Abstract
The Mus81–Eme1 complex is a structure-specific endonuclease that plays an important role in rescuing stalled replication forks and resolving the meiotic recombination intermediates in eukaryotes. We have determined the crystal structure of the Mus81–Eme1 complex. Both Mus81 and Eme1 consist of a central nuclease domain, two repeats of the helix–hairpin–helix (HhH) motif at their C-terminal region, and a linker helix. While each domain structure resembles archaeal XPF homologs, the overall structure is significantly different from those due to the structure of a linker helix. We show that a flexible intradomain linker that formed with 36 residues in the nuclease domain of Eme1 is essential for the recognition of DNA. We identified several basic residues lining the outer surface of the active site cleft of Mus81 that are involved in the interaction with a flexible arm of a nicked Holliday junction (HJ). These interactions might contribute to the optimal positioning of the opposite junction across the nick into the catalytic site, which provided the basis for the “nick and counternick” mechanism of Mus81–Eme1 and for the nicked HJ to be the favored in vitro substrate of this enzyme.
[Keywords: Structure-specific endonuclease, nicked Holliday junction, substrate preference, Mus81–Eme1, crystal structure]
The efficient and faithful repair of DNA damage is essential for the maintenance of genomic integrity and cell survival. Homologous DNA recombination (HR) provides a main pathway that functions in the repair of DNA double-stranded breaks (DSBs) and interstrand cross-links (ICLs). HR is also believed to play an important role in the recovery of stalled or broken replication forks (RFs) during DNA replication, and contributes to tolerance of DNA damage (Li and Heyer 2008). In addition to a central role in DNA repair, HR provides critical support for the correct segregation of homologous chromosomes during the first meiotic division. In all these events, a major step is the formation of Holliday junctions (HJs), four-way branched DNA intermediates, which must be resolved to separate repaired chromosomes (Holliday 1964; Schwacha and Kleckner 1995; Cox et al. 2000; Lilley and White 2001; McGlynn and Lloyd 2003). It also has been proposed that regressed RFs, which adopt a four-way structure, may be subject to cleavage as a mechanism of bypassing DNA lesions such as ICLs (Cox et al. 2000; Rothstein et al. 2000; Heyer et al. 2003; McGlynn and Lloyd 2003; Michel et al. 2004). HJs and related four-way DNA structures are resolved through coordinated single-stranded cleavages across the junction by the junction-resolving enzymes (Lilley and White 2001).
The Mus81–Eme1 complex (referred to as the MUS81 complex throughout the text) is a structure-specific nuclease that is a member of the XPF family of structure-specific endonucleases, which share a highly conserved motif, ERKX3D, at the catalytic site (Boddy et al. 2001; Lilley and White 2001; Ciccia et al. 2003; Ogrunc and Sancar 2003; Hollingsworth and Brill 2004). It is believed that Mus81 forms a partner with Eme1 in fission yeast or human (or Mms4 in budding yeast), cleaves a variety of fork structures that may arise during RF arrest or recombination, and rescues the stalled fork through a DSB (De Laat et al. 1998; Wood 1997; Boddy et al. 2001; Abraham et al. 2003). Several in vivo and in vitro studies have indicated that the primary role for MUS81 is to resolve HJs (Boddy et al. 2001; Chen et al. 2001; Doe et al. 2002; Bastin-Shanower et al. 2003; Gaillard et al. 2003; Odagiri et al. 2003; Blais et al. 2004; Gaskell et al. 2007). However, biochemical studies showed that Mus81–Eme1 cleaves four-way DNA junctions that contain an exposed 5′ DNA strand end at or close to the junction crossover point. For instance, the eukaryal MUS81 complex most efficiently cleaves nicked HJs (nHJs) and D-loops, followed by 3′ flaps and RFs (Gaillard et al. 2003; Osman et al. 2003; Whitby et al. 2003; Fricke et al. 2005). The eukaryal MUS81 complex is known to cleave the four-way HJs by a coupled “nick and counternick” mechanism, whereby the cleavage introduced on one strand is followed by a second cleavage at the opposite strand with a larger rate enhancement of the second resolving cut (Gaillard et al. 2003). The MUS81 complex recognizes the branched DNA intermediates and typically cleaves 3–6 base pairs (bp) of the 5′ regions of the junction crossover point, and the junction specificity is conserved among different organisms (Bastin-Shanower et al. 2003; Roberts and White 2005a).
Several observations have suggested that Mus81 plays an important role in the repair of “replicative-specific damages” that arise during the progression of the RFs or during replication restarts. (1) Genetic analysis in yeast showed that mus81, eme1, and mms4 mutants exhibit hypersensitivity to various agents that cause replication arrests, but not to ionizing radiation (Boddy et al. 2000; Doe et al. 2002; Bastin-Shanower et al. 2003; Hiyama et al. 2006); (2) immunofluorescence indicated that Mus81 localizes to regions of DNA damage in human S-phase cells (Gao et al. 2003); and (3) deletion of Mus81 (or Mms4) and Sgs1 (Top3) DNA helicase in yeast led to synthetic lethality, suggesting a functional link between Mus81 and Sgs1 helicase in the late steps of recombination (Boddy et al. 2000; Kaliraman et al. 2001; Mullen et al. 2001). Furthermore, Mus81 physically interacts with Cds1 (Rad53/Chk2) in fission yeast and Rad54 in budding yeast, which implicated potential roles for the MUS81 complex in cell cycle checkpoints and recombination repair, as well as replication processes (Boddy et al. 2000; Kai et al. 2005). Loss of Mus81, MMS4, or Eme1 in yeast leads to meiotic failure and severe reductions in spore viability (Boddy et al. 2001; de los Santos et al. 2001; Kaliraman et al. 2001).
Despite accumulating evidence that Mus81 and Eme1 play critical roles in the processing of aberrant RF structures, the molecular mechanisms underlying the recognition and cleavage of substrate DNA remain elusive. The formation of a complex between Mus81 and Eme1 is essential for the processing of branched DNA intermediates during replication (Boddy et al. 2001; Fu and Xiao 2003). However, it is not yet understood how eukaryal Mus81 specifically binds to Eme1. The structures of Aeropyrum pernix (Ap) XPF and Pyrococcus furiosus (Pf) Hef nuclease were elucidated recently (Nishino et al. 2003, 2005; Newman et al. 2005). These proteins contain a central nuclease domain and two pairs of helix–hairpin–helix (HhH) motifs at the C-terminal region. It has been speculated that the overall domain organizations of Mus81 and Eme1 may be different from that of archaeal XPF or Hef since the sequence alignment analysis predicted the presence of only one HhH motif in the C-terminal region of eukaryal Mus81 and no HhH motif in the C-terminal region of eukaryal Eme1 (Ciccia et al. 2003; Ogrunc and Sancar 2003; Roberts and White 2005b; Hartung et al. 2006). It is unclear how the MUS81 complex achieves a “nick and counternick” mechanism to produce linear duplex products and preferentially selects nHJs over other substrates. It is also unclear how the MUS81 complex specifically recognizes its cleavage site from the junction. In addition, despite the importance of Eme1 in the maintenance of genomic stability, the primary role of Eme1 remains unresolved. In this study, we provide answers to these important questions using an integrated approach of structural biology and biochemistry.
Results
Rationale and structure determination
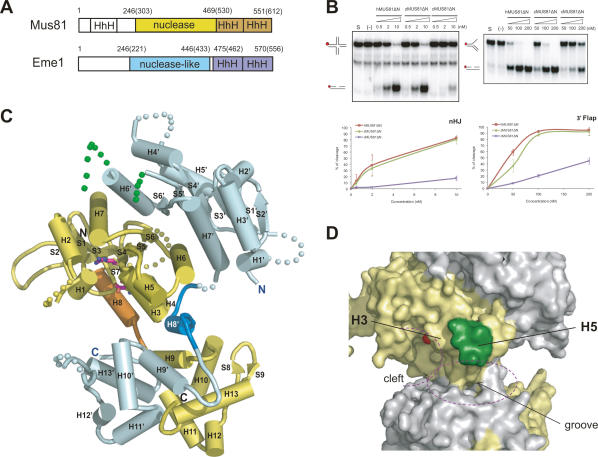
Coexpression of full-length human Mus81 and Eme1 in Escherichia coli leads to the aggregation of proteins, and only a small quantity of proteins can be purified. To isolate a protein complex suitable for crystallization, we constructed a truncated form of human Mus81 (residues 246–551; hMus81ΔN) and Eme1 (residues 246–570, hEme1ΔN) in which the N-terminal region was removed from both proteins (Fig. 1A).
Figure 1.
Structural and functional features of the cMUS81ΔN complex (A) Schematic representation of Mus81 and Eme1. Residue numbers of zMus81 are shown in parentheses. The colored region represents the construct used for crystallization. (B) Comparison of nuclease activities of the human, zebrafish, and chimeric MUS81ΔN complexes. The nHJ (left) or 3′ flap (right) DNA substrates (62.5 nM) were incubated for 60 min at 37°C with increasing amounts of each protein (nanomolar) as indicated above. Reactions with GST protein (10 nM and 200 nM for nHJ and 3′ flap) used for negative control (−) are shown in the second lane. Reaction mixtures were analyzed by 10% native PAGE. (Top) Schematic diagrams of each substrates and product DNA structures with a 32P-labeled 5′ end (red circle) are displayed on the left side. (Bottom) The total amount of cleavage products was quantified by PhosphorImager analysis and expressed as a percentage of total radiolabel. Results are shown with means ± standard error of the mean (SEM) from three independent experiments. (The quantized data are also shown in Fig. 5B and Supplemental Figs. 1, 5, and 7C,D). (C) The overall structure of the cMUS81ΔN complex determined at 2.7 Å resolution. zMus81ΔN is colored in yellow, with the active site residues and the interdomain linker (helix H8) between the nuclease and HhH2 domain shown in magenta and orange, respectively. hEme1ΔN is colored in cyan, a disordered 36R linker is shown in dotted green, and the interdomain linker (H8′) is shown in blue. (D) Surface representation displaying the deep cleft and a central groove of the cMUS81ΔN complex. The region that functions for the bump (helices H3 and H5) is marked and colored in green (helix H5). The active site cleft and groove are marked.
Full-length hMus81–Eme1 was shown previously to cleave various DNA substrates, including RFs and 3′ flap DNA, but not splayed-arm DNA (Constantinou et al. 2002; Ciccia et al. 2003). In our kinetic and activity assays, the hMus81ΔN–Eme1ΔN complex (hMUS81ΔN) cleaves nHJs, RFs, and 3′ flap DNA as efficiently as that of full-length hMUS81, which suggests that the removal of the N-terminal regions of hMus81 and hEme1 did not affect the nuclease activities of the hMUS81 complex (Table 1; Supplemental Fig. 1). Furthermore, our analysis shows that the hMUS81ΔN complex cleaves nHJs more efficiently than the RF or 3′ flap substrate (Table 1; Supplemental Fig. 1). When we reduce the assay time to 30 min, the results of nuclease activity assays were similar to those with reactions for 60 min (data not shown). In addition, while the hMUS81ΔN complex can cleave a regressed leading strand replication fork (RLe), it does not adequately process the regressed lagging strand replication fork (RLa), double flap (DF), splayed-arm, and 5′ flap substrates (Supplemental Fig. 1).
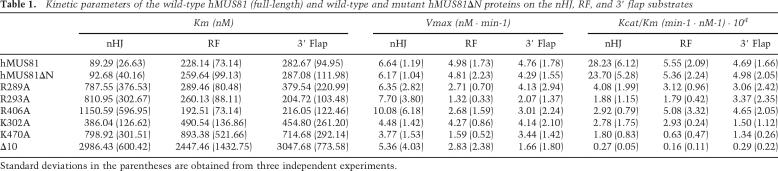
Table 1.
Kinetic parameters of the wild-type hMUS81 (full-length) and wild-type and mutant hMUS81ΔN proteins on the nHJ, RF, and 3′ flap substrates
Standard deviations in the parentheses are obtained from three independent experiments.
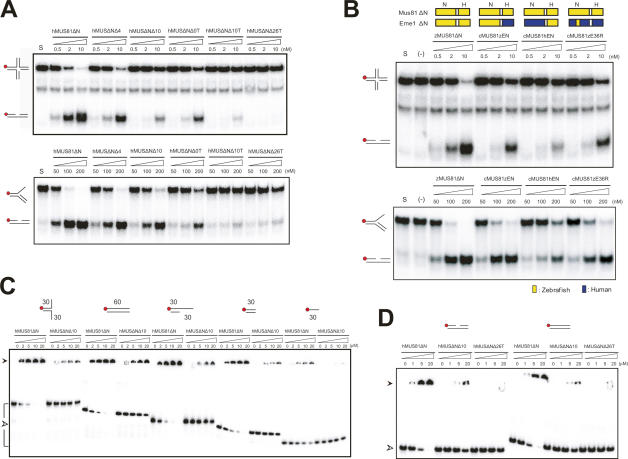
Initially, we obtained the crystals of the hMUS81ΔN complex. The crystals belong to space group P3121 with a = b = 85.8 Å, c = 176.4 Å, and contained one complex molecule in an asymmetric unit. Although the crystals exhibited an anisotropic problem that showed reasonable diffraction qualities in only one direction and diffracted poorly (∼8 Å) in another direction, we managed to collect the data at 3.5 Å resolution. However, because of the weak anomalous signals at resolution shells between 3.5 and 5.5 Å, we could not determine the structure. To overcome this problem, we attempted to crystallize Mus81 and Eme1 homologs from other species. Zebrafish Mus81ΔN (zMus81ΔN) and Eme1ΔN (zEme1ΔN) share 61% and 47% sequence identity with those of hMus81ΔN and hEme1ΔN, respectively, and the complex displays the same substrate preference compared with that of hMUS81ΔN (Fig. 1B; Supplemental Figs. 1, 2). The overall nuclease activities of zMUS81ΔN (residues 303–612 in zMus81ΔN, and 221–556 in zEme1ΔN) are comparable with those of hMUS81ΔN on a variety of substrates.
Although we could not obtain the crystal of the zMUS81ΔN complex, we successfully grew the crystals of chimeric Mus81ΔN–Eme1ΔN (cMUS81ΔN), in which zMus81ΔN was combined with hEme1ΔN. Despite of the different crystallization conditions, the space group and unit cell dimensions are identical to those of hMUS81ΔN. The crystals of the cMUS81ΔN complex diffracted to 2.7 Å resolution and we successfully determined the structure of this complex (Supplemental Table 1). The nuclease activities of this cMUS81ΔN complex on a variety of substrates are significantly reduced compared with those of zMUS81ΔN or hMUS81ΔN (Fig. 1B). Interestingly, upon replacement of an intradomain linker intervening between the residues at 368 and 403 (36R linker) in a nuclease-like domain of hEme1 to the corresponding region of zEme1, the nuclease activities of cMUS81ΔN are recovered to a level comparable with those of zMUS81ΔN (see below). Finally, using the structure of cMUS81ΔN, we also determined the structures of hMUS81ΔN and a 36R linker mutant of cMUS81ΔN [cMUS81ΔN(zE36R)] in which the nuclease activity is significantly recovered compared with cMUS81ΔN, at 3.5 Å and 2.7 Å resolutions, respectively (Supplemental Fig. 3; Supplemental Table 1).
Overall structure of the MUS81ΔN complex
The overall structures of hMUS81ΔN, cMUS81ΔN, and cMUS81ΔN(zE36R) are virtually identical, with root mean squares deviation (RMSD) values of 1.0–1.4 Å for 457 aligned Cα atoms (Supplemental Fig. 3). Because of the limited resolution and accuracy of the hMUS81ΔN structure compared with those of the structure of the cMUS81ΔN complex, we will focus primarily on the description of the cMUS81ΔN structure throughout the text. Both zMus81ΔN and hEme1ΔN are formed with an α/β nuclease domain at the N-terminal region and the two repeats of the HhH motif at the C-terminal region (HhH2 domain) (Fig. 1C; Supplemental Fig. 2). In zMus81, the two domains are connected by a helix H8 in a nuclease domain and a loop with four residues, thus forming a compact structure, whereas in hEme1, they are connected by an extended linker with a helix H8′ and two loops, forming a more extended structure. Although each of the nuclease domains and HhH2 domains of zMus81 and hEme1 are similar, the overall structures of zMus81 and hEme1 are notably different because of the difference in the linker regions of the two proteins and form an asymmetric dimer.
Figure 1C shows the overall arrangement of the cMUS81ΔN complex, in which the nuclease domain of zMus81 is surrounded by a nuclease-like domain of the hEme1 and HhH2 domains of cMUS81ΔN. While no direct contact is observed between the nuclease domain and the HhH2 domain of zMus81, the nuclease domain of zMus81 is packed tightly against the nuclease-like domain, a linker helix, and the HhH2 domain of hEme1 (Fig. 1C; Supplemental Fig. 3). The nuclease domain of zMus81 is formed with a seven-stranded β-sheet surrounded by three helices on both sides (H2, H7, and H6′ on one side, and H8, H4, and H6 on another side; H denotes a helix of Mus81, and H′ denotes a helix of Eme1). The nuclease-like domain of Eme1 is formed with a six-stranded sheet, and helices H6′, H4′, and H5′ are packed on one side of the sheet and H3′, H7′, and H6 are packed on another side of the sheet. The structurally similar nuclease domain of Mus81 and nuclease-like domain of hEme1 (RMSD of 2.5 Å for 131 Cα atoms) interact through two helices (H6 and H7) and a strand (S6) in a pseudo-twofold symmetry.
The C-terminal domains of both Mus81 and Eme1 are formed with five helices (H9–H13) with a pair of HhH motifs. Mus81 contains an additional hairpin with two strands, and the two structurally similar HhH2 domains (RMSD of 1.4 Å for 79 Cα atoms) are arranged in a pseudo-twofold symmetry. The domains interact through three helices (H9, H11, and H13) and three loops (H8–H9, H10–H11, and a C-terminal loop).
A shallow groove with a height of 10 Å and a depth of 5 Å, which is sufficient to accommodate a ssDNA, runs across the central region between the nuclease and the HhH2 domains of the cMUS81 complex (Fig. 1D). At each end of the groove, a deep cleft with a diameter of ∼15 Å is formed between the nuclease domain of zMus81 and the HhH2 domain of hEme1, and between the nuclease-like domain of hEme1 and the HhH2 domain of zMus81. The top wall of the central groove is formed by helices H3 and H5 and a loop H5–H6 of Mus81, and the bottom wall is formed by helices H9′ and H10′ of Eme1 (Fig. 1D). We presume that the binding of substrate DNA is achieved, in part, by this central groove and a deep cleft of Mus81 (see below).
One of the interesting features of the cMUS81ΔN complex structure is the 36R linker that connects strand S5′ and helix H6′ of Eme1. This region is disordered in both structures of the hMUS81ΔN and cMUS81ΔN complexes, and appears to be highly flexible, as indicated by the susceptibility to protease (Supplemental Fig. 4). An equivalent region is not found in the structure of Mus81 and, as will be described later, this region plays a critical role in substrate recognition, as the removal of this region abrogates the DNA-binding and nuclease activity of the hMUS81ΔN complex.
The largest differences between the structures of hMUS81ΔN and cMUS81ΔN are observed near the two loops S3–S4 and H7–S7. Since these regions are distant from the active site, it is unlikely that these structural differences contribute to the differences of nuclease activities between these two complexes.
Comparison with other structures
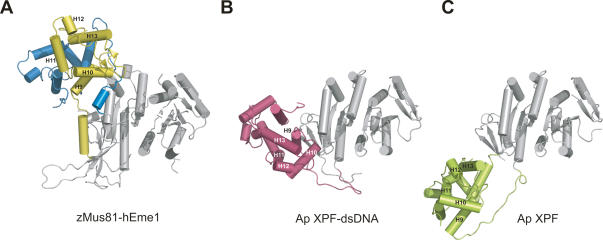
The tertiary and quaternary structures of the nuclease domain of cMUS81 closely resemble those of other nucleases. For example, the Cα backbone atoms of Mus81 or Eme1 can be superimposed very well with those of Pf Hef (1.7 Å for 131 atoms), Ap XPF (1.9 Å for 130 atoms), and the central domain of ERCC1 (2.2 Å for 118 atoms), with RMSD values of 1.7–2.2 Å for 118–130 atoms (Nishino et al. 2003; Newman et al. 2005; Tsodikov et al. 2005). Although the nuclease domains of both zMus81 and hEme1 are similar to those of the members of the XPF family, hEme1 is relatively more similar to these proteins. In zMus81, the linker helix H8, surrounding strands (S1, S3, and S4), and loops (N terminus, S3–S4) are either absent or significantly shortened in other nuclease domains (Figs. 1C, 2). The HhH2 domains of zMus81 and hEme1 are also very similar to the equivalent domain structures compared above, with RMSD values of 2.0–2.3 Å for 57–64 Cα atoms. Interestingly, the structures of the HhH2 domains of zMus81 and hEme1 resemble each other most closely (1.4 Å) compared with other HhH2 domains listed above, whereas the nuclease domains between Mus81 and Eme1 are the most distantly related compared with other XPF family members.
Figure 2.
Structural comparison between cMUS81ΔN and Ap XPF. Overall structures of the cMUS81ΔN complex (A), the Ap XPF-dsDNA complex (B), and apo Ap XPF (C) are shown. The nuclease domains (gray) from three structures are in the same orientation, and the rest of the structures including the HhH2 domains of each structure are colored yellow and blue (zMus81ΔN and hEme1ΔN), pink (Ap XPF-dsDNA) and green (apo Ap XPF). Equivalent secondary structures among three structures are numbered from H9 to H13.
The overall arrangement of the nuclease domain and HhH2 domain in MUS81 is significantly different from that of apo Ap XPF or Ap XPF bound to DNA, which is the only other nuclease–HhH2-linked structure at present (Fig. 2; Newman et al. 2005). When the nuclease domain is superimposed on the equivalent domain of DNA-bound Ap XPF, the HhH2 domain is rotated and twisted by 45° and 30°, respectively. Compared with that of apo Ap XPF, the HhH2 domain of cMUS81 is rotated by 60°.
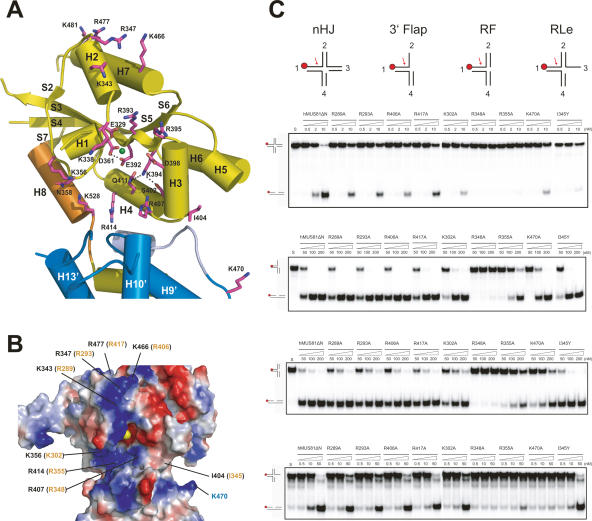
Structure of an active site
Mus81 shares a GDXnERKX3D signature motif with the XPF family members. These conserved residues are located at the center of the deep cleft of zMus81, which is formed by a helix (H8) and two loops (H1–H2 and S2–S3) on the left, a helix (H3) and loops (H5–H6, H7–S6, and H3–H4) of Mus81 on the right, and a helix (H13′) and loops (H12′–H13′ and H9′–H10′) of Eme1 at the bottom (Fig. 3A,B). The catalytically important residues, Glu392 and Asp398, are positioned at the bottom of the pocket. The side chain of Glu392 is within the H-bond distance of Asp361. The side chain of Lys394 interacts with Ser402 and Gln411, whereas the side chains of Arg393 and Arg395 are exposed to the surface. In addition to these residues, a number of basic residues are clustered around the cleft. Lys338, Lys343, Arg347, Lys466, Arg477, and Lys481 are present on the top, outer surface of the cleft, and Lys356, Asn358, Arg407, Gln411, Arg414, and Lys528 are located in the inner side of the cleft (Fig. 3A,B). The side chains of these residues are exposed to the surface, raising the possibility that they might be involved in DNA binding. To test this possibility, we mutated these residues to alanine and analyzed their nuclease activities on various DNA substrates, including nHJ, 3′ flap, RF, and RLe substrates (Fig. 3C; Supplemental Fig. 5). For the nuclease activity analysis, we used hMUS81ΔN protein rather than cMUS81ΔN protein, because it exhibits better nuclease activity. Mutations on the residues including Arg289 (Lys343 of zMus81), Arg293 (Arg347), Arg406 (Lys466), and Arg417 (Arg477) that were located at the top, outer surface of the deep cleft of Mus81 did not affect the nuclease activities on 3′ flap and RF substrates, which suggested that this region is not important for the cleavage of 3′ flap and RF substrates. However, these mutant proteins exhibit significantly decreased activities on an nHJ compared with those of wild-type hMUS81ΔN, which suggests that the flexible arm 3 of an nHJ might be involved in the interaction with residues on this surface region (Fig. 3C; Supplemental Fig. 5). In contrast, the cleavage activities of these mutant proteins on the RLe substrate are very similar to those of the wild-type hMUS81ΔN protein. Since the arm 3 of RLe is a flexible ssDNA, we presume that the arm 3 of a flexible four-way branched DNA must be a well-structured DNA to specifically interact with the surface patch of the cleft (Fig. 3C).
Figure 3.
Active site structure of cMUS81ΔN. (A) Active site of the cMUS81ΔN complex. The side chains of zMus81 are colored in magenta, and the green sphere indicates modeled Mn++ from the structure of the Hef nuclease domain (PDB ID: 1J25). Oxygen and nitrogen atoms are shown in red and blue, respectively. The dotted black lines indicate hydrogen bonds. (B) Electrostatic surface representation of active site in the cMUS81ΔN complex. Basic and acidic surfaces are colored in blue and red, respectively. Mn++ (yellow) was modeled from the structure of the Hef nuclease domain (PDB ID: 1J25). Residue numbers and positions that are mutated in our analysis are shown in black (zMus81) and blue (hEme1). The residues that are equivalent in hMus81 are shown in orange. (C) Cleavage activities of active site mutant proteins of hMUS81ΔN on branched DNA substrates. (Top) A schematic of each substrate is shown with a red circle to indicate the position of its radiolabel at the 5′ end of arm 1. A cleavage site is shown with a red arrow. The nHJ, 3′ flap, RF, and RLe DNA substrates were incubated with increasing amounts (nanomolar) of wild-type or point mutants as indicated in the top lane. For the activity assays, native or mutant hMUS81ΔN proteins were used and reaction products were analyzed on a 10% native gel. Schematic diagrams of DNA structures with a 32P-labeled 5′ end (red circle) are shown at the left. Reaction mixture in the absence of native or mutant hMUS81ΔN is shown on the left side of the gel and is labeled S.
To further analyze if these surface residues are involved in substrate binding, we measured the kinetic constants of Arg289Ala, Arg293Ala, and Arg406Ala mutants and compared them with the values of the wild-type hMUS81ΔN complex (Table 1). The hMUS81ΔN complex exhibits about threefold decreased Km on an nHJ compared with that on RF or 3′ flap DNA, which suggests that the hMUS81ΔN complex binds more tightly to the nHJ compared with RF or 3′ flap DNA. While the Km values of three hMUS81ΔN mutant proteins on an nHJ are increased by ninefold to 13-fold compared with that of the wild-type hMUS81ΔN, the Km values of the surface mutant proteins on RF or 3′ flap are similar to that of the wild-type hMUS81ΔN. These data suggest that the three residues—Arg289, Arg293, and Arg406—are specifically involved in the interactions with the flexible arm of the nHJ.
In contrast to these surface residues, mutations of the residues that are in a deep cleft (Lys302 and Arg355, Lys356 and Arg414 of zMus81) significantly affect the nuclease activities on both nHJ and 3′ flap substrates. Two hMus81ΔN mutants on a central groove—Arg348Ala (Arg407 of zMus81) and Ile345Tyr (I404 of zMus81)—also nearly abrogated the nuclease activities of hMUS81ΔN. The Km values of the mutant proteins in which each of these residues were replaced are increased on nHJ, 3′ flap, and RF by threefold to ninefold compared with the wild type hMUS81ΔN, which suggests that the central groove is involved in the recognition of various substrates. In archaeal XPF, the equivalent residues are either hydrophobic or acidic residues. In hEme1ΔN, the equivalent residues of ERK in Mus81 are hydrophobic residues, LVL (313–315), and are packed by the hydrophobic environment.
Interface between Mus81 and Eme1
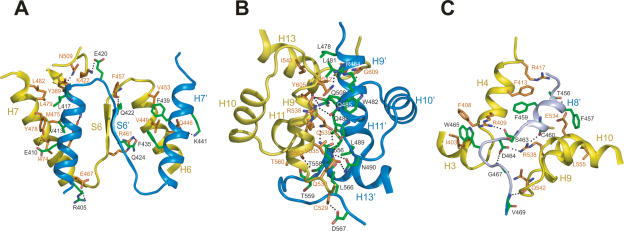
Mus81 interacts with Eme1 primarily through three interfaces involving a buried surface area of 7094 Å (Fig. 4A–C). These interfaces include one between the nuclease domain of zMus81 and the nuclease-like domain of hEme1 (buried surface area of 2240 Å), HhH2 domains between zMus81 and hEme1 (2446 Å), and one between the nuclease and HhH2 domains of zMus81 and a linker region of hEme1 (2408 Å). The three interfaces appear to be equally important for dimerization. The nuclease interface is formed by interactions between helices H7 and H6′, between helices H7′ and H6, and between strands S6 and S6′. This interface is formed primarily through hydrophobic interactions (Fig. 4A). Val413 and Leu417 of Eme1 make van der Waals contacts to the side chains of Tyr389, Met475, Tyr478, and Leu479 of Mus81. Phe435 and Phe439 of Eme1 also form hydrophobic interactions with Val449 and Val453 of Mus81. In addition, two ion pairs and seven H-bonds further stabilize this interface.
Figure 4.
Interfaces between zMus81ΔN and hEme1ΔN. Close-up view of the interface between the nuclease domain of zMus81ΔN and the nuclease-like domain of hEme1ΔN (A), between the HhH2 domain of zMus81ΔN and the HhH2 domain of hEme1ΔN (B), and between the linker region of zMus81ΔN and the linker region of hEme1ΔN (C). The side chains of zMus81ΔN and hEme1ΔN are colored in orange and green, respectively. Oxygen and nitrogen atoms are shown in red and blue, respectively. The dotted lines indicate intermolecular hydrogen bonds and ion pairs between zMus81 and hEme1.
In the HhH2 interface, helices H9, H11, and H13 of Mus81 interact with helices H9′, H11′, and H13′ of Eme1 (Fig. 4B). In addition, the C-terminal loops of Mus81 and Eme1 further stabilize the interface by interacting with helix H9′ of Eme1 and helix H9 and loop H8–H9 of Mus81, respectively. Both the H-bonds (12 H-bonds) and the van der Waals interactions contribute significantly to this interface. In Pf Hef and Ap XPF, hydrophobic interactions are predominant in the HhH2 interface (Newman et al. 2005; Nishino et al. 2005).
The linker region of Eme1 forms a stable helix. The two loops flanking the H8′ helix are also well structured, and are packed against the polar environment formed by the nuclease and HhH2 domains of Mus81 through 14 H-bonds and an ion pair (Fig. 4C). The H8′ helix of Eme1 interacts with helices H4, H9, and H10 of Mus81. The key features include ring stacking between the Phe459 ring of Eme1 and the Phe413 ring of Mus81, and between the Phe457 ring of Eme1 and the side chain of Arg533 of Mus81. The H8′–H9′ loop of Eme1 interacts with loops H3–H4, and helices H3 and H4. In particular, Trp465 of Eme1 is surrounded by Ile403, Phe408, and Arg409 of Mus81.
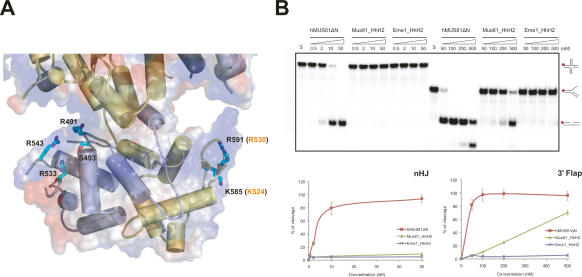
Importance of the HhH2 domain in substrate binding
Biochemical analysis of archaeal Hef, XPF, and human ERCC1 revealed that dsDNA or ssDNA binds to their HhH2 domains (Newman et al. 2005; Nishino et al. 2005; Tsodikov et al. 2005). Furthermore, the crystal structure of the Ap XPF–DNA complex indicated that the minor groove of DNA is recognized by the two hairpin regions of the HhH2 domains (Newman et al. 2005). Thus, we examined whether the HhH2 domains of Mus81 and Eme1 also contribute to DNA binding. We superimposed the structure of the HhH domain of the Ap XPF–DNA complex onto the corresponding portion of zMus81 or hEme1. In Ap XPF, positively charged residues in the two hairpin regions are involved in the DNA binding. The corresponding hairpins are much longer in zMus81 and hEme1, and hydrophobic residues are located at equivalent positions in zMus81 and hEme1. Nevertheless, several basic residues are present in these hairpin regions (loop H9–H10 and strands S8 and S9), and thus we constructed two mutant proteins in which the residues of hMus81ΔN or hEme1ΔN were mutated simultaneously (Fig. 5A). We replaced Lys524 and Arg530 (Lys585 and Arg591 of zMus81) in strands S8 and S9 of hMus81 simultaneously with glutamate. We also simultaneously replaced Arg491, Ser493, Arg533, and Arg543 in hEme1 with glutamate. We used wild-type and mutant hMUS81ΔN proteins for the analyses of nuclease activity (Fig. 5B). Judging from circular dichroism analysis, these multiple mutations did not perturb the overall structures of hMus81ΔN and hEme1ΔN (Supplemental Fig. 6). Each mutation nearly abrogated the nuclease activity of hMUS81ΔN on nHJ and 3′ flap DNA. Because of the low nuclease activities of these mutant proteins, we could not determine their kinetic parameters (data not shown). Since the mutated residues are distant from the catalytic site, it is likely that these residues are involved in the binding of a substrate DNA (Fig. 5B).
Figure 5.
Analysis between HhH2 domains of cMUS81 and DNA substrates. (A) Structure of the two HhH2 domains of cMUS81. Secondary structures of zMus81ΔN are colored in yellow, and hEme1ΔN are light blue. The side chains of zMus81ΔN and hEme1ΔN that are mutated to glutamate in our analysis are colored in cyan. Equivalent residue numbers of hMus81ΔN are shown in orange. (B, top) Nuclease activities of wild-type and HhH2 mutant proteins of hMus81ΔN and hEme1ΔN. For Mus81_HhH2, Lys524 and Arg530 of hMus81ΔN were replaced with Glu simultaneously. For Eme1_HhH2, Arg491, Ser493, Arg533, and Arg543 of hEme1ΔN were replaced with Glu simultaneously. The nHJ and 3′ flap DNA substrates reacted with increasing amount of wild-type or HhH2 domain mutant hMUS81ΔN. Numbers at the top of the lanes indicate protein concentrations (nanomolar). DNA structures with a 32P-labeled 5′ end (red circle) are shown in the right column. (Bottom) The total amount of cleavage products was quantified by PhosphorImager analysis and expressed as a percentage of total radiolabel.
The 36R intradomain linker of Eme1 is essential for DNA binding
The limited proteolytic digestion analysis suggested that residues between 368 and 403 in hEme1 are connected by a highly flexible linker, which we presumed to be a loop region (Supplemental Fig. 4). Even without this 36R linker, zMus81ΔN and hEme1ΔN (or hMus81ΔN and hEme1ΔN) form a stable complex, which suggests that this linker is not critical for the dimerization between Mus81ΔN and Eme1ΔN. However, the ternary complex did not display any nuclease activities (Fig. 6A; Supplemental Fig. 7C). Interestingly, removal of this linker dramatically reduced the DNA-binding activity in EMSA (Supplemental Fig. 7A). We assessed the importance of 36R linker by changing the length of the linker. Figure 6A shows that the nuclease activity of hMUS81ΔN decreased significantly when the length of the linker was decreased. Shortening the 36R linker length by four or 10 residues decreased the nuclease activities of hMUS81ΔN on the nHJ and 3′ flap DNA (Fig. 6A; Supplemental Fig. 7C). Cleavage of the 36R linker of hEme1 into two parts also significantly decreased the nuclease activity and the DNA-binding affinity of the hMUS81ΔN complex. Reducing the size of this cleaved linker of hEme1 by 10 or 26 residues abrogated the DNA-binding and nuclease activities of hMUS81ΔN on 3′ flap and nHJ substrates, demonstrating the importance of this 36R linker in DNA binding and cleavage.
Figure 6.
Roles of the 36R intradomain linker of Eme1. (A) Nuclease activity of wild-type hMUS81ΔN and the 36R linker deletion mutant proteins. The 60mer nHJ and 3′ flap DNA substrates (62.5 nM) were incubated for 60 min at 37°C with increasing amounts (nanomolar) of wild-type or mutant hMUS81ΔN. The top panel shows protein constructs with the numbers of deleted residues and cleavage of a 36R linker. For instance, hMUSΔNΔ4 represents a mutant in which four residues of the 36 linker are deleted, and hMUSΔNΔ10T (ternary complex) represents a mutant in which the 36 linker is cleaved into two parts and 10 residues are deleted. DNA structures with a 32P-labeled 5′ end (red circle) are shown at the left side. Reaction products were analyzed by 10% native polyacrylamide gel. (B) DNA cleavage assay of chimeric Eme1 mutants. A schematic diagram shows the construct of each mutant. (Top) Components from zebrafish Mus81 (or Eme1) and human Mus81 (or Eme1) are colored in yellow and blue, respectively. cMUS81zEN indicates an Eme1 construct with a zebrafish nuclease domain (zEN) with a human HhH domain, and cMUS81hEN represents an Eme1 construct with a human nuclease domain (hEN) and a zebrafish HhH domain. cMUS81(zE36R) indicates hEme1 mutant in which 36R linker is substituted with an equivalent region of zEme1. (C) Binding assays between 36R linker deletion mutant proteins and various DNA substrates. Substrate structures used for analysis are shown on the top. Numbers indicate the lengths of nucleotides. Red circles show the 32P-labeled 5′ DNA end. Reaction mixtures were analyzed on 8% native gel. (D) Binding assays between 36R linker deletion mutant proteins and a duplex DNA or a nicked DNA. Substrate structures used for analysis are shown at the top. Red circles show the 32P-labeled 5′ DNA end. Reaction mixtures were analyzed on 8% native gel.
Since it is possible that the 36R linker may form an arch through which DNA might pass, as in 5′ flap endonuclease-1 (Hwang et al. 1998), we attached the biotin-streptavidin group at the 3′ end of the branched region of an nHJ substrate and measured the cleavage activity of the hMUS81ΔN complex (Supplemental Fig. 7B). The hMUS81ΔN complex cleaves the biotin-streptavidin-attached nHJ substrate as efficiently as an nHJ, which suggests that the 36R linker is unlikely to form an arch for the passage of a substrate.
Because the cMUS81ΔN complex displays significantly decreased nuclease activities compared with those of zMUS81ΔN on various substrates, we hypothesized that hEme1ΔN might affect the cleavage activity of cMUS81ΔN. To examine which part of hEme1ΔN was critical to the reduced activity of the cMUS81ΔN complex compared with zMUS81ΔN, we first constructed two chimeric Eme1 mutant proteins (Fig. 6B; Supplemental Fig. 7D). In one construct (cMUS81zEN: zEme1 at the N-terminal region of the nuclease-like domain), we fused the N-terminal portion (residues 221–438) of zEme1 to the C-terminal portion (residues 452–570) of hEme1. In another (cMUS81hEN: hEme1 at N-terminal region of the nuclease-like domain), the N-terminal portion of hEme1 (246–451) was fused to the C-terminal portion of zEme1 (439–556). While cMUS81zEN exhibited significant nuclease activities against nHJ and 3′ flap DNA compared with zMUS81ΔN, cMUS81hEN (hEme1 at the N-terminal region) shows activities similar to those of cMUS81ΔN, which suggested that the critical region required for the decreased DNA-binding and nuclease activity of cMUS81ΔN is present in the N-terminal portion of hEme1 (Figs. 1B, 6B). We then replaced the 36R linker of hEme1 with the equivalent region (residues 345–390) of zEme1 [cMUS81(zE36R)] and measured the nuclease activities of this cMUS81ΔN linker mutant. The activities of this mutant were improved significantly, which suggests that the 36R linker plays a key role in substrate-binding and nuclease activity (Fig. 6B; Supplemental Fig. 7D).
Possible role of the 36R intradomain linker of Eme1
Since our data revealed that the 36R linker is critical in recognizing substrate DNA, we wondered which portion of the substrate DNA interacts with the 36R linker. First, kinetic analysis revealed that the Km values of the hMUS81ΔNΔ10 complex on nHJ, RF, and 3′ flap substrates are 10- to 35-fold higher than those of the wild-type hMUS81ΔN protein (Table 1). We then compared the binding affinities of hMUS81ΔN and the hMUS81ΔN mu-tant in which the 36R linker region was partly deleted on several DNA substrates, including nHJ, 3′ flap, nicked duplex, splayed-arm, 3′-overhang, single-stranded, and duplex DNA. The hMUS81ΔN complex binds to branched, nicked, and duplex DNA with similar affinity (Fig. 6C; Supplemental Fig. 7A). Also, compared with hMUS81ΔNΔ10 or hMUS81ΔNΔ26T mutant, hMUS81ΔN exhibited tighter binding to nicked DNA. These data suggest that the 36R linker is at least involved in dsDNA binding (Fig. 6D). Third, while hMUS81ΔN could interact with the splayed-arm DNA, the hMUS81ΔNΔ10 mutant protein bound poorly to splayed-arm DNA (Fig. 6C). Similarly, the hMUS81ΔNΔ10 mutant protein weakly interacted with the 3′ overhang DNA. These findings suggest that if the 36R linker of the hMUS81ΔNΔ10 mutant protein does not interact properly with the duplex region of the splayed-arm or 3′ overhang DNA, the HhH2 motif or other regions of the hMUS81ΔN complex may not interact tightly with the ssDNA region of a substrate DNA. While the hMUS81ΔN complex bound poorly to 30-nucleotide (nt)-long ssDNA, it interacted with 30-nt-long dsDNA. However, deletion of the 36R linker abrogated the interaction with 30-nt-long dsDNA, whereas it interacted with 60-nt duplex DNA, although the binding affinity was slightly decreased (Fig. 6C). Since the hMUS81ΔN mutant binds to nicked DNA and dsDNA with similar affinity, this suggests that the 36R linker is not specific for the recognition of the 5′ end of the junction. Taken together, we propose that the primary role of the 36R linker is to recognize the duplex portion of a substrate DNA and to stabilize the interaction between the MUS81ΔN complex and the substrate DNA, which appears to be required for efficient cleavage.
Discussion
In the present study, we described the structures of the cMUS81ΔN and hMUS81ΔN complexes, which are virtually identical. Previously, on the basis of sequence comparison analysis, it has been speculated that the Mus81 and Eme1 proteins might have different domain organizations from archaeal XPF family members (Ciccia et al. 2003; Ogrunc and Sancar 2003; Roberts and White 2005b; Hartung et al. 2006). However, the crystal structures of the cMUS81ΔN and hMUS81ΔN complexes reveal that both Mus81ΔN and Eme1ΔN are formed with nuclease and HhH2 domains, displaying similar domain organization to that of XPF family members. These findings demonstrated the significance of the structural conservation of these nuclease family members in the processing of branched DNA substrates. However, despite the similarities in domain organization and the structures of each nuclease and HhH2 domain with other XPF family members, the structures of the interdomain linker regions of Mus81 and Eme1 are significantly different from those of archaeal XPF, Hef, or eukaryal ERCC1 (Nishino et al. 2003, 2005; Newman et al. 2005; Tsodikov et al. 2005). While archaeal XPF, Hef, and human ERCC1 have a flexible interdomain linker, the equivalent region of Mus81 and Eme1 is formed with a helix flanked by rigid loops (Supplemental Fig. 4) and, together with differences in its surrounding structure, these differences lead to different arrangement of the nuclease and HhH domains in overall structure.
In our kinetic analysis, the hMUS81ΔN complex exhibits about fivefold higher kcat/Km value toward an nHJ over RF or 3′ flap substrate. Previously, it has been reported that the MUS81 complex from Saccharomyces cerevisiae exhibits about threefold higher kcat/Km value toward 3′ flap substrate over nHJ (Fricke et al. 2005). It is unclear why the MUS81 complex proteins from human and S. cerevisiae have different substrate preferences. One possibility is that there might be some functional differences between the MUS81 complex proteins from different species. It has been proposed that the MUS81 complex proteins from Schizosaccharomyces pombe and S. cerevisiae might have such functional differences (Bastin-Shanower et al. 2003; Gaillard et al. 2003; Osman et al. 2003). We note that the sequence identities between hMus81 and S. cerevisiae Mus81 and between hEme1 and S. cerevisiae Eme1 are only ∼22% and 13%, respectively, and there might be some correlations between the low sequence identity and the possible functional differences between the hMUS81 and S. cerevisiae MUS81 proteins.
Although Eme1 is an important component in the maintenance of genomic stability, the role of Eme1 in the MUS81 complex remains elusive. (Fu and Xiao 2003; Abraham et al. 2003). How would Eme1 contribute to the substrate cleavage through dimerization with Mus81? We assessed which portions of Eme1 were involved in substrate recognition or cleavage using chimeric Eme1 proteins. While cMUS81hEN (in which the nuclease domain of hEme1ΔN is fused to the HhH2 domain of zEme1ΔN) exhibited reduced nuclease activities like cMUS81ΔN, the nuclease activities of cMUS81zEN (in which the nuclease domain of zEme1ΔN and the HhH2 domain of hEme1ΔN are fused) and cMUS81z36R (in which a 36R linker of hEme1 is replaced with an equivalent linker of zEme1) are significantly recovered (Fig. 6B). Furthermore, EMSA suggests that the 36R linker plays a crucial role in stable binding to a substrate DNA, presumably through the recognition of the duplex portion. These findings suggest that there might be a threshold for binding affinity in terms of the efficient cleavage of branched substrates, and such affinity can be achieved only in the presence of the intact 36R linker. While the intradomain linker of hEme1 is comprised of 36 residues, the equivalent region of zEme1 is formed with 46 residues (Supplemental Fig. 2). Previous studies showed that a variant of hEme1 with a 13-residue insertion at the 36R linker exhibited abrogated nuclease activity (Blais et al. 2004). These findings suggest that there is a certain range for the length of the 36R linker of Eme1 required for stable interaction with substrates and the nuclease activities of the MUS81 complex. The 36R linker contains several basic residues that might be involved in the interactions with substrate DNA. Our data exclude the possibility that the 36R linker may form an arch through which a substrate might be threaded. Instead, we expect that the 36R linker may form a specific conformation upon binding to DNA substrates, and the cleavage of this linker might perturb the conformation of the 36R linker. Thus, we propose that one crucial role of Eme1 in the maintenance of genomic stability is to provide a stable interaction between the MUS81 complex and a DNA substrate. We note that other regions of Eme1 may also contribute to the binding and cleavage of a substrate DNA, since the replacement of the 36R linker in chimeric Eme1 did not fully recover the activities to the level of the zMUS81 complex.
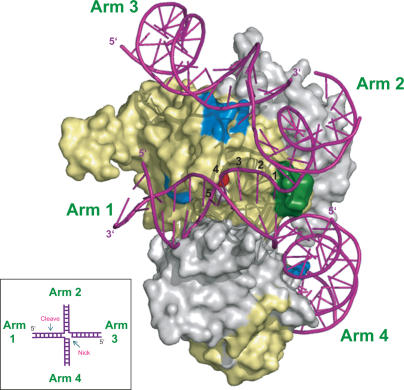
Based on the crystal structure and mutation analysis, we built a complex model between the MUS81ΔN and an nHJ (Fig. 7). It has been shown that the MUS81 complex initially cuts the junction of HJs and then cleaves the opposing strand with high efficiency, generating linear duplex strands (Gaillard et al. 2003). The nHJ is a favored substrate, whereas the HJ cannot be processed efficiently, presumably because of the restraint at the junctions between the arms of the HJ. In our model, introduction of a nick at the junction of HJ would release the restraint at the junction between arms 3 and 4 (Fig. 7). We propose that, upon cleavage, arm 3 near the nick becomes flexible and undergoes substantial movement to interact with the surface patch that contains a series of basic residues at the outer surface of an active site. These interactions might lead to optimal positioning of the junction between arms 1 and 2 into the catalytic site. It is possible that these interactions may be assisted by the possible hinge-bending movement between the nuclease and HhH2 domains of MUS81ΔN upon binding of a substrate DNA, as observed in the structure of the Ap XPF–DNA complex (Newman et al. 2005).
Figure 7.
A proposed model for the Mus81–Eme1 complex bound to an nHJ DNA substrate. A surface representation of cMUS81ΔN and a DNA substrate (magenta) is shown. The red sphere indicates modeled Mn++ from the structure of Hef nuclease domain (PDB ID: 1J25). A schematic diagram for the nHJ is shown in the inset. The modeled bases at the catalytic site are numbered.
The nHJ is a favored in vitro substrate of hMUS81 and fission yeast MUS81 over 3′ flap and RF substrates. Kinetic analysis showed that an nHJ binds to hMUS81ΔN more tightly compared with the flexible three-way branched DNA. However, upon mutation of the residues at a surface patch of hMUS81ΔN, the binding affinity of an nHJ decreased to 13-fold compared with the binding affinity to the wild-type hMUS81ΔN, whereas the binding affinities of the 3′ flap and RF substrates did not change. Furthermore, the nuclease activities of hMUS81ΔN on 3′ flap, RF, and RLe are similar, which suggests that the interaction between the flexible arm with a duplex DNA (Fig. 7, arm 3) and the surface patch outside the cleft plays a key role in substrate preference of nHJs over RF, 3′ flap, and RLe (Supplemental Fig. 1).
We showed that both HhH2 domains of Mus81 and Eme1 must bind to a substrate DNA simultaneously. The minor groove of the duplex DNA is bound to the hairpin loop of the HhH domain in the Ap XPF–DNA complex structure (Newman et al. 2005). Thus, the HhH2 domains of the Mus81 and Eme1 proteins provide the platform in which a DNA substrate binds. Furthermore, our EMSA shows that the ssDNA binds weakly to hMUS81ΔN compared with the dsDNA. Thus, the ssDNA portion of the splayed-arm, 3′-overhang, and 5′ flap substrates may not form sufficiently stable interactions with the HhH2 domains of hMUS81ΔN, which might explain why these substrates are not processed efficiently by the hMUS81ΔN complex.
The MUS81 complex cleaves several bases adjacent to the 5′ junction of a substrate DNA (Bastin-Shanower et al. 2003; Roberts and White 2005a). How would MUS81 select the specific cleavage sites? We propose that two factors are important for this selection. First, our model suggests that the substrate DNA cannot bind simultaneously to both HhH2 domains of the Mus81 and Eme1 proteins without bending, which might lead to the distortion of the cleavage sites of a substrate. It has been proposed that the substrate DNA undergoes bending and distortion upon binding to Ap XPF or Pf Hef (Newman et al. 2005; Nishino et al. 2005). However, it is unclear if the bending of the substrate DNA would be as substantial as that proposed in the archaeal XPF–DNA complex, since the length and geometry of the two hairpin loops of the HhH2 domains are somewhat different in MUS81ΔN and archaeal XPF. Secondly, the top wall of the central groove (helices H3 and H5) could function as a bump that directs the cleaved strand into the catalytic site (Figs. 1D, 7). In our model, the binding of a substrate to the HhH2 domains of MUS81ΔN, in conjunction with strand distortion, would position the junction between arms 1 and 4 into the central groove of the MUS81 complex (Fig. 7). The mutational studies demonstrated the importance of the central groove in the binding of a substrate DNA (Table 1; Fig. 3C). Subsequently, flexible arms 3 and/or 2 might collide with the top wall of the groove and undergo gross movement to interact with the surface patch and active site cleft, respectively. The distance from the bump of the wall to the catalytic residues at the active site is ∼3–5 bases (Fig. 7). Thus, the combined function of strand distortion and the blocking by the top wall might cause the unstacking of base pairs around the junction center and locate the scissile phosphodiester bond to the catalytic site, which might explain why the cleavage occurs on the several bases adjacent to the 5′ junction (Bastin-Shanower et al. 2003).
In conclusion, we described the crystal structures of both hMUS81ΔN and cMUS81ΔN, and showed the structural and functional conservation of eukaryal MUS81 to the archaeal XPF family members in branched DNA processing. We provided the structural basis of hMUS81 for the substrate preference toward nHJs over three-way branched DNA substrates. The basic residues at the surface patch of hMUS81 appear to be critical for interaction with a flexible arm near the nick of an nHJ. In addition, we identified and elucidated the biochemical function of the 36R linker, an essential component of Eme1 in the processing of branched DNA, which might explain the role of Eme1 in the maintenance of genomic stability. The MUS81 complex structure and mutational analysis described here provide an important step toward understanding the cleavage mechanism for the flexible branched DNA substrates.
Materials and methods
Construction, expression, and purification
His-tagged hEme1ΔN (residues 246–551) and hMus81ΔN (246–570) were amplified by PCR from each plasmid containing full-length hEme1 and hMus81. The truncated zebrafish Mus81 (zMus81ΔN, 303–612) and Eme1 (zEme1ΔN, 221–556) were cloned from zebrafish cDNA, and were then inserted into a pCDF-duet vector (NdeI and XhoI sites) and pET-duet vector (EcoRI and HindIII sites), respectively. hMUS81ΔN, zMUS81ΔN, and cMUS81ΔN were induced in E. coli BL21 (DE3) and purified by a Ni++-column, a cation exchange column (Mono S), and a gel filtration column (Superdex 75), respectively. The protein complex was concentrated to 12 mg/mL by ultrafiltration and stored at −80°C. A selenomethionine (Se-Met)-substituted protein complex of cMUS81ΔNΔ10 was cultured by growing the plasmids in E. coli strain B834 (DE3) in M9 minimal medium. The protein complex was purified by the same method used for the purification of native proteins.
Crystallization and data collection
Crystals of the cMUS81ΔN complex were grown by the hanging drop vapor diffusion method against a crystallization buffer containing 0.1 M Na-Acetate (pH 5.0) and 2.2–2.5 M ammonium chloride at 4°C. Crystals of the hMUS81ΔN complex were grown against a crystallization buffer containing 0.1M bis-tris propane-HCl (BTP) (pH 6.5), 1.2–1.3 M Li2SO4, 100 mM MgCl2, and 2 mM CdCl2 at 18°C. Diffraction data were collected at −170°C, with crystals flash-frozen in crystallization buffer containing 30% glycerol. Diffraction data from native crystals of cMUS81ΔN were collected at 1.0000 Å, and data from selenium-derivatized crystals of the cMUS81ΔN complex or the hMUS81ΔN complex were collected at peak absorption, 0.9794 Å, on beamline 4A at the Pohang Advanced Light Source. The cMUS81ΔN crystals formed in space group P3121 with a = 89.4 Å, b = 89.4 Å, c = 169.3 Å, and the hMUS81ΔN crystals formed in the same space group with a = 85.8 Å, b = 85.8 Å, c = 176.4 Å and contained one complex molecule in an asymmetric unit. Diffraction data integration, scaling, and merging were performed using HKL2000 (Otwinowski and Minor 1997).
Structure determination and refinement
The structure of the cMUS81ΔNΔ10 complex was determined by single anomalous scattering dispersion (SAD) method using Se-Met-derivatized crystals. The 18 selenium sites in the asymmetric unit were calculated in SHELXC and SHELXD (Schneider and Sheldrick 2002). Phase determination was carried out with SOLVE. After the flattening of solvent, a high-quality electron density map with a resolution of 2.9 Å was obtained using the RESOLVE program. The electron density was interpreted and traced using Coot (Emsley and Cowtan 2004) and was refined with CNS (Brünger et al. 1998). The structures of cMUS81ΔN, hMUS81ΔN, cMUS81ΔNΔ33T and cMUS81ΔNzE36R complex were determined by difference Fourier methods using CNS (Brünger et al. 1998).
Endouclease assays
Cleavage reaction mixtures (20 μL) contained 62.5 nM 32P-labeled substrate DNA and enzyme (as indicated) in reaction buffer (25 mM Tris-HCl at pH 8.0, 1 mM dithiothreitol, 100 μg/mL bovine serum albumin, 6% glycerol, 10 mM MgCl2). Reaction mixtures were incubated for 60 min at 37°C, and were stopped by the addition of one-fifth volume of stop mixture (2.5% SDS, 200 mM EDTA, 10 mg/mL proteinase K), followed by 30 min incubation at 37°C. Reaction products were resolved on 10% native polyacrylamide gel in 1× TBE buffer. Gels were run for 120 min at 13 Vcm−1, and the level of cleavage was then quantified by a PhosphorImager using MultiGauge version 3.0 (FujiFilm). In all analyses, we performed three independent experiments to measure the nuclease activities.
Acknowledgments
We thank Chul Hee Kim for providing zebrafish cDNA, Jin Seok Kim and Chang Wook Lee for help with protein purification, and Heung Soo Lee, Kyung Jin Kim, and Kyung Hwa Kim for help with data collection (PAL4A and PAL6C). This work was supported by funds from the National Creative Research Initiatives (Ministry of Science and Technology) and the BK21 program (Ministry of Education).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1618708.
References
- Abraham J., Lemmers B., Hande M.P., Moynahan M.E., Chahwan C., Ciccia A., Essers J., Hanada K., Chahwan R., Khaw A.K., et al. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 2003;22:6137–6147. doi: 10.1093/emboj/cdg580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastin-Shanower S.A., Fricke W.M., Mullen J.R., Brill S.J. The mechanism of Mus81–Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1–Rad10. Mol. Cell. Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais V., Gao H., Elwell C.A., Boddy M.N., Gaillard P.H., Russel P., McGowan C.H. RNA interference inhibition of Mus81 reduces mitotic recombination in human cells. Mol. Biol. Cell. 2004;15:552–562. doi: 10.1091/mbc.E03-08-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M.N., Lopez-Girona A., Shanahan P., Interthal H., Heyer W.D., Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M.N., Gaillard P.-H.L., McDonald W.H., Shanahan P., Yates J.R., Russell P. Mus81–Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crstallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chen X.B., Melchionna R., Denis C.M., Gaillard P.H., Blasina A., de Van Weyer I., Boddy M.N., Russell P., Vialard J., McGowan C.H. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- Ciccia A., Constantinou A., West S.C. Identification and characterization of the human mus81–eme1 endonuclease. J. Biol. Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- Constantinou A., Chen X.-B., McGowan C.H., West S.C. Holliday junction resolution in human cells: Two junction endonucleases with distinct substrate specificities. EMBO J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- De Laat W.L., Appeldoorn E., Jaspers N.G.J., Hoeijmakers J.H.J. DNA structural elements required for ERCC1–XPF endonuclease activity. J. Biol. Chem. 1998;273:7835–7842. doi: 10.1074/jbc.273.14.7835. [DOI] [PubMed] [Google Scholar]
- de los Santos T., Loidl J., Larkin B., Hollingsworth N.M. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics. 2001;159:1511–1525. doi: 10.1093/genetics/159.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe C.L., Ahn J.S., Dixon J., Whitby M.C. Mus81–Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fricke W.M., Bastin-shanower S.A., Brill S.J. Substrate specificity of the Saccharomyces cerevisiae Mus81–Mms4 endonuclease. DNA Repair (Amst.) 2005;4:243–251. doi: 10.1016/j.dnarep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Fu Y., Xiao W. Functional domains required for the Saccharomyces cerevisiae Mus81–Mms4 endonuclease complex formation and nuclear localization. DNA Repair (Amst.) 2003;2:1435–1447. doi: 10.1016/j.dnarep.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Gaillard P.H., Noguchi E., Shanahan P., Russell P. The endogenous Mus81–Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- Gao H., Chen X.B., McGowan C.H. Mus81 endonuclease localizes to nucleoli and to regions of DNA damage in human S-phase cells. Mol. Biol. Cell. 2003;14:4826–4834. doi: 10.1091/mbc.E03-05-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaskell L.J., Osman F., Gilbert R.J.C., Whitby M.C. Mus81 cleavage of Holliday junctions: A failsafe for processing meiotic recombination intermediates? EMBO J. 2007;26:1891–1901. doi: 10.1038/sj.emboj.7601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F., Suer S., Bergmann T., Puchta H. The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res. 2006;34:4438–4448. doi: 10.1093/nar/gkl576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W.D., Ehmsen K.T., Solinger J.A. Holliday junctions in the eukaryotic nucleus: Resolution in sight? Trends Biochem. Sci. 2003;28:548–557. doi: 10.1016/j.tibs.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Hiyama T., Katsura M., Yoshihara T., Ishida M., Kinomura A., Tonda T., Asahara T., Miyagawa K. Haploinsufficiency of the Mus81–Eme1 endonuclease activates the intra-S-phase and G2/M checkpoints and promotes rereplication in human cells. Nucleic Acids Res. 2006;34:880–892. doi: 10.1093/nar/gkj495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R.A. A mechanism for gene conversion in fungi. Genet. Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- Hollingsworth N.M., Brill S.J. The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes & Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang K.Y., Baek K., Kim H.Y., Cho Y. The crystal structure of flap endonuclease-1 from Methanococcus jannaschii. Nat. Struct. Biol. 1998;5:707–713. doi: 10.1038/1406. [DOI] [PubMed] [Google Scholar]
- Kai M., Boddy M.N., Russell P., Wang T.S. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes & Dev. 2005;19:919–932. doi: 10.1101/gad.1304305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman V., Mullen J.R., Fricke W.M., Bastin-Shanower S.A., Brill S.J. Functional overlap between Sgs1–Top3 and the Mms4–Mus81 endonuclease. Genes & Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Heyer W.-D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D.M., White M.F. The junction-resolving enzymes. Nat. Rev. Mol. Cell Biol. 2001;2:433–443. doi: 10.1038/35073057. [DOI] [PubMed] [Google Scholar]
- McGlynn P., Lloyd R.G. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 2003;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- Michel B., Grompone G., Flores M.J., Bidnenko V. Multiple pathways process stalled replication forks. Proc. Natl. Acad. Sci. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen J.R., Kaliraman V., Ibrahim S.S., Brill S.J. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. 2001. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M., Murray-Rust J., Lally J., Rudolf J., Fadden A., Knowles P.P., White M.F., McDonald N.Q. Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition. EMBO J. 2005;24:895–905. doi: 10.1038/sj.emboj.7600581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T., Komori K., Ishino Y., Morikawa K. X-ray and biochemical anatomy of an archaeal XPF/Rad1/Mus81 family nuclease: Similarity between its endonuclease domain and restriction enzymes. Structure. 2003;11:445–457. doi: 10.1016/s0969-2126(03)00046-7. [DOI] [PubMed] [Google Scholar]
- Nishino T., Komori K., Ishino Y., Morikawa K. Structural and functional analyses of an archaeal XPF/Rad1/Mus81 nuclease: Asymmetric DNA binding and cleavage mechanisms. Structure. 2005;13:1183–1192. doi: 10.1016/j.str.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Odagiri N., Seki M., Onoda F., Yoshimura A., Watanabe S., Enomoto T. Budding yeast mms4 is epistatic with rad52 and the function of Mms4 can be replaced by a bacterial Holliday junction resolvase. DNA Repair (Amst.) 2003;2:347–358. doi: 10.1016/s1568-7864(02)00234-3. [DOI] [PubMed] [Google Scholar]
- Ogrunc M., Sancar A. Identification and characterization of human MUS81–MMS4 structure-specific endonuclease. J. Biol. Chem. 2003;278:21715–21720. doi: 10.1074/jbc.M302484200. [DOI] [PubMed] [Google Scholar]
- Osman F., Dixon J., Doe C.L., Whitby M.C. Generating crossovers by resolution of nicked Holliday junctions: A role for Mus81–Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Roberts J.A., White M.F. DNA end-directed and processive nuclease activities of the archaeal XPF enzyme. Nucleic Acids Res. 2005a;33:6662–6670. doi: 10.1093/nar/gki974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J.A., White M.F. An archaeal endonuclease displays key properties of both eukaryal XPF–ERCC1 and Mus81. J. Biol. Chem. 2005b;280:5924–5928. doi: 10.1074/jbc.M412766200. [DOI] [PubMed] [Google Scholar]
- Rothstein R., Michel B., Gangloff S. Replication fork pausing and recombination or ‘gimme a break.’. Genes & Dev. 2000;14:1–10. [PubMed] [Google Scholar]
- Schneider T.R., Sheldrick G.M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- Schwacha A., Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Tsodikov O.V., Enzlin J.H., Scharer O.D., Ellenberger T. Crystal structure and DNA binding functions of ERCC1, a subunit of the DNA structure-specific endonuclease XPF–ERCC1. Proc. Natl. Acad. Sci. 2005;102:11236–11241. doi: 10.1073/pnas.0504341102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby M.C., Osman F., Dixon J. Cleavage of model replication forks by fission yeast Mus81–Eme1 and budding yeast Mus81–Mms4. J. Biol. Chem. 2003;278:6928–6935. doi: 10.1074/jbc.M210006200. [DOI] [PubMed] [Google Scholar]
- Wood R.D. Nucleotide excision repair in mammalian cells. J. Biol. Chem. 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]