity is required for the essential function of Rat1. The plasmid-expressed proteins carry a triple-HA tag (Kim et al. 2004), and Western blotting showed that the wild-type Rat1 and Rat1D235A have similar expression levels (Fig. 7B). Northern analysis for the 5.8S rRNA confirmed that wild-type Rat1, but not Rat1D235A, complemented the phenotype of Rat1 depletion for 5.8SS and 5.8SL synthesis (Fig. 7C).
Figure 7.
The catalytically inactive mutant Rat1D235A does not correct defects in 5.8S rRNA maturation and Pol I termination when Rat1 is depleted. (A) PMET3-RAT1 strains transformed with plasmid without insert (vector), with RAT1 (wild type [WT]), or with rat1 (D235A) were tested for growth at 30°C on SD-methionine or SD + 5 mM methionine. (B) Western blot for detection of tagged Rat1 or Rat1D235A with HA3. Nop1 detection was used as a loading control. (C) Northern analysis of 5.8S rRNA maturation. RNAs were extracted as described in Figure 1B. Total RNA was resolved on an 8% polyacrylamide/urea gel. Mature 5.8SS and 5.8SL rRNAs were detected with probe 017 (see Fig. 1A; Supplemental Table S2). (D–F) TRO analysis over the rDNA repeats in strains PMET3-RAT1 + vector (D), PMET3-RAT1 + p-RAT1 (E), and PMET3-RAT1 + p-rat1 (D235A) (F). Primers and quantifications are as in Figure 5. The mean of three independent experiments is shown with standard deviation. (G–I) Profiles of Pol I cross-linking over the intergenic region of rDNA repeats in strains PMET3-RAT1 + vector (G), PMET3-RAT1 + p-RAT1 (H), and PMET3-RAT1 + p-rat1 (D235A) (I). Primers and quantifications are as described in Figure 6. A representative graph of two independent experiments is shown.
The ability of Rat1D235A to support Pol I termination was tested by TRO (Fig. 7D–F) and Pol I ChIP (Fig. 7G–I). In TRO analyses, Rat1 depletion for 12 h strongly enhanced read-through over probes r3–r7 (Fig. 7D). This increase was suppressed by plasmid expression of wild-type Rat1 (Fig. 7E) but not by expression of Rat1D235A (Fig. 7F). In ChIP analyses, depletion of Rat1 displaced the Pol I peak from primers igs1.3 (T1) to igs1.2 (T2/RFB) (Fig. 7G). This displacement was suppressed by plasmid expression of wild-type Rat1 (Fig. 7H) but not by expression of Rat1D235A (Fig. 7I).
These data show that catalytically inactive Rat1 does not function in 5.8Ss rRNA maturation or in Pol I termination at T1. This resembles the phenotypes of strains lacking Rai1, which show reduced Rat1 activity (Fig. 5; Supplemental Fig. 1). These data strongly support the proposed function of Rat1 as a “torpedo” for Pol I.
Discussion
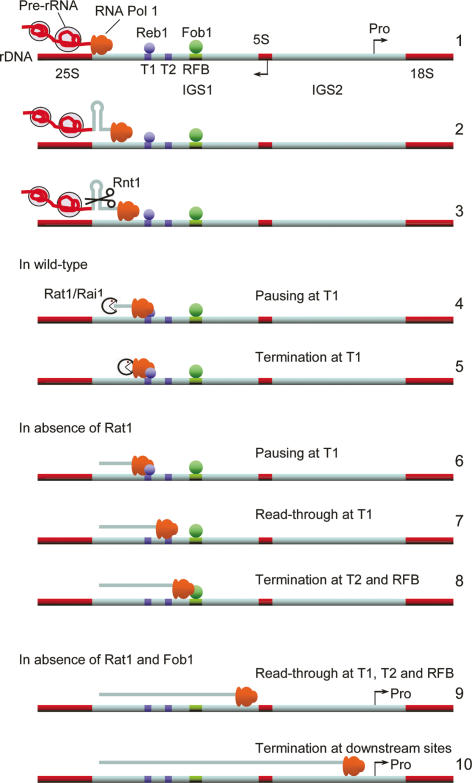
In this study, we demonstrate that the 5′-to-3′ exonuclease Rat1 plays a role in Pol I transcription termination in S. cerevisiae. The model illustrated in Figure 8 resembles the torpedo model described for mRNA transcription termination (Kim et al. 2004; West et al. 2004). The 35S pre-rRNA is cleaved cotranscriptionally by Rnt1 across a stem–loop structure, at positions +14/15 and +49/50, 3′ to the 25S rRNA. These cleavages form the entry sites for Rat1, which attacks the nascent RNA downstream from the cleavage sites. Rat1 potentially catches up with the polymerase, which is predicted to be paused at the major, “Reb1-dependent” T1 terminator (Reeder and Lang 1997). Cotranscriptional degradation by Rat1 might trigger the release of Pol I from the DNA template through the destabilization of the 8- to 9-nt RNA/DNA hybrid and/or the disruption of RNA–protein interactions implicated in the stabilization of Pol I complexes. Previous analyses suggested the existence of a transcript release factor for Pol I termination in yeast (Reeder and Lang 1997; Tschochne and Milkereit 1997; Jansa and Grummt 1999), which might correspond to Rat1.
Figure 8.
Model for the role of Rat1 as a torpedo in Pol I transcription termination. (1) The elongating Pol I carries the nascent pre-rRNA, which assembles cotranscriptionally with ribosome synthesis factors. (2) Pol I transcribes through Rnt1 cleavage sites in IGS1. (3) The pre-rRNA is cleaved cotranscriptionally across a stem–loop structure by Rnt1. (4,5) In wild-type cells, these cleavages form the entry site for the Rat1/Rai1 complex. This complex degrades the nascent transcript and catches the polymerase, which is predicted to be paused at site T1, stimulating transcription termination. (6–8) In the absence of Rat1, the polymerase largely transcribes through T1 but terminates at T2 and the RFB. (9,10) In the absence of both Rat1 and Fob1, RNA polymerases can transcribe through T2 and the RFB and terminate upstream of the 5S gene or in IGS2.
The Rat1-interacting protein Rai1 is required for efficient 5′ maturation of 5.8S rRNA and termination of Pol II transcription (Xue et al. 2000; Kim et al. 2004), and was also required for efficient Pol I termination. Rai1 was, however, dispensable for pre-rRNA transcript degradation following Rnt1 cleavage. Conversely, Xrn1 was able to degrade the cleaved IGS1 transcripts in the absence of Rat1, but did not participate in transcription termination. Xrn1 is reported to be cytoplasmic exclusively (Johnson 1997), and it may therefore be unable to interact with nuclear termination factors and/or fail to degrade the pre-rRNA cotranscriptionally. We speculate that increased Pol I read-through in the rai1Δ strain may reflect slowed loading or reduced processivity of Rat1 in the absence of its cofactor, preventing it from catching the polymerase within this short time window. This model was strengthened by the observation that catalytically inactive Rat1D235A fails to complement Pol I termination defects seen on depletion of wild-type Rat1. These features resemble pre-rRNA processing, where loss of either Rat1 or Rai1 impairs processing of the 5′-extended precursor to 5.8S rRNA. This leads to a decrease in 5.8SS rRNA relative to the alternative 5.8SL species, consistent with a short time window for productive processing to 5.8SS. In contrast, Xrn1 degrades 5′-extended 5.8S rRNA species that would otherwise accumulate in the absence of Rat1 activity, but does not contribute to 5.8SS maturation.
In ChIP analyses, levels of Rat1 over the rDNA 3′-ETS region were lower than over the 3′ regions of the Pol II genes ADH1 and CYC1. In the case of Pol II termination, Rat1 interacts with CTD-binding protein Rtt103 and other 3′-end processing factors (Kim et al. 2004; Luo et al. 2006). This might strengthen the cross-linking of Rat1 to the Pol II transcribed DNA. In contrast, Rat1 has not been reported to interact directly with RNA Pol I, so positive ChIP signals on the rDNA require cross-linking of Rat1 to the nascent transcript, bound to the transcribing polymerase, bound to the DNA. The relatively low Rat1 ChIP signal over the rDNA IGS1 region is also likely to reflect its very short residency times, consistent with the very rapid and efficient rRNA processing pathway. Pol I requires only 1–2 sec to transcribe the 44 nt from the major Rnt1 cleavage site (+50 relative to 25S sequence) to T1 (+93). The “Reb1-dependent” terminator may pause Pol I (Reeder et al. 1999; Prescott et al. 2004), allowing more time for termination to occur, but the high rate of rDNA transcription (around one transcript is completed every 3 sec on each active rDNA repeat) suggests that the time available will still be very limited. Note also that ChIP signals derived from rDNA transcription units will be diluted by the nontranscribed rDNA repeats, which account for half of the rDNA array.
Our ChIP data localized Rat1 to both IGS1 and the 5′ region of the 35S rDNA, possibly reflecting looping of the rDNA as proposed previously (Johnson and Warner 1989; Kulkens et al. 1992). This might arise through oligomerization of Reb1, since this binds the rDNA at T1 in IGS1 and just upstream of the Pol I promoter in IGS2 (Reeder and Lang 1997). Such loops have been detected recently between the 5′ and 3′ ends of protein-coding genes (O’Sullivan et al. 2004; Ansari and Hampsey 2005). However, Rat1 also functions in the 5′ maturation of the 5.8S and 25S rRNAs, and in the rapid degradation of other excised pre-rRNA spacer fragments (Henry et al. 1994; Petfalski et al. 1998; Geerlings et al. 2000). It is therefore also possible that Rat1 is loaded cotranscriptionally onto the nascent pre-rRNA to promote efficient pre-rRNA processing, as has been reported for other ribosome synthesis factors (Gallagher et al. 2004). Moreover, processing in ITS1, where Rat1 functions in 5.8S synthesis, and processing in IGS1 are reported to be coupled (Allmang and Tollervey 1998), suggesting further interactions.
In addition to the Rat1/Rai1 complex, other factors presumably play roles in Pol I termination. The small Pol I subunit Rpa12 can interact directly with Rnt1 (Catala et al. 2008) and the absence of Rpa12 or Rnt1 leads to Pol I read-through over IGS1 (Reeder et al. 1999; Prescott et al. 2004; Catala et al. 2008). The absence of Rpa12 might interfere directly with cotranscriptional cleavage by Rnt1, preventing loading of the Rat1/Rai1 complex. However, nascent IGS1 transcripts were not observed in rpa12Δ strains in Miller spreads (Prescott et al. 2004), suggesting that degradation of the transcripts is ongoing. Other possible models for read-through in rpa12Δ strains include a failure of the defective Pol I to pause at termination sites or alterations in Pol I conformation that render it more refractory to the torpedo activity of Rat1/Rai1. Notably, efficient termination of mRNA synthesis requires both a change in Pol II conformation downstream from the cleavage/polyadenylation site and the degradation of the 3′ transcript by Rat1/Rai1 (Luo et al. 2006; Rosonina et al. 2006).
In the absence of Rat1, Pol I reads through the major T1 terminator but stops at T2 and the RFB. The absence of the RFB-binding protein Fob1 alone had no effect on termination at T1, but in the absence of both Rat1 and Fob1 a substantial fraction of Pol I transcribed the entire IGS1 region. Thus, in addition to its key roles in rDNA metabolism, the RFB serves as an extra barrier for Pol I transcription. T2 and the “Fob1-dependent” terminator might cooperate to ensure that any Pol I complexes that escape T1 do not collide with rDNA replication forks moving toward the RFB in opposite orientation to Pol I during S phase (Takeuchi et al. 2003).
The loss of Fob1 from the Rat1-depleted strain also reduced but did not abolish termination at T2. The sequence of T2, which is extremely dT-rich, probably leads to slippage of Pol I, with reiterative synthesis leading to formation of long poly(U) tracts as seen previously in vitro (Jeong et al. 1996). It is therefore possible that T2 possesses some intrinsic termination activity. Elongation of Pol I through T2 (and elsewhere downstream in IGS1 and IGS2) might also be impaired by formation of R-loops, generated by interactions between the IGS1 transcripts, which may not be packaged by the ribosome synthesis machinery, and the rDNA (Tous and Aguilera 2007).
Other factors that potentially contribute to termination at T2 include the Nrd1–Nab3 complex (M. Koper and J. Kufel, unpubl.) or the TRAMP and exosome complexes. These function in the termination and degradation of noncoding RNAs that are transcribed through the RFB/T2 region by Pol II in the opposite orientation to Pol I (Houseley et al. 2007; Vasiljeva et al. 2008). The noncoding RNAs are generated at levels very much lower than the pre-rRNA, but might also influence Pol I termination at T2 and the RFB via effects on chromatin structure.
In mammals, the Pol I terminator is an 18-base-pair (bp) sequence motif called the Sal box that is repeated 10 times (sites T1–T10) downstream from the 3′ end of the pre-RNA coding region. The first eight Sal boxes are recognized by TTF-1, which mediates both RNA Pol I termination (Kuhn and Grummt 1989) and rDNA replication fork stalling (Gerber et al. 1997). However, TTF-1 blocks rDNA replication only when bound to Sal box T2 (Gerber et al. 1997), which lies 122 nt away from the major site of transcription arrest at Sal box T1, physically separating these processes. Yeast Reb1 is homologous with TTF-1, since the C-terminal halves of both proteins share homology with the DNA-binding domain of the proto-oncoprotein c-Myb (for review, see Reeder and Lang 1997), whereas Fob1 is functionally related to TTF-1 in blocking replication fork progression. Thus, in both systems the major site of Pol I termination and the RFB are physically separated, although there are mechanistic differences. No RNase III cleavage site has yet been identified at the 3′ end of the mammalian 28S rRNA, but a 3′ exonuclease trims the pre-28S after release of RNA Pol I (Kuhn and Grummt 1989). The release factor implicated in Pol I termination in mammals is PTRF, which is unrelated to Rat1 (Jansa and Grummt 1999). The involvement of mammalian Xrn2 in Pol I termination has not been addressed, but these observations suggest that it may not have the same role as yeast Rat1.
The model proposed here for transcription termination by Pol I shows striking similarities to current models for Pol II termination on protein-coding genes. These, too, invoke cotranscriptional cleavage, providing an entry site for Rat1/Rai1 or Xrn2 (Kim et al. 2004; West et al. 2004; Kaneko et al. 2007), which then catch the polymerase at downstream pause sites (Gromak et al. 2006). The results reveal potentially important links between the transcriptional and post-transcriptional steps in rRNA synthesis, and between ribosome biogenesis and mRNA synthesis.
Materials and methods
Strains, plasmids, and growth conditions
Yeast strains and plasmids used in this study are listed in Supplemental Table S1. The PMET3-RAT1 strain was constructed by one-step PCR strategy using plasmid pTLN28ff (van Nues and Beggs 2001). xrn1Δ, rai1Δ, or fob1Δ deletions (template plasmids pFA6a-kanMX6/NatMX6) and the C-terminal 13Myc construct (template plasmid pFA6a-13MYC∷Hph; a generous gift from R. Grainger) were performed as described (Longtine et al. 1998).
Growth and handling of S. cerevisiae were by standard techniques. For Rat1 depletion, cells (PMET3-RAT1 strains) were grown at 30°C to OD600 ∼0.3–0.4 in SD medium lacking methionine, then were transferred to the same prewarmed medium supplemented with methionine (5 mM). The growth was continued for several hours and maintained in exponential phase by dilution with prewarmed SD medium with methionine.
RNA analyses
RNA extractions, Northern hybridization, and primer extension were as described (Thomson and Tollervey 2005). Standard 1.2% agarose/glyoxal gel or 6% or 8% acrylamide/8.3 M urea gels were used to separate high- or low-molecular-weight RNAs. Primers for RNA analysis are listed in Supplemental Table S2. To detect RAT1 mRNA (3.02 kb), a random priming probe was prepared according to Strip-EZTM (Ambion) using primers Rat1mRNA-F and Rat1mRNA-R (Supplemental Table S2). Northern hybridization was done overnight at 42°C using ULTRAHyb buffer (Ambion), and stringent washes were done at 58°C with SSC 0.2× SDS 0.1%.
Western blots
Total protein extracts and Western blot analysis were performed using standard procedures. Nop1p was detected with a mouse anti-Nop1 antibody (kindly provided by J. Aris, University of Florida). Rat1-HA3 tag was detected with mouse anti-HA sc-7392 antibody (Santa Cruz Biotechnology).
ChIP
Yeast cells (75 mL of 0.5–0.7 A600/mL; 12.5 mL per immunoprecipitation) were cross-linked with formaldehyde (1% final) for 15 min (TAP-tagged proteins) or 30 min (Pol I), quenched with glycine (137.5 mM) for 10 min, and washed with ice-cold PBS 1×. Cells were resuspended with 350 μL of lysis buffer (50 mM HEPES-KOH at pH 7.5, 140 mM NaCl, 1 mM EDTA at pH 8, 1% Triton X-100, 0.1% w/v sodium deoxycholate, plus CPI 1× [Roche Protease inhibitor cocktail tablets containing EDTA]), mixed with 500 μL of glass beads (Sigma, G8772), and vortexed (Vortex Genie 2T, Scientific Industries) for 45 min at full speed at 4°C. Glass beads were removed and cross-linked chromatin was recovered by centrifugation at full speed for 10 min at 4°C (supernatant discarded). Six-hundred microliters of lysis buffer were added on the top of the pellet. Sonication of chromatin was performed for 1 min 30 sec (10 sec ON, 15 sec OFF, 20% amplitude; Branson Digital Sonifier) to yield an average DNA fragment size of ∼500 bp. “Cells” were spun for 30 min at full speed at 4°C. The supernatant (∼600 μL) was diluted 1:1 with lysis buffer supplemented with glycerol 10%. Two-hundred microliters of lysate were mixed with 20 μL (bed volume) of protein A sepharose Cl-4B beads (GE Healthcare) (for rabbit antibody) or Gammabind G sepharose beads (GE Healthcare) (for mouse antibody) and cleared for 1 h at 4°C. Ten microliters were kept for Input DNA. Immunoprecipitations were performed by adding 12.5 μL of anti-Myc mAb9E10 (Santa Cruz Biotechnology) for Rpa34-13MYC ChIPs, or 4 μL of anti-Rpa190 (generous gift from Michel Riva, CEA-Saclay-France; or our affinity-purified rabbit antibody from Eurogentec) for Rpa190 ChIPs. To assess contribution of background, a “beads-only” internal control was usually prepared in parallel to immunoprecipitated samples but without addition of any antibody. Beads, antibody, and “cleared-sonicated chromatin” were mixed on a rotating wheel overnight at 4°C. Beads were recovered and washed successively with lysis buffer, lysis buffer with 500 mM NaCl, wash buffer (10 mM Tris-HCl at pH 8, 0.25 M LiCl, 0.5% NP-40, 0.5% w/v sodium deoxycholate, 1 mM EDTA at pH 8, plus CPI 1×), and TE (100 mM Tris-Cl at pH 8, 10 mM EDTA at pH 8) at 4°C. Cross-link reversal was done by incubating the washed beads overnight at 65°C in 125 μL of TE buffer containing 1% SDS. After proteinase K treatment (900 μg/mL) for 3 h at 55°C, DNA was purified using Qiagen PCR purification kit and eluted with 70 μL of buffer EB containing RNase A (0.5 μg/mL). In the case of TAP-tagged proteins, immunoprecipitations were performed as above but using sepharose CL-4B beads (Sigma) for the clearing step and IgG sepharose 6 Fast Flow beads (GE Healthcare) for the immunoprecipitations. To assess the background, the nontagged strain was used in parallel as an external control.
Quantitative PCRs (qPCRs) were performed in triplicate with SYBR green JumpStart Taq ReadyMix (Sigma) and a Stratagene MX3005P real-time PCR machine. Reaction volumes were 10 μL: 5 μL of Mix, 0.05 μL of Rox, 0.03 μL of each primer (300 nM), and 1 μL of template. Cycling parameters were 2 min at 95°C, then 40 cycles of 10 sec at 95°C, 10 sec at 55°C, and 15 sec at 72°C. Primers used for real-time PCR are listed in Supplemental Table S3.
Values for ChIPs were determined using the formula ΔΔCt = 2−(ΔCt IP − ΔCt Background). “Ct IP” is the cycle number for immunoprecipitate, and “Ct Background” is the cycle number for external control (isogenic nontagged strain in the case of TAP-tagged proteins in Fig. 4) or the cycle number for internal control (without antibody in the case of Pol I in Figs. 6, 7). ΔCt IP = Ct IP − Ct Input DNA. ΔCt Background = Ct Background − Ct Input DNA. In the case of Pol I ChIPs (Figs. 6, 7), ΔΔCt for ADH1-1 primers (ADH1 is a Pol II transcribed gene) was subtracted from ΔΔCt for each rDNA primer set.
TRO analysis
TRO assay was performed essentially as described (Birse et al. 1997) with the following modifications. Fifty milliliters of cells were harvested, washed, and permeabilized in 0.5% sarkosyl for 20 min at 4°C. Cells were resuspended in transcription buffer with the addition of 2 mM DTT and 1.33 mM each ATP, GTP, and CTP. Transcription was allowed to proceed for 5 min at 30°C in the presence of 30–60 μCi of [α32P] UTP (3000 Ci mMol−1; Hartmann Analytic). Cells were then washed with AE buffer (NaAc 50 mM, EDTA 10 mM) and RNA was extracted using GTC/phenol and the zirconia-silica beads method. RNA was partially hydrolyzed with 0.2 M NaOH for 5 min at 4°C and neutralized by addition of 0.4 vol of 0.5 M Tris-HCl. Single-stranded M13 phage constructs were used for hybridization as described (Prescott et al. 2004). Probes (5 μg per slot) were immobilized on Bright-Star nylon membrane (Ambion) using Bio-Dot Microfiltration Apparatus (Bio-Rad) and UV-cross-linked. Filters were hybridized overnight at 42°C in 5× SSC, 50% formamide, 0.05% SDS, 10× Denhardt’s solution. Autoradiographs were visualized using FujiFilm FLA-7000 scanner and quantified using FujiFilm Multi Gauge version 3.0 software.
Acknowledgments
We are indebted to Nick Proudfoot for M13 phage constructs, Michel Riva for anti-Rpa190, and Steven Buratowski and Minkyu Kim for pRS315 plasmids. We thank Bernhard Dichtl, Maria Vogelauer, Kim Kotovic, Martin Kos, Emma Thomson, Steve Innocente, Richard Grainger, David Barrass, Olivier Cordin, and Aracelli Castillo for advice and/or critical reading of manuscript. We thank Kawa.Ska Sp. Z o.o. (Zalesie Gorne, Poland) for the FujiFilm FLA-7000 system. A.E.H. was the recipient of an EMBO fellowship. This work was supported by EU grant LSHG-CT-2005-518280 and the Wellcome Trust.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.463708.
References
- Alen C., Kent N.A., Jones H.S., O’Sullivan J., Aranda A., Proudfoot N.J. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol. Cell. 2002;10:1441–1452. doi: 10.1016/s1097-2765(02)00778-5. [DOI] [PubMed] [Google Scholar]
- Allmang C., Tollervey D. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J. Mol. Biol. 1998;278:67–78. doi: 10.1006/jmbi.1998.1693. [DOI] [PubMed] [Google Scholar]
- Amberg D.C., Goldstein A.L., Cole C.N. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Ansari A., Hampsey M. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes & Dev. 2005;19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awrey D.E., Weilbaecher R.G., Hemming S.A., Orlicky S.M., Kane C.M., Edwards A.M. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem. 1997;272:14747–14754. doi: 10.1074/jbc.272.23.14747. [DOI] [PubMed] [Google Scholar]
- Birse C.E., Lee B.A., Hansen K., Proudfoot N.J. Transcriptional termination signals for RNA polymerase II in fission yeast. EMBO J. 1997;16:3633–3643. doi: 10.1093/emboj/16.12.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala M., Tremblay M., Samson E., Conconi A., Abou Elela S. Deletion of Rnt1p alters the proportion of open versus closed rDNA repeats in yeast. Mol. Cell. Biol. 2008;28:619–629. doi: 10.1128/MCB.01805-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher J.E., Dunbar D.A., Granneman S., Mitchell B.M., Osheim Y., Beyer A.L., Baserga S.J. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes & Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerlings T.H., Vos J.C., Raue H.A. The final step in the formation of 25S rRNA in Saccharomyces cerevisiae is performed by 5′ → 3′ exonucleases. RNA. 2000;6:1698–1703. doi: 10.1017/s1355838200001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J.K., Gogel E., Berger C., Wallisch M., Muller F., Grummt I., Grummt F. Termination of mammalian rDNA replication: Polar arrest of replication fork movement by transcription termination factor TTF-I. Cell. 1997;90:559–567. doi: 10.1016/s0092-8674(00)80515-2. [DOI] [PubMed] [Google Scholar]
- Gromak N., West S., Proudfoot N.J. Pause sites promote transcriptional termination of mammalian RNA polymerase II. Mol. Cell. Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henras A.K., Bertrand E., Chanfreau G. A cotranscriptional model for 3′-end processing of the Saccharomyces cerevisiae pre-ribosomal RNA precursor. RNA. 2004;10:1572–1585. doi: 10.1261/rna.7750804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Y., Wood H., Morrissey J.P., Petfalski E., Kearsey S., Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J., Kotovic K., El Hage A., Tollervey D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J. 2007;26:4996–5006. doi: 10.1038/sj.emboj.7601921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Brito I.L., Villen J., Gygi S.P., Amon A., Moazed D. Inhibition of homologous recombination by a cohesin-associated clamp complex recruited to the rDNA recombination enhancer. Genes & Dev. 2006;20:2887–2901. doi: 10.1101/gad.1472706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansa P., Grummt I. Mechanism of transcription termination: PTRF interacts with the largest subunit of RNA polymerase I and dissociates paused transcription complexes from yeast and mouse. Mol. Gen. Genet. 1999;262:508–514. doi: 10.1007/s004380051112. [DOI] [PubMed] [Google Scholar]
- Jeong S.W., Lang W.H., Reeder R.H. The yeast transcription terminator for RNA polymerase I is designed to prevent polymerase slippage. J. Biol. Chem. 1996;271:16104–16110. doi: 10.1074/jbc.271.27.16104. [DOI] [PubMed] [Google Scholar]
- Johnson A.W. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 1997;17:6122–6130. doi: 10.1128/mcb.17.10.6122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.P., Warner J.R. Unusual enhancer function in yeast rRNA transcription. Mol. Cell. Biol. 1989;9:4986–4993. doi: 10.1128/mcb.9.11.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H.S., Kawauchi J., Braglia P., Alen C.M., Kent N.A., Proudfoot N.J. RNA polymerase I in yeast transcribes dynamic nucleosomal rDNA. Nat. Struct. Mol. Biol. 2007;14:123–130. doi: 10.1038/nsmb1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S., Rozenblatt-Rosen O., Meyerson M., Manley J.L. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes & Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Krogan N.J., Vasiljeva L., Rando O.J., Nedea E., Greenblatt J.F., Buratowski S. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- Kufel J., Dichtl B., Tollervey D. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA. 1999;5:909–917. doi: 10.1017/s135583829999026x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Grummt I. 3′-end formation of mouse pre-rRNA involves both transcription termination and a specific processing reaction. Genes & Dev. 1989;3:224–231. doi: 10.1101/gad.3.2.224. [DOI] [PubMed] [Google Scholar]
- Kulkens T., van der Sande C.A., Dekker A.F., van Heerikhuizen H., Planta R.J. A system to study transcription by yeast RNA polymerase I within the chromosomal context: Functional analysis of the ribosomal DNA enhancer and the RBP1/REB1 binding sites. EMBO J. 1992;11:4665–4674. doi: 10.1002/j.1460-2075.1992.tb05568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang W.H., Reeder R.H. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:649–658. doi: 10.1128/mcb.13.1.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Luo W., Johnson A.W., Bentley D.L. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: Implications for a unified allosteric-torpedo model. Genes & Dev. 2006;20:954–965. doi: 10.1101/gad.1409106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygerou Z., Allmang C., Tollervey D., Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- O’Sullivan J.M., Tan-Wong S.M., Morillon A., Lee B., Coles J., Mellor J., Proudfoot N.J. Gene loops juxtapose promoters and terminators in yeast. Nat. Genet. 2004;36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- Petfalski E., Dandekar T., Henry Y., Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell Biol. 1998;18:1181–1189. doi: 10.1128/mcb.18.3.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott E.M., Osheim Y.N., Jones H.S., Alen C.M., Roan J.G., Reeder R.H., Beyer A.L., Proudfoot N.J. Transcriptional termination by RNA polymerase I requires the small subunit Rpa12p. Proc. Natl. Acad. Sci. 2004;101:6068–6073. doi: 10.1073/pnas.0401393101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder R.H., Lang W. The mechanism of transcription termination by RNA polymerase I. Mol. Microbiol. 1994;12:11–15. doi: 10.1111/j.1365-2958.1994.tb00989.x. [DOI] [PubMed] [Google Scholar]
- Reeder R.H., Lang W.H. Terminating transcription in eukaryotes: Lessons learned from RNA polymerase I. Trends Biochem. Sci. 1997;22:473–477. doi: 10.1016/s0968-0004(97)01133-x. [DOI] [PubMed] [Google Scholar]
- Reeder R.H., Guevara P., Roan J.G. Saccharomyces cerevisiae RNA polymerase I terminates transcription at the Reb1 terminator in vivo. Mol. Cell. Biol. 1999;19:7369–7376. doi: 10.1128/mcb.19.11.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosonina E., Kaneko S., Manley J.L. Terminating the transcript: Breaking up is hard to do. Genes & Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- Stevens A., Poole T.L. 5′-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′-exonuclease-1. J. Biol. Chem. 1995;270:16063–16069. doi: 10.1074/jbc.270.27.16063. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y., Horiuchi T., Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes & Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson E., Tollervey D. Nop53p is required for late 60S ribosome subunit maturation and nuclear export in yeast. RNA. 2005;11:1215–1224. doi: 10.1261/rna.2720205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tous C., Aguilera A. Impairment of transcription elongation by R-loops in vitro. Biochem. Biophys. Res. Commun. 2007;360:428–432. doi: 10.1016/j.bbrc.2007.06.098. [DOI] [PubMed] [Google Scholar]
- Tschochne H., Milkereit P. RNA polymerase I from S. cerevisiae depends on an additional factor to release terminated transcripts from the template. FEBS Lett. 1997;410:461–466. doi: 10.1016/s0014-5793(97)00636-4. [DOI] [PubMed] [Google Scholar]
- van der Sande C.A., Kulkens T., Kramer A.B., de Wijs I.J., van Heerikhuizen H., Klootwijk J., Planta R.J. Termination of transcription by yeast RNA polymerase I. Nucleic Acids Res. 1989;17:9127–9146. doi: 10.1093/nar/17.22.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues R.W., Beggs J.D. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001;157:1451–1467. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L., Kim M., Terzi N., Soares L.M., Buratowski S. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol. Cell. 2008;29:313–323. doi: 10.1016/j.molcel.2008.01.011. [DOI] [PubMed] [Google Scholar]
- West S., Gromak N., Proudfoot N.J. Human 5′ → 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- Xue Y., Bai X., Lee I., Kallstrom G., Ho J., Brown J., Stevens A., Johnson A.W. Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol. 2000;20:4006–4015. doi: 10.1128/mcb.20.11.4006-4015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]