Abstract
Hematopoietic stem cells (HSCs) are multipotent progenitors that give rise to all types of blood cells. In the present study, we document that HSC development and functions are negatively regulated by the E3 ubiquitin ligase c-Cbl (casitas B-cell lymphoma). HSCs of c-Cbl−/− mice exhibit augmented pool size, hyperproliferation, greater competence, and enhanced long-term repopulating capacity. Our mechanistic studies identified that c-Cbl−/− HSCs are hyperresponsive to thrombopoietin (TPO) and display elevated levels of STAT5 phosphorylation, thus leading to increased c-Myc expression. In essence, our data unequivocally identify c-Cbl as a novel negative regulator of developmental and functional properties of HSCs.
[Keywords: Hematopoietic stem cells, c-Cbl, thrombopoietin, STAT5, self-renewal]
Hematopoietic stem cells (HSCs) have the ability to strike a balance between self-renewal and lineage commitment (Morrison et al. 1995). A constellation of intrinsic and extrinsic cellular mechanisms regulate self-renewal versus differentiation of HSCs (Phillips et al. 2000). Even though the phenotypic and functional properties of HSCs have been characterized extensively, almost nothing is known about the molecular machinery that governs self-renewal of HSCs (Phillips et al. 2000). To date, many candidate genes crucial for HSC self-renewal have been identified. These include transcriptional factors, cell cycle regulators, signaling molecules, surface receptors, and cytokines (Attar and Scadden 2004). However, thus far, the importance of ubiquitylation pathways and E3 ligases in HSC self-renewal remains elusive.
The c-Cbl (casitas B-cell lymphoma) proto-oncogene is the cellular homolog of v-Cbl, the retroviral transforming gene of the Cas NS-1 murine leukemia virus (Langdon et al. 1989). c-Cbl functions mainly as a negative regulator of signal transduction pathways, largely through its E3 ubiquitin ligase activity (Levkowitz et al. 1999; Thien and Langdon 2001; Duan et al. 2004; Schmidt and Dikic 2005; Ryan et al. 2006). The c-Cbl protein has been suggested to execute degradation of various cellular proteins, receptors, and signaling molecules including STAT5, Notch1, and c-Kit (Goh et al. 2002; Jehn et al. 2002; Zeng et al. 2005). c-Cbl−/− mice are viable, fertile, and outwardly normal in appearance; nevertheless, an increase in positive selection of thymic T cells and enhanced ductal density and branching of mammary glad were observed (Murphy et al. 1998; Naramura et al. 1998). Despite the vast knowledge available on the biochemistry of c-Cbl, the contribution of c-Cbl to the development of the immune system remains largely unknown. In the present study, we hypothesized that c-Cbl might play a pivotal role in the development and functions of HSCs.
Results and Discussion
c-Cbl deficiency leads to augmented HSC numbers
To determine whether c-Cbl is expressed in HSCs, defined cell fractions that include long-term HSCs (LT-HSCs; Lin−Sca1+c-Kit+CD34−Flt3−), short-term HSCs (ST-HSCs; Lin−Sca1+c-Kit+CD34+Flt3−), and multipotent progenitors (Lin−Sca1+c-Kit+CD34+Flt3+) were sorted from the bone marrow (BM) of C57BL/6 mice and real-time PCR analysis was performed. Although the expression of c-Cbl mRNA was detected in all HSC subsets, maximal expression of c-Cbl was found in the LT-HSCs (Supplemental Fig. 1A).
To understand the role of c-Cbl in HSC development, we first studied the phenotype of HSCs in c-Cbl−/− mice. In contrast to the recent report demonstrating the role of c-Cbl in down-regulating c-Kit receptor expression in mast cell lines (Zeng et al. 2005), our analysis of HSCs revealed similar expression levels of c-Kit and Sca1 in c-Cbl−/− and c-Cbl+/+ mice. Next, we estimated the pool size of LSK (Lin−Sca1+c-Kit+) cells in the BM. Interestingly, we found a significant increase in both relative and absolute numbers of the LSK fraction in c-Cbl−/− mice compared with the wild-type littermates (Fig. 1A; Supplemental Fig. 1B). To further understand whether the increased LSK pool is due to an increased frequency of LT-HSCs, we enumerated the cell numbers of HSC fractions. While the relative proportions of LT-HSCs and ST-HSCs remain similar in c-Cbl-deficient and wild-type mice (Fig. 1B, top panel), their absolute numbers were significantly increased in c-Cbl−/− mice (Fig. 1B, bottom panel). To corroborate these findings, an alternative immunophenotyping approach (Kiel et al. 2005) that identifies the most primitive HSCs based on the “SLAM code” (CD150+CD48−) was adopted. In line with our previous results, the numbers of CD150+CD48− cells were increased in c-Cbl−/− mice compared with the controls (Fig. 1C; Supplemental Fig. 1C). Regardless of the increased HSC numbers, analysis of the differentiated hematopoietic lineages revealed comparable numbers of B (CD19+), DC (CD11c+), NK (NK1.1+), T (CD3e), and erythroid (TER119+) lineage cells in c-Cbl mutant and wild-type mice (data not shown). Nevertheless, a modest increase of myeloid cell (CD11b+) numbers was observed in c-Cbl−/− mice (C. Rathinam and R.A. Flavell, unpubl.).
Figure 1.
Augmented size of HSC pool in c-Cbl−/− BM. (A) Total BM cells of c-Cbl+/+ and c-Cbl−/− were prepared and stained with antibody cocktail that recognizes lineage markers (CD11b, Gr1, B220, CD3e, and TER119), Sca1, and c-Kit, and were analyzed by flow cytometry. Lin− cells were pregated (top panels) and further analyzed for Sca1 and c-Kit expression (middle panels). (Bottom panels) Absolute numbers of LSK cells, as average per animal (two tibia and fibula), were determined. Each group contains n = 5 mice, age 4 wk. Data are representative of seven independent experiments. (B) Relative (top) and absolute (bottom) numbers of LT-HSCs, ST-HSCs, and MPPs were determined, as average per animal (two tibia and fibula), by discriminating LSK cells based on CD34 and Flt3 expression. Each group contains n = 5 mice, age 4 wk. Data are representative of seven independent experiments. (C, bottom panel) Absolute numbers of CD150+CD48− cells, as average per animal (two tibia and fibula), were determined from the BM (n = 5 mice). Data are representative of five independent experiments.
c-Cbl−/− HSCs are more potent and competent than wild-type HSCs in reconstituting the hematopoietic system
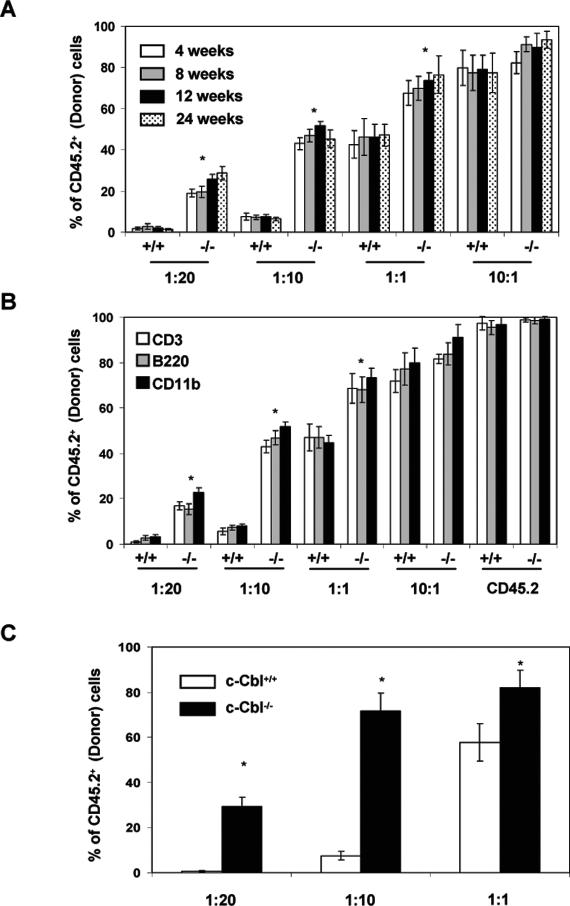
In an attempt to assess the functional properties of c-Cbl−/− HSCs, particularly in a quantitative setting, competitive repopulation experiments were performed. Limiting dilutions of LSK cells derived from either c-Cbl−/− or wild-type mice (both CD45.2) were mixed with defined numbers of wild-type competitor (CD45.1) LSK cells and were transplanted into lethally irradiated congenic (CD45.1) recipients. After 4, 8, 12, and 24 wk of transplantation, recipient mice were bled, and the percentage of donor-derived (CD45.2+) blood cells was calculated (Supplemental Fig. 2). Strikingly, the proportion of donor-derived hematopoiesis was consistently higher, at all indicated time points, in groups of mice injected with c-Cbl−/− LSK cells, when compared with the groups that received wild-type LSK cells (Fig. 2A). Next, to assess donor-derived multilineage reconstitution, the percentage of CD45.2-derived lympho-myeloid lineage cells, in the peripheral blood of recipient mice, was calculated. Analysis revealed a comparable efficiency in multilineage differentiation of LSK cells obtained from both c-Cbl−/− and wild-type mice. However, the proportion of donor-derived lympho-myeloid cells was much higher in recipients injected with c-Cbl−/− LSK cells (Fig. 2B).
Figure 2.
c-Cbl−/− HSCs are more efficient and competent in reconstituting the hematopoietic system. (A,B) Competitive repopulation experiment. Varying numbers of CD45.2+ c-Cbl−/− and c-Cbl+/+ LSK cells were mixed with defined numbers of CD45.1+ (competitor) LSK cells and subsequently transplanted into lethally irradiated (11 Gy) CD45.1+ congenic recipients (N = 10 mice). After 4, 8, 12, and 24 wk of transplantation, recipient mice were bled and the total percentage of donor-derived (CD45.2+) blood cells (A) and their lympho-myeloid differentiation capacities (B) were determined. (B) Recipients transplanted with donor LSK cells in the absence of competitor cells (last set of bars) served as controls. Data are representative of two independent experiments. (C) Secondary transplantation experiment. RBC-depleted BM cells (2 × 106) from the indicated groups of primary recipients (of the competitive repopulation experiments) were injected into lethally irradiated CD45.1+ congenic recipients (N = 10 mice). Three months after secondary transplantation, recipient mice were bled and the proportion of donor-derived cells was calculated. Data are representative of two independent experiments.
To corroborate these findings, we performed similar competitive repopulation experiments using RBC-depleted total BM cells. BM cells of either c-Cbl−/− or wild-type mice were mixed with competitor BM (CD45.1) cells at defined ratios (1:10, 1:1, and 10:1, respectively) and transplanted into lethally irradiated CD45.1 congenic recipients.
After 5, 10, and 20 wk of transplantation, recipient mice were bled, and the frequency of donor-derived hematopoiesis and multilineage reconstitution was assessed. As expected, the donor-derived hematopoiesis was higher in recipients injected with c-Cbl−/− BM cells, compared with the recipients that received wild-type BM cells (Supplemental Fig. 3). Thus, the data of total BM transplantation experiments rule out the possibility that c-Cbl-deficient HSCs “simply changed” their surface marker phenotype.
To evaluate the long-term repopulating abilities of c-Cbl−/− HSCs under competitive conditions, serial transplantation experiments were performed. BM cells (2 × 106) from the selected groups (1:20, 1:10, and 1:1) of primary recipients were transplanted into lethally irradiated CD45.1 secondary recipients. Three months after transplantation, peripheral blood of the recipients was analyzed for CD45.2-derived hematopoiesis (Supplemental Fig. 4). In contrast to the c-Cbl+/+ recipient groups, c-Cbl−/− recipient groups displayed an increase of donor (CD45.2)-derived hematopoiesis over time (Fig. 2C; Supplemental Fig. 5).
To confirm that competition occurred at the HSC level and did not reflect combined effects in the differentiated lineages, BM LSK cells from the secondary recipients were gated and their CD45 isotype expression was analyzed. The increase in donor-derived hematopoiesis in recipients that received c-Cbl-deficient cells was directly proportional to the increased frequency of CD45.2 LSK cells in the BM (Supplemental Fig. 6A). To rule out the possibility that the increased repopulation efficiency of c-Cbl-deficient LSK cells might be due to increased homing properties, CFSE-labeled LSK cells of c-Cbl−/− and wild type were transplanted into lethally irradiated wild-type (CD45.1) recipients. Analysis of CFSE+CD45.2+ LSK cells in the BM of recipients, after 12 h of transplantation, suggested a comparable homing efficiency between c-Cbl−/− and wild-type cells (Supplemental Fig. 6B).
Taken together, these results clearly demonstrate that the c-Cbl−/− HSCs are more potent than the wild-type HSCs in repopulating the hematopoietic system.
Recently, osteoclasts (cells involved in bone resorption) have been shown to play a crucial role in regulating HSC mobilization (Kollet et al. 2006). In addition, c-Cbl−/− mice exhibit defects in osteoclast functions (Chiusaroli et al. 2003). Thus, to exclude the possible involvement of the “stem cell niche” in regulating the pool size of HSCs (Moore and Lemischka 2006; Scadden 2006), wild-type (CD45.1) LSK cells were transplanted into either lethally irradiated c-Cbl−/− or wild-type recipient (CD45.2) mice. Three months later, donor-derived hematopoiesis in c-Cbl−/− and wild-type recipients was confirmed (Supplemental Fig. 7). Analysis of LSK cells of donor origin (CD45.1) indicated a similar HSC pool size in both c-Cbl−/− and wild-type recipients (Fig. 3A). Alternatively, c-Cbl mutant LSK cells were injected into wild-type (CD45.1) recipients, and analysis of LSK cells after 3 mo of transplantation revealed an increase in frequency of c-Cbl−/−-derived LSK cells in wild-type recipients (Fig. 3B). However, the multilineage differentiation capacity of c-Cbl−/− and wild-type donor cells was comparable (Supplemental Fig. 8).
Figure 3.
Increased HSC pool size of c-Cbl−/− mice is cell-intrinsic. (A) Sorted LSK cells from c-Cbl+/+ (CD45.1) mice were transplanted into lethally irradiated (11 Gy) c-Cbl−/− or c-Cbl+/+ (CD45.2) congenic recipients. (Top panels) Three months after transplantation, donor-derived (CD45.1+) lin− cells were gated and analyzed for Sca1 and c-Kit expression. (Bottom panel) Absolute numbers of donor-derived LSK cells were determined from the BM of both hind limbs (n = 5 mice). Data are representative of two independent experiments. (B) Sorted LSK cells from either c-Cbl−/− or c-Cbl+/+ (CD45.2) mice were transplanted into lethally irradiated (11 Gy) wild-type congenic (CD45.1) recipients. (Top panels) Three months after transplantation, donor-derived (CD45.2+) lin− cells were gated and analyzed for Sca1 and c-Kit expression. (Bottom panel) Absolute numbers of donor-derived LSK cells were determined from the BM of both hindlimbs (n = 5 mice). Data are representative of two independent experiments.
c-Cbl mutant HSCs show hyperproliferation in vivo and are hyperresponsive to thrombopoietin (TPO) in vitro
To elucidate whether the increased HSC pool size in c-Cbl-deficient mice is due to their accelerated proliferation rates in vivo, BrdU labeling experiments were performed. A comparison between wild-type and c-Cbl mutant cells identified an increased proliferation of c-Cbl−/− HSCs (Fig. 4A). Next, we analyzed the in vitro proliferative potential of c-Cbl-deficient LSK cells, using CFSE dilution experiments, in response to a HSC cytokine cocktail (IL3, SCF, IL6, TPO, and Flt3L). While wild-type cells showed modest proliferation rates, an augmented proliferation was noticed with c-Cbl−/− cells (Supplemental Fig. 9). Next, the ex vivo expansion potential of c-Cbl−/− LSK cells was quantified. Although both c-Cbl mutant and wild-type cells were equally viable (>85%) for up to 10 d of in vitro culture, a sustained ex vivo expansion was observed only with c-Cbl−/− HSCs (Fig. 4B).
Figure 4.
c-Cbl−/− HSCs exhibit accelerated proliferation rates and hyperresponsiveness to TPO signals. (A) In vivo BrdU incorporation experiment. The in vivo proliferation potential of c-Cbl+/+ and c-Cbl−/− LSK cells was measured by flow cytometry after feeding BrdU through drinking water for 2 d. LSK cells were pregated and histograms were generated for quantifying the incorporated BrdU. Data are representative of two independent experiments. (B) Ex vivo expansion of LSK cells of c-Cbl+/+ and c-Cbl−/− mice. Cells (1 × 104) were cultured in the presence of cytokine cocktail. At indicated time points, aliquots of cells were counted and the absolute numbers were calculated. Shown are the mean values of duplicate samples. Data are representative of three independent experiments. (C) CD34−LSK cells (1 × 104) of c-Cbl+/+ and c-Cbl−/− mice were cultured in the presence of indicated cytokines. After 4 d of in vitro culture, cells were counted and the absolute numbers were calculated. Shown are the mean values of triplicate samples. Data are representative of two independent experiments. (D) Viability of HSCs in response to TPO. LSK cells from c-Cbl+/+ and c-Cbl−/− mice were cultured in the presence of either cytokine cocktail or TPO for 48 h. Cells were stained with propidium iodide and analyzed by flow cytometry. Cells cultured in the absence of cytokines served as negative control. Data are representative of three independent experiments. (E) c-Myc mRNA expression in LSK cells on day 0 and day 3 of in vitro culture in the presence of cytokine cocktail. Input cDNA quantity was normalized according to HPRT expression levels. Shown are the mean values of duplicate samples. Data are representative of three independent experiments.
To identify the specific cytokine(s) responsible for the increased ex vivo expansion of c-Cbl-deficient HSCs, purified LSK cells of c-Cbl−/− and wild-type mice were cultured in the presence of individual cytokines, and their expansion rates were quantified. While both c-Cbl−/− and wild-type HSCs cultured in the presence of other cytokines showed similar rates of ex vivo expansion, c-Cbl−/− HSCs cultured in the presence of TPO only, SCF + TPO, and cytokine cocktail showed accelerated ex vivo expansion (Fig. 4C). Next, we assessed the viability of c-Cbl−/− HSCs in the presence of TPO. Interestingly, the viability of c-Cbl-deficient LSK cells was much higher when compared with the wild-type cells (Fig. 4D; Supplemental Fig. 10). To assess whether c-Cbl is phosphorylated in HSCs upon addition of TPO, either CD34− LSK or lin− cells of wild-type mice were stimulated with TPO, and phospho-c-Cbl levels were measured. Interestingly, c-Cbl underwent phosphorylation in response to TPO (Supplemental Fig. 11). To explain the hyperproliferative phenotype of c-Cbl−/− LSK cells at the molecular level, the expression status of various candidate molecules, implicated previously in HSC proliferation (Attar and Scadden 2004; Satoh et al. 2004), was quantified through real-time PCR. Despite similar expression levels of p18, p21, and p27 in c-Cbl wild-type and mutant cells (Supplemental Fig. 12), increased levels of c-Myc transcripts were found in c-Cbl-deficient HSCs (Fig. 4E).
Ablation of c-Cbl leads to hyperphosphorylation and accumulation of STAT5 protein in HSCs
The members of the Cbl family of proteins are highly conserved and contain a tyrosine kinase-binding (TKB) domain and a RING finger domain. While the TKB domain of c-Cbl is involved in binding to specific phosphotyrosine motifs on signaling proteins, the RING finger domain is vital for its E3 ligase activity (Thien and Langdon 2001; Thien et al. 2001; Duan et al. 2004; Schmidt and Dikic 2005; Ryan et al. 2006). A point mutation that substitutes cysteine with an alanine at the 379 position of the c-Cbl RING finger domain abolishes its E3 ligase function (Joazeiro et al. 1999; Ota et al. 2000; Thien and Langdon 2001; Schmidt and Dikic 2005; Ryan et al. 2006). Recently, knock-in mice (referred to as c-CblA/−) with a Cys-to-Ala substitution at position 379 in the RING finger domain of c-Cbl have been generated and reported (Thien et al. 2005). Although E3 ligase activity is ablated, expression levels of the mutant c-Cbl protein remain unaffected in c-CblA/− mice, as do adaptor-binding functions and the integrity of its TKB domain. To evaluate whether the increased HSC pool in c-Cbl-deficient mice is due to the lack of E3 ligase function of c-Cbl, the HSC compartment in c-CblA/− mice was analyzed. As noticed in c-Cbl−/− mice, the frequencies of LT-HSCs, ST-HSCs, and MPPs were significantly increased in c-CblA/− mice when compared with their wild-type littermates (Fig. 5A). Next, we checked whether HSCs of c-CblA/− mice show hyperproliferative responses to TPO. As expected, HSCs of c-CblA/− mice exhibited increased in vitro proliferation in the presence of TPO (Fig. 5B). Based on these results, we hypothesized that c-Cbl-mediated ubiquitylation is critical for normal TPO signaling in HSCs.
Figure 5.
Increased HSC numbers in c-CblA/− mice and enhanced STAT5 phosphorylation in c-Cbl mutant HSCs. (A) Total BM cells of c-Cbl+/+ and c-CblA/− were prepared and stained with antibody cocktail that recognizes lineage markers, Sca1, and c-Kit, and were analyzed by flow cytometry. (Top) Lin− cells were pregated and further analyzed for Sca1 and c-Kit expression. Relative (middle) and absolute (bottom) numbers of LT-HSCs, ST-HSCs, and MPPs of G1 were determined, as average per animal (two tibia and fibula), by discriminating LSK cells based on CD34 and Flt3 expression. Each group contains n = 5 mice, age 4 wk. Data are representative of two independent experiments. (B) CD34−LSK cells (1 × 104) of c-Cbl+/+ and c-CblA/− mice were cultured in the presence of indicated cytokines. After 4 d of in vitro culture, cells were counted and the absolute numbers were calculated. Shown are the mean values of triplicate samples. Data are representative of two independent experiments. (C) Phospho-STAT5 detection in HSCs. Sorted CD34−LSK cells from c-Cbl+/+ and c-Cbl−/− mice were stimulated with TPO for indicated time points. Cells were fixed, permeabilized, treated with PE-conjugated phospho-STAT5 antibodies, and analyzed by flow cytometry. Shaded histograms represent c-Cbl+/+ cells and open histograms represent c-Cbl−/− cells. Data are representative of three independent experiments. (D) Detection of intracytoplasmic STAT5 protein levels. Sorted LSK cells of c-Cbl+/+ and c-Cbl−/− mice were fixed, permeabilized, and stained with primary antibodies specific for STAT5 protein. After incubation with fluorochrome-conjugated secondary antibodies, cells were washed and analyzed by flow cytometry. Cells treated with isotype (filled histograms) served as controls. Data are representative of two independent experiments. (E) c-Cbl interacts with STAT5. Lin− cells of wild-type BM were transduced with retroviruses encoding c-Cbl-myc-IRES-EGFP and STAT5-IRES-Neomycin. Thirty-six hours after transduction, GFP+ cells were sorted and cultured in the presence of G418 for 3 d. Cells were harvested and lysed, and the cell lysates were subjected to coimmunoprecipitation using myc antibodies. Precipitates were subjected to immunoblotting and were detected using c-Cbl and STAT5 antibodies. Cells transduced with only c-Cbl-IRES-EGFP and backbone (IRES-EGFP) served as controls. (F) LSK cells of c-Cbl−/− mice were transduced with retroviruses either encoding STAT5-specific shRNA1 or control backbone. Thirty-six hours after transduction, GFP+ cells were sorted and cultured either in the presence (black) or the absence (white) of TPO. After 4 d of in vitro culture, cells were counted and the absolute numbers were calculated. Shown are the mean values of triplicate samples. Data are representative of two independent experiments.
TPO exerts its biological effects through its receptor, c-Mpl (Kimura et al. 1998). Stimulation of c-Mpl with TPO results in the activation of JAK2, which in turn phosphorylates STAT5, causing its nuclear translocation (Kato et al. 2005). In order to identify the specific target that is ubiquitylated by c-Cbl during TPO signaling, first we quantified the protein levels of c-Mpl and Jak2 in c-Cbl-deficient HSCs. Our results indicated similar expression profiles of c-Mpl and Jak2 in c-Cbl-deficient HSCs when compared with wild-type cells (Supplemental Fig. 13). Next, we evaluated the phosphorylation levels of STAT5 protein in c-Cbl-mutant HSCs in response to TPO. Interestingly, an augmented STAT5 phosphorylation was observed in c-Cbl mutant HSCs (Fig. 5C). Analysis of the total STAT5 protein in c-Cbl-deficient HSCs suggested that the increased phosphorylation is due to either increased synthesis or accumulation of STAT5 protein in c-Cbl-deficient cells (Fig. 5D). These results were confirmed independently by Western blotting using lin− cells (Supplemental Fig. 14). Next, we quantified STAT5 mRNA through real-time PCR. Surprisingly, STAT5 mRNA levels were comparable between c-Cbl−/− and wild-type LSK cells (Supplemental Fig. 15), thus suggesting a post-translational deregulation of STAT5 in c-Cbl-deficient cells. To demonstrate the interaction between c-Cbl and STAT5, coimmunoprecipitation experiments were performed. Immunoblotting analysis of the precipitates demonstrated an interaction between c-Cbl and STAT5 in primary hematopoietic progenitor cells (Fig. 5E). These data suggest that c-Cbl might be involved in STAT5 degradation in HSCs.
To prove that the hyperproliferative phenotype of c-Cbl−/− HSCs is due to accumulated STAT5 protein levels, LSK cells were transduced with lentiviruses encoding shRNA specific for STAT5. The efficiency of STAT5 knockdown in LSK and lin− cells was documented (Supplemental Fig. 16A,B). Upon STAT5 knockdown, c-Cbl−/− LSK cells showed a reduction in in vitro proliferation only in the presence of TPO (Fig. 5F), but not in the presence of either SCF alone or cytokine cocktail (Supplemental Fig. 16C). Taken together, these data undoubtedly support a role for c-Cbl in the development and maintenance of HSCs.
In essence, the present study documents that c-Cbl deficiency in HSCs leads to increased STAT5 phosphorylation in response to TPO and results in enhanced c-Myc expression (Supplemental Fig. 17). Interestingly, a direct correlation between increased STAT5 activation and augmented c-Myc expression has been observed previously (Matikainen et al. 1999; Lord et al. 2000; Tsuruyama et al. 2002), and constitutive activation of STAT5 in HSCs leads to leukemic transformation (Kato et al. 2005). The HSC phenotype of c-Cbl−/− mice such as hyperproliferation and increased competence could be explained, at least partly, by the increased c-Myc expression in HSCs. In keeping with our findings, retroviral-mediated expression of either constitutively active STAT5 or c-Myc results in hyperproliferation and augmented self-renewal of HSCs (Satoh et al. 2004; Kato et al. 2005). However, we cannot exclude the possibility that there are additional mechanisms through which c-Cbl regulates HSC functions. Nevertheless, our data provide novel molecular insights into the negative regulation of HSC development.
Over the past few decades, it has become evident that protein ubiquitylation plays a vital role in a variety of cellular processes such as morphogenesis of neuronal networks, long-term memory, circadian rhythms, regulation of the immune and inflammatory responses, and biogenesis of organelles (Hershko and Ciechanover 1998; Ciechanover and Schwartz 2002). Abnormalities in the ubiquitylation system have been shown to cause pathological responses including malignant transformation and several genetic diseases (Ciechanover and Schwartz 2002). In the immune system, knowledge of ubiquitylation pathways—in particular, the role of E3 ligases—is largely confined to lymphocyte functions (Liu 2004; Mueller 2004). The study presented here highlights, for the first time, the importance of E3 ligases and ubiquitylation pathways in the control of HSC development and function.
Further understanding of the roles played by c-Cbl in cytokine signaling might be useful for manipulating stem cells for tissue engineering and cell-based therapies.
Materials and methods
Mice
c-Cbl−/− and c-CblA/− mice were generated as reported previously (Naramura et al. 1998; Thien et al. 2005). c-Cbl−/−, c-Cbl+/+, c-CblA/−, and CD45.1 mice were kept under specific pathogen-free conditions in the animal care facility at Yale University. All mouse experiments were approved by the Institutional Animal Care and Use Committee of Yale University.
Cell culture
In vitro cultures were performed using purified CD34− LSK or LSK cells in the presence of either all or some of the following recombinant cytokines: 10 ng/mL rm-IL3, 10 ng/mL rm-IL6, 50 ng/mL rm-SCF, 10 ng/mL rm-TPO, and 50 ng/mL rh-Flt3L (all from Peprotech). Cells were cultured in IMDM medium supplemented with 10% FCS, 2 mM L-glutamine, 1% penicillin–streptomycin and 1 mM nonessential amino acids. To assess viability, cells were harvested, stained with 1 μg/mL propidium iodide (Sigma Aldrich), and analyzed by flow cytometry.
Western blotting and immunoprecipitation
For Western blot analysis, 20 μg of protein was loaded on a 3%–8% TA gel (Invitrogen), separated by electrophoresis, and blotted onto nylon membranes. The membranes were treated with primary antibodies specific for STAT5, phospho-STAT5, c-Cbl, and phospho-c-Cbl (all from BD), and GAPDH (Santa Cruz Biotechnologies) followed by staining with secondary antibodies conjugated to horseradish peroxidase. The enzymatic reaction was visualized using SuperSignal West Pico Chemiluminescent Substrate Kit (Pierce).
Immunoprecipitation was performed with Myc antibodies using a commercially available kit (Upstate Biotechnology) according to the manufacturer’s instructions.
Statistical analysis
Data are presented as mean ± SEM. Statistical significance was assessed using a two-sided Student’s t-test. P values of >0.05 were considered to be nonsignificant (NS), and P values of <0.05 were represented as an asterisk (*).
Acknowledgments
We acknowledge Ronald Baron and Cecile Itzstein for supplying a breeding pair of c-Cbl−/− and c-Cbl+/A mice. We sincerely thank Badri J. Narayanan for the necessary help. We also thank Martin Kriegel for helpful discussions. We are indebted to Elizabeth Eynon for expert advice. We are very thankful to Frances Manzo for the help with manuscript submission. We acknowledge the extended support of the Yale Cell Sorter Facility. R.A.F. is an investigator of the Howard Hughes Medical Institute. C.R. is funded by a grant from the Bill and Melinda Gates foundation through the Grand Challenges in Global Health initiative.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1651408.
References
- Attar E.C., Scadden D.T. Regulation of hematopoietic stem cell growth. Leukemia. 2004;18:1760–1768. doi: 10.1038/sj.leu.2403515. [DOI] [PubMed] [Google Scholar]
- Chiusaroli R., Sanjay A., Henriksen K., Engsig M.T., Horne W.C., Gu H., Baron R. Deletion of the gene encoding c-Cbl alters the ability of osteoclasts to migrate, delaying resorption and ossification of cartilage during the development of long bones. Dev. Biol. 2003;261:537–547. doi: 10.1016/s0012-1606(03)00299-9. [DOI] [PubMed] [Google Scholar]
- Ciechanover A., Schwartz A.L. Ubiquitin-mediated degradation of cellular proteins in health and disease. Hepatology. 2002;35:3–6. doi: 10.1053/jhep.2002.30316. [DOI] [PubMed] [Google Scholar]
- Duan L., Reddi A.L., Ghosh A., Dimri M., Band H. The Cbl family and other ubiquitin ligases: Destructive forces in control of antigen receptor signaling. Immunity. 2004;21:7–17. doi: 10.1016/j.immuni.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Goh E.L., Zhu T., Leong W.Y., Lobie P.E. c-Cbl is a negative regulator of GH-stimulated STAT5-mediated transcription. Endocrinology. 2002;143:3590–3603. doi: 10.1210/en.2002-220374. [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Jehn B.M., Dittert I., Beyer S., von der Mark K., Bielke W. c-Cbl binding and ubiquitin-dependent lysosomal degradation of membrane-associated Notch1. J. Biol. Chem. 2002;277:8033–8040. doi: 10.1074/jbc.M108552200. [DOI] [PubMed] [Google Scholar]
- Joazeiro C.A., Wing S.S., Huang H., Leverson J.D., Hunter T., Liu Y.C. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- Kato Y., Iwama A., Tadokoro Y., Shimoda K., Minoguchi M., Akira S., Tanaka M., Miyajima A., Kitamura T., Nakauchi H. Selective activation of STAT5 unveils its role in stem cell self-renewal in normal and leukemic hematopoiesis. J. Exp. Med. 2005;202:169–179. doi: 10.1084/jem.20042541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel M.J., Yilmaz O.H., Iwashita T., Yilmaz O.H., Terhorst C., Morrison S.J. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kimura S., Roberts A.W., Metcalf D., Alexander W.S. Hematopoietic stem cell deficiencies in mice lacking c-Mpl, the receptor for thrombopoietin. Proc. Natl. Acad. Sci. 1998;95:1195–1200. doi: 10.1073/pnas.95.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollet O., Dar A., Shivtiel S., Kalinkovich A., Lapid K., Sztainberg Y., Tesio M., Samstein R.M., Goichberg P., Spiegel A., et al. Osteoclasts degrade endosteal components and promote mobilization of hematopoietic progenitor cells. Nat. Med. 2006;12:657–664. doi: 10.1038/nm1417. [DOI] [PubMed] [Google Scholar]
- Langdon W.Y., Hartley J.W., Klinken S.P., Ruscetti S.K., Morse H.C. v-cbl, an oncogene from a dual-recombinant murine retrovirus that induces early B-lineage lymphomas. Proc. Natl. Acad. Sci. 1989;86:1168–1172. doi: 10.1073/pnas.86.4.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levkowitz G., Waterman H., Ettenberg S.A., Katz M., Tsygankov A.Y., Alroy I., Lavi S., Iwai K., Reiss Y., Ciechanover A., et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol. Cell. 1999;4:1029–1040. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- Liu Y.C. Ubiquitin ligases and the immune response. Annu. Rev. Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- Lord J.D., McIntosh B.C., Greenberg P.D., Nelson B.H. The IL-2 receptor promotes lymphocyte proliferation and induction of the c-myc, bcl-2, and bcl-x genes through the trans-activation domain of Stat5. J. Immunol. 2000;164:2533–2541. doi: 10.4049/jimmunol.164.5.2533. [DOI] [PubMed] [Google Scholar]
- Matikainen S., Sareneva T., Ronni T., Lehtonen A., Koskinen P.J., Julkunen I. Interferon-α activates multiple STAT proteins and upregulates proliferation-associated IL-2Rα, c-myc, and pim-1 genes in human T cells. Blood. 1999;93:1980–1991. [PubMed] [Google Scholar]
- Moore K.A., Lemischka I.R. Stem cells and their niches. Science. 2006;311:1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- Morrison S.J., Uchida N., Weissman I.L. The biology of hematopoietic stem cells. Annu. Rev. Cell Dev. Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- Mueller D.L. E3 ubiquitin ligases as T cell anergy factors. Nat. Immunol. 2004;5:883–890. doi: 10.1038/ni1106. [DOI] [PubMed] [Google Scholar]
- Murphy M.A., Schnall R.G., Venter D.J., Barnett L., Bertoncello I., Thien C.B., Langdon W.Y., Bowtell D.D. Tissue hyperplasia and enhanced T-cell signalling via ZAP-70 in c-Cbl-deficient mice. Mol. Cell. Biol. 1998;18:4872–4882. doi: 10.1128/mcb.18.8.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramura M., Kole H.K., Hu R.J., Gu H. Altered thymic positive selection and intracellular signals in Cbl-deficient mice. Proc. Natl. Acad. Sci. 1998;95:15547–15552. doi: 10.1073/pnas.95.26.15547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota S., Hazeki K., Rao N., Lupher M.L., Andoniou C.E., Druker B., Band H. The RING finger domain of Cbl is essential for negative regulation of the Syk tyrosine kinase. J. Biol. Chem. 2000;275:414–422. doi: 10.1074/jbc.275.1.414. [DOI] [PubMed] [Google Scholar]
- Phillips R.L., Ernst R.E., Brunk B., Ivanova N., Mahan M.A., Deanehan J.K., Moore K.A., Overton G.C., Lemischka I.R. The genetic program of hematopoietic stem cells. Science. 2000;288:1635–1640. doi: 10.1126/science.288.5471.1635. [DOI] [PubMed] [Google Scholar]
- Ryan P.E., Davies G.C., Nau M.M., Lipkowitz S. Regulating the regulator: Negative regulation of Cbl ubiquitin ligases. Trends Biochem. Sci. 2006;31:79–88. doi: 10.1016/j.tibs.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Satoh Y., Matsumura I., Tanaka H., Ezoe S., Sugahara H., Mizuki M., Shibayama H., Ishiko E., Ishiko J., Nakajima K., et al. Roles for c-Myc in self-renewal of hematopoietic stem cells. J. Biol. Chem. 2004;279:24986–24993. doi: 10.1074/jbc.M400407200. [DOI] [PubMed] [Google Scholar]
- Scadden D.T. The stem-cell niche as an entity of action. Nature. 2006;441:1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- Schmidt M.H., Dikic I. The Cbl interactome and its functions. Nat. Rev. Mol. Cell Biol. 2005;6:907–918. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- Thien C.B., Langdon W.Y. Cbl: Many adaptations to regulate protein tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2001;2:294–307. doi: 10.1038/35067100. [DOI] [PubMed] [Google Scholar]
- Thien C.B., Walker F., Langdon W.Y. RING finger mutations that abolish c-Cbl-directed polyubiquitylation and downregulation of the EGF receptor are insufficient for cell transformation. Mol. Cell. 2001;7:355–365. doi: 10.1016/s1097-2765(01)00183-6. [DOI] [PubMed] [Google Scholar]
- Thien C.B., Blystad F.D., Zhan Y., Lew A.M., Voigt V., Andoniou C.E., Langdon W.Y. Loss of c-Cbl RING finger function results in high-intensity TCR signaling and thymic deletion. EMBO J. 2005;24:3807–3819. doi: 10.1038/sj.emboj.7600841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruyama T., Nakamura T., Jin G., Ozeki M., Yamada Y., Hiai H. Constitutive activation of Stat5a by retrovirus integration in early pre-B lymphomas of SL/Kh strain mice. Proc. Natl. Acad. Sci. 2002;99:8253–8258. doi: 10.1073/pnas.112202899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng S., Xu Z., Lipkowitz S., Longley J.B. Regulation of stem cell factor receptor signaling by Cbl family proteins (Cbl-b/c-Cbl) Blood. 2005;105:226–232. doi: 10.1182/blood-2004-05-1768. [DOI] [PubMed] [Google Scholar]