Abstract
The apoptosis-modulating protein Daxx functions as a transcriptional repressor that binds to and suppresses the activity of nuclear factor-κB member RelB, among other transcription factors. The mechanism by which Daxx represses RelB target genes remains elusive. In this report, we demonstrate that Daxx controls epigenetic silencing of RelB target genes by DNA methylation. Daxx potently represses the RelB target genes dapk1, dapk3, c-flip, and birc3 (ciap2) at both the mRNA and protein levels. Recruitment of Daxx to target gene promoters, and its ability to repress them, is RelB-dependent, as shown by experiments using relB−/− cells. Importantly, methylation of target promoters is decreased in daxx−/− cells compared with daxx+/+ cells, and stable transfection of daxx−/− cells with Daxx restores DNA methylation. Furthermore, Daxx recruits DNA methyl transferase 1 (Dnmt1) to target promoters, resulting in synergistic repression. The observation that Daxx functions to target DNA methyltransferases onto RelB target sites in the genome provides a rare example of a gene-specific mechanism for epigenetic silencing. Given the documented role of several of the RelB-regulated genes in diseases, particularly cancer, the findings have implications for developing therapeutic strategies based on epigenetic-modifying drugs.
[Keywords: Daxx, RelB, DNMT1, transcription, DNA methylation, epigenetics]
Substantial evidence suggests that epigenetic mechanisms control the genome. The epigenome is responsible for establishing gene expression profiles in various tissues and cell types in vertebrates (D’Alessio and Szyf 2006). DNA methylation at CpG dinucleotides is the most intensely investigated epigenetic modification (Baylin 2005; Fraga and Esteller 2007) and is typically associated with transcriptional repression (Baylin 2005; Baylin and Ohm 2006). However, the mechanisms by which DNA methylation patterns are established are only partially understood. Identification of DNA methyl transferases (Dnmts) has provided important initial insights, yet knowledge of how specific CpG methylation patterns are established by Dnmts in a gene-selective manner is still limited.
Daxx is predominantly a nuclear protein, often localized within subnuclear compartments called PML oncogenic domains, or PODs (Torii et al. 1999; Maul et al. 2000; Michaelson 2000; Kawai et al. 2003; Takahashi et al. 2004). Two recurring themes continue to emerge throughout the literature on Daxx: (1) regulation of transcription, and (2) regulation of apoptosis. At one level, Daxx is a repressor of gene expression that binds histone deacetyl transferases (Hdacs) (Hollenbach et al. 2002; Ecsedy et al. 2003), Dnmts (Michaelson et al. 1999; Muromoto et al. 2004), and chromatin-modifying proteins (Hollenbach et al. 2002; Xue et al. 2003; Tang et al. 2004). On another level, Daxx is an apoptosis-modulating protein implicated in certain types of cell death pathways (Torii et al. 1999; Michaelson 2000; Kawai et al. 2003; Salomoni and Khelifi 2006). How, and if, these two themes are inextricably linked, however, is unclear.
RelB is a member of the nuclear factor (NF)-κB transcription factor family that plays crucial roles in regulating innate and adaptive immunity, cell differentiation, and apoptosis. Generation of mice deficient for each Rel family protein has revealed that individual NF-κB family members have distinct functions (Burkly et al. 1995; Alcamo et al. 2002). RelB is essential for the development of thymic medullary epithelium, dendritic cell function, and secondary lymphoid tissue organization (Burkly et al. 1995). RelB-deficient mice commonly succumb to multiorgan inflammation early in life (Hoffmann and Baltimore 2006). Because RelB acts as both a transcriptional activator and as a repressor of NF-κB-dependent gene expression (Burkly et al. 1995; Hoffmann and Baltimore 2006), it may have both inflammatory and anti-inflammatory activities in vivo.
Recent data from our laboratory showed that Daxx interacts with and suppresses the ability of RelB to activate target genes (Croxton et al. 2006). Based on the ability of RelB to bind and recruit Daxx to RelB target sites in the genome, we hypothesized that Daxx may do more than simply inhibit RelB, but may also actively repress target genes, effectively converting RelB from a transactivator to a repressor. Further, based on previous knowledge that Daxx interacts with Dnmts (Michaelson et al. 1999; Muromoto et al. 2004), we hypothesized that Daxx exerts its transcriptional-repressive actions, at least in part, through DNA methylation. In this report, we demonstrate that Daxx functions to target Dnmt1 onto RelB target sites in the genome, resulting in their DNA hypermethylation and epigenetic silencing. Furthermore, Daxx’s repressive function is RelB-dependent, with Daxx failing to target and silence these genes in relB−/− cells. The RelB/Daxx interaction thus represents a paradigm for explaining how a single transcription factor (RelB) can operate as either an activator or repressor of target genes. Our study may also reconcile some ambiguities about Daxx’s apoptotic functions by discovering that Daxx can regulate both pro- and anti-apoptotic genes, thus suggesting a connection between its transcriptional and apoptotic activities.
Results
Expression of Daxx-repressed genes is increased in daxx−/− cells
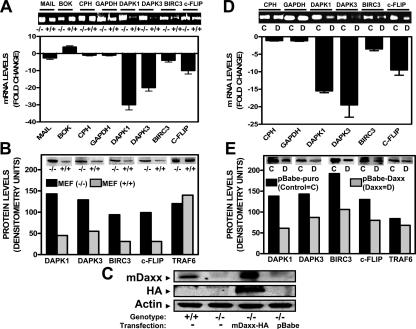
We reported previously that gene transfer-mediated overexpression of Daxx represses expression of a subset of apoptosis-modulating genes regulated by NF-κB, including the genes encoding Birc3 (cIAP2), Survivin, c-Flip, and Dapk3 (Croxton et al. 2006). To extend these results, we used Daxx-deficient mouse embryonic fibroblasts (MEFs), analyzing by quantitative real-time PCR (Q-PCR) the expression of some of the candidate target genes that appeared to be repressed by Daxx overexpression in various human epithelial cancer cell lines. In addition to these genes, we also examined the gene encoding Dapk1, because preliminary experiments suggested that its expression is suppressed by Daxx overexpression in several human tumor cell lines and because of its close relation to Dapk3. Our a priori expectation was that expression of the Daxx-repressed genes would be increased in daxx−/− cells. In agreement with this prediction, we observed approximately fivefold higher levels of Birc3 (cIAP2) mRNA, ∼10-fold higher c-Flip (Cflar) mRNA, ∼20-fold higher Dapk3 (Zipk) mRNA, and ∼30-fold higher Dapk1 mRNA levels in daxx−/− compared with daxx+/+ MEFs (Fig. 1A). Survivin (Birc5) mRNA levels were also fivefold higher in daxx−/− compared with daxx+/+ MEFs (Supplemental Fig. S2A). In contrast, Cph and Gapdh mRNA levels were similar in daxx−/− and daxx+/+ MEFs. Protein analysis by immunoblotting corroborated mRNA expression data, showing increases in c-Flip, Birc3, Dapk1, Dapk3 (Fig. 1B), and Survivin (Supplemental Fig. S2B) protein levels in Daxx-deficient cells, but not Traf6 (Fig. 1B) or β-actin (data not shown).
Figure 1.
Expression of Daxx-repressed genes is increased in Daxx−/− MEFs. (A) Analysis of Mail, Bok, Cph, Gapdh, Dapk1, Dapk3, Birc3, and c-Flip mRNA levels. daxx+/+ and daxx−/− MEFs were used for expression analysis of RelB candidate target genes (mail, bok, dapk1, dapk3, birc3, and c-flip) or control housekeeping genes (cph and gapdh). RNA (1 μg) was converted to cDNA by RTase and amplified by Q-PCR (40 cycles) using SYBR Green fluorescence to quantify DNA products. Data for daxx+/+ cells are expressed as a fold change (based on 2ΔΔCt values) relative to daxx−/− cells (mean ± SD; n = 3). (B) Analysis of Dapk1, Dapk3, Birc3, c-Flip, and Traf6 protein levels. Lysates from daxx+/+ and daxx−/− MEFs were normalized for total protein content (60 μg) and analyzed by SDS-PAGE and immunoblotting using antibodies specific for mouse Dapk1, Dapk3, Birc3, c-Flip, and β-Actin, and results were quantified by densitometry. (C) Analysis of Daxx, HA-Daxx, and β-Actin protein levels in Daxx-reconstituted cells. mDaxx (HA-tagged) was reconstituted into daxx−/− as described in the Materials and Methods. Lysates from four different cell types (daxx+/+, daxx−/−, daxx−/− + mDaxx-pBabe, and daxx−/− + pBabe) were processed as described in B and then blotted with anti-Daxx, anti-HA, or anti-β-Actin antibodies. (D) Analysis of Cph, Gapdh, Dapk1, Dapk3, Birc3, and c-Flip mRNA levels in Daxx-reconstituted cells. MEF daxx−/− + pBabe (control) and daxx−/− + mDaxx-pBabe were used for expression analysis of RelB candidate target genes (dapk1, dapk3, birc3, and c-flip) or control housekeeping genes (cph and gapdh) similarly to A. (E) Analysis of Dapk1, Dapk3, Birc3, c-Flip, and Traf6 protein levels in Daxx-reconstituted cells. The daxx−/− + pBabe control lysates and daxx−/− + mDaxx-pBabe lysates were processed as in B and then immunoblotted with the corresponding antibodies.
Because our previous analysis of Daxx-overexpressing epithelial cancer cell lines indicated increased expression of the genes encoding Bok and Mail (Croxton et al. 2006), we also evaluated expression of these genes in daxx−/− and daxx+/+ MEFs. Consistent with previous results, Bok mRNA levels were approximately fourfold higher in daxx+/+ cells. Mail mRNA levels were only slightly decreased (Fig. 1A). Thus, Daxx also may collaborate with other transcription factors to increase expression of some genes, but these effects could also be indirect and, due to repression of repressors that control expression of genes such as bok−, a topic that will not be addressed further here.
Cell cycle analysis and comparisons of cell cycle proteins showed that the differences in gene expression observed in daxx−/− and daxx+/+ cells were not due to variations in cell proliferation rates (Supplemental Fig. S1). Altogether, these data obtained using Daxx-deficient murine cells complement previous results obtained using Daxx-overexpressing human cells (Croxton et al. 2006) and strengthen evidence that Daxx regulates a subset of apoptosis-relevant genes.
Daxx reconstitution in daxx−/− MEFs restores wild-type expression of RelB target genes
To confirm that Daxx is directly involved in repression of NF-κB-regulated target genes in MEFs, we reconstituted Daxx protein expression in daxx−/− MEFs by retroviral infection. Accordingly, daxx−/− MEFs were reconstituted with empty vector (pBabe-puro, control) versus mouse Daxx vector (pBabe-mDaxx, HA-tagged). Immunoblot analysis using anti-HA antibody confirmed expression of Daxx-HA (∼120 kDa) in the stably transfected cells. Immunoblot analysis with anti-Daxx antibody suggested that the level of Daxx protein in Daxx-reconstituted daxx−/− MEFs was twofold to threefold higher than that of endogenous Daxx protein in the wild-type MEFs. Actin served as a loading control (Fig. 1C), validating these comparisons of protein levels.
Both mRNA (Fig. 1D) and protein analysis (Fig. 1E) revealed that Daxx reconstitution into Daxx-deficient cells caused a significant decrease in expression of Dapk1, Dapk3, c-Flip, and Birc3. In contrast, expression of several control genes measured at the mRNA level (Cph and Gapdh) or at the protein level (Traf6) was unchanged, demonstrating the specificity of these reconstitution experiments. Thus, these experiments confirm that Daxx is directly responsible for the repression of RelB-responsive genes.
Recruitment of Daxx to target gene promoters is RelB-dependent
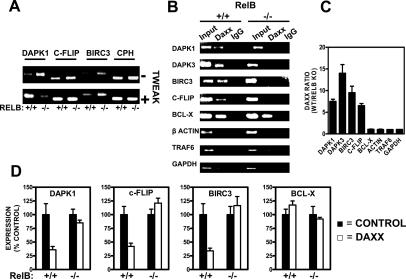
To confirm that expression of the analyzed gene set is RelB-dependent, relB+/+ and relB−/− MEFs were treated with the cytokine TWEAK, which is known to stimulate RelB activity (Varfolomeev et al. 2007). TWEAK induced increases in the expression of the candidate target genes encoding Dapk1, c-Flip, and Birc3 but not of the Cph control in wild-type (relB+/+) MEFs, confirming RelB responsiveness (Fig. 2A). In contrast, TWEAK stimulated either no increase (c-Flip), only a slight increase (Birc3), or a decrease (Dapk1) in mRNA expression in relB−/− cells, demonstrating that RelB modulates the activities of these genes in TWEAK-stimulated cells (Fig. 2A). Furthermore, evidence that RelB regulates the activity of these genes was observed at basal conditions, based on comparisons of unstimulated relB+/+ versus relB−/− cells (Fig. 2A, top panel). In unstimulated cells, levels of Dapk1, c-Flip, and Birc3 mRNAs were lower in relB+/+ compared with relB−/− cells, suggesting that RelB mediates repression of these genes under usual culture conditions.
Figure 2.
RelB-dependence of Daxx association with target gene promoters. (A) Expression analysis of RelB candidate target genes (dapk1, c-flip, and birc3) or control housekeeping gene (cph) in resting and cytokine-stimulated relB+/+ and relB−/− cells. MEFs (relB+/+ and relB−/−) were cultured without or with the cytokine TWEAK (25 ng/mL) for 24 h. Total RNA was then isolated; RNA (1 μg) was converted to cDNA by RTase and amplified by Q-PCR (40 cycles) using SYBR Green fluorescence to quantify DNA products. PCR products were analyzed by agarose gel electrophoresis. (B) Comparison of relB+/+ and relB−/− MEFs to determine association of Daxx with RelB candidate target gene promoters (dapk1, dapk3, birc3, and c-flip) and control gene promoters (bcl-x, β-actin, traf6, and gapdh) in vivo, using ChIP assays. Chromatin was immunoprecipitated with anti-Daxx or control IgG antibodies. Target genes were amplified by Q-PCR (36 cycles) using primers that encompassed the NF-κB-binding sites of each promoter (Supplemental Fig. S7), and then was analyzed by gel electrophoresis. Input represents 10% of the chromatin specimen, subjected directly to Q-PCR without IP. (C) ChIP analysis reveals differences in RelB dependence of Daxx’s recruitment to various promoters. Values represent averages of two duplicate Q-PCR reactions from two ChIP experiments. (D) Daxx represses RelB target gene promoters. MEFs (relB+/+ and relB−/−; 85%–90% confluency in 12-well plates) were transiently transfected with three different plasmids: mDaxx-pEBB, renilla luciferase reporter (pRL-TK), and either Dapk1-pLuc-pGIB, Flip-pGL3-1500, Birc3-pGL2-1400, or Bcl-X-pGL2-1200, for a total of 1.6 μg of DNA per well. The ratio of experimental vector:coreporter vector (renilla luciferase) was 10:1. Luciferase assays were performed at 24 h post-transfection. All assays were performed in triplicate.
To explore whether Daxx associates directly with RelB candidate target gene promoters, chromatin immunoprecipitation (ChIP) followed by Q-PCR analysis was performed, comparing relB+/+ and relB−/− MEFs. The promoter regions amplified for these experiments are provided as Supplemental Material (Supplemental Fig. S7). ChIP assays showed that Daxx associates with dapk1, dapk3, birc3, and c-flip promoters in RelB-containing cells but not in RelB-deficient cells (Fig. 2B), suggesting that Daxx associates with these genes through its interaction with RelB. Q-PCR analysis, performed to quantify the extent to which RelB is required for Daxx’s recruitment to the target genes, revealed significant RelB dependence (Fig. 2C). As controls, ChIP assays with traf6, gapdh, bcl-x, and β-actin promoters (RelB-unresponsive genes) revealed specificity of Daxx protein binding to RelB-responsive genes, in that Daxx was not associated with the traf6, gapdh, or β-actin gene promoters. Note, however, that Daxx association with the bcl-x promoter was detected in both relB+/+ and relB−/− cells, which may be due to the presence of a HSF1-binding site in the region of the bcl-x gene promoter assayed (Supplemental Fig. S7). In this regard, HSF1 has been reported to bind Daxx (Boellmann et al. 2004).
Daxx repression of target gene promoters is RelB-dependent
Because we found that the Daxx protein associates with target genes in a RelB-dependent manner, we next investigated whether Daxx’s repressional activity is also RelB-dependent. We performed reporter gene assays using relB+/+ and relB−/− cells. Transient transfection of dapk1, c-flip, or birc3 promoter–luciferase reporter gene constructs together with mouse Daxx (mDaxx) led to potent suppression of these promoters in RelB-containing but not in RelB-deficient cells (Fig. 2D). In contrast, mDaxx did not repress the bcl-x promoter–luciferase reporter gene regardless of RelB status (Fig. 2D, right-most panel), confirming the specificity of these results. Note that the promoter regions cloned into these luciferase reporter gene plasmids contain the regions assayed for DNA methylation plus adjacent regions that constitute the entirety of the presumed promoters (see Supplemental Fig. S8). Together, these data suggest that Daxx associates with and represses target promoters through RelB.
Daxx associates with Dnmt family members in vivo
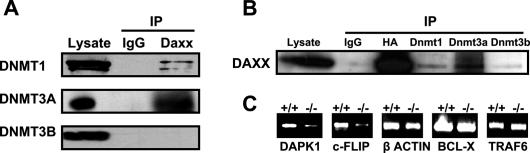
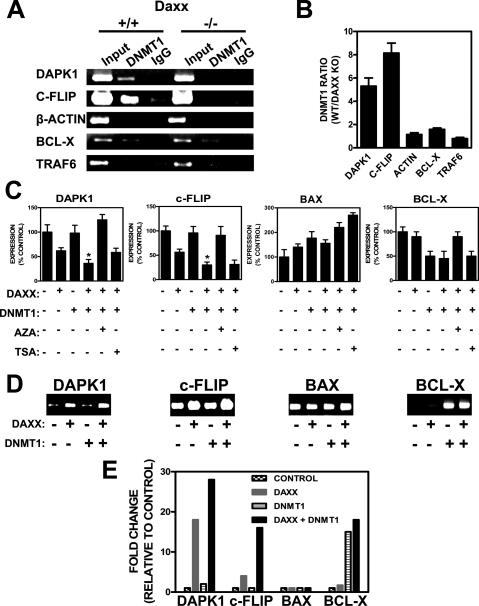
To determine whether Daxx’s mechanism of repression of RelB-responsive genes involves Dnmts, we first analyzed whether Daxx interacts with Dnmts in vivo in MEFs. Accordingly, coimmunoprecipitation experiments were performed using HA-Daxx-expressing MEFs (Fig. 3A,B). Using anti-Daxx antibody for immunoprecipitations, we detected via immunoblotting that endogenous Dnmt1 and Dnmt3a interact with Daxx (Fig. 3A). No interaction was detected with the Dnmt family member Dnmt3b (Fig. 3A). Reciprocally, using anti-Dnmt1, anti-Dnmt3a, and anti-Dnmt3b for immunoprecipitations (IPs), Daxx association was detected with endogenous Dnmt1 and Dnmt3a, but not Dnmt3b (Fig. 3B). These data thus extend a prior report of Dnmt1 association with Daxx (Muromoto et al. 2004), demonstrating that endogenous Dnmt1 and Dnmt3a associate with Daxx in MEFs.
Figure 3.
Daxx associates with Dnmts in vivo and regulates methylation of RelB target genes: analysis by MeCP2 capture. (A) Daxx-reconstituted MEF lysates were subjected to IP with anti-Daxx antibody or control IgG. The resulting immune complexes were analyzed by SDS-PAGE, followed by immunoblotting with antibodies specific for Dnmt1, Dnmt3a, or Dnmt3b. Lysates (same total protein content used for IP) were also run directly in gels, as a control. (B) Daxx-reconstituted MEF lysates were subjected to IP with anti-Dnmt1, Dnmt3a, Dnmt3b, or control IgG. The resulting immune complexes were analyzed by SDS-PAGE, followed by immunoblotting with antibodies specific for Daxx. Lysates were also analyzed as a control. Immunoblotting with anti-HA was performed to confirm effective IP of HA-Daxx (reconstituted Daxx). (C) An affinity capture method employing an immobilized MeCP protein that binds methylated DNA was used to assess the DNA methylation status of RelB candidate target gene (dapk1 and c-flip) and control promoters (β-actin, bcl-x, and traf6) in daxx+/+ versus daxx−/− MEFs. Genomic DNA (4 μg) was digested with MseI; fragments of MseI-digested DNA retaining CpG islands were purified through a DNA purification column and then incubated with immobilized MeCP2. Captured DNA fragments were amplified by Q-PCR. PCR products were analyzed by agarose gel electrophoresis.
Methylation of target promoters is decreased in daxx−/− cells
To explore whether DNA methylation participates in Daxx-mediated repression of target genes, we investigated the methylation status of the RelB target promoters using daxx+/+ and daxx−/− MEFs. Two approaches were utilized—namely, (1) affinity capture of methylated DNA using the CpG-binding protein MeCP2 (Fig. 3C), and (2) bisulfite treatment and sequencing (Fig. 4). Using the MeCP2 capture method, increased methylation of the RelB-regulated promoters of the dapk1 and c-flip genes was observed in Daxx-containing relative to Daxx-deficient cells (Fig. 3C). To confirm specificity for RelB target genes, we also analyzed β-actin, bcl-x, and traf6 promoters as controls. These promoters do not bind RelB and are insensitive to Daxx-mediated repression. No Daxx-related change in methylation of the β-actin, bcl-x, and traf6 promoters was detected (Fig. 3C).
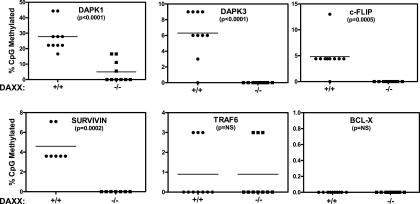
Figure 4.
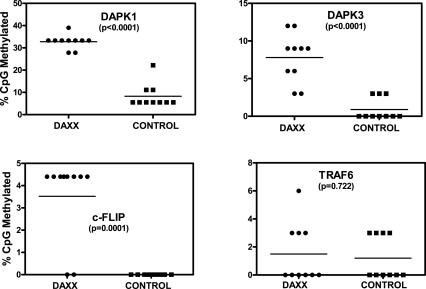
Daxx regulates methylation of RelB target genes: bisulfite sequencing method. Genomic DNA from daxx+/+ (circles) and daxx−/− (squares) MEFs was treated with bisulfite, and 0.5 μg was amplified by PCR using primers specific for the indicated gene promoters. Amplified products were cloned and 10–16 clones were sequenced per target promoter. The methylation profile of the promoters was determined by comparing the sequence of bisulfite-converted DNA with unmodified DNA. The percentage of methylated CpG sites are shown for each promoter examined. Promoters are listed in order of methylation change between daxx+/+ and daxx−/− MEFs from highest to lowest. The difference in percentage of methylation between daxx+/+ and daxx−/− MEFs was as follows: dapk1, 23%; dapk3, 6.3%; c-flip, 5%; survivin, 4.6%; traf6, 0%; and bcl-x, 0%.
These results were confirmed independently by the bisulfite sequencing method (Fig. 4), showing extensive methylation differences between daxx+/+ and daxx−/− MEFs in the RelB target gene promoters. DNA methylation differences were especially striking in the dapk1 promoter, where the percentage of CpG dinucleotide methylation was 28% in daxx+/+ MEFs versus ∼5% in daxx−/− MEFs. The promoters of the dapk3, c-flip, and survivin genes also showed Daxx-dependent differences in DNA methylation patterns, with significant decreases in promoter methylation in daxx−/− MEFs (Fig. 4; Supplemental Fig. S2C). Although we assayed only a portion of these promoters where CpG-rich regions are found, we also performed DNA methylation analysis of adjacent regions for two of the promoters (dapk1 and c-flip) and observed similar differences in CpG methylation in daxx+/+ and daxx−/− cells (Supplemental Figs. S4, S5), consistent with the notion that epigenetic silencing and DNA hypermethylation commonly span large regions of DNA (Frigola et al. 2006). In contrast, no methylation changes were observed in the bcl-x and traf6 control gene promoters (Fig. 4; Supplemental Fig. S3). These data demonstrate that Daxx modulates DNA methylation of the promoters of RelB target genes.
Daxx reconstitution in daxx−/− MEFs restores DNA methylation
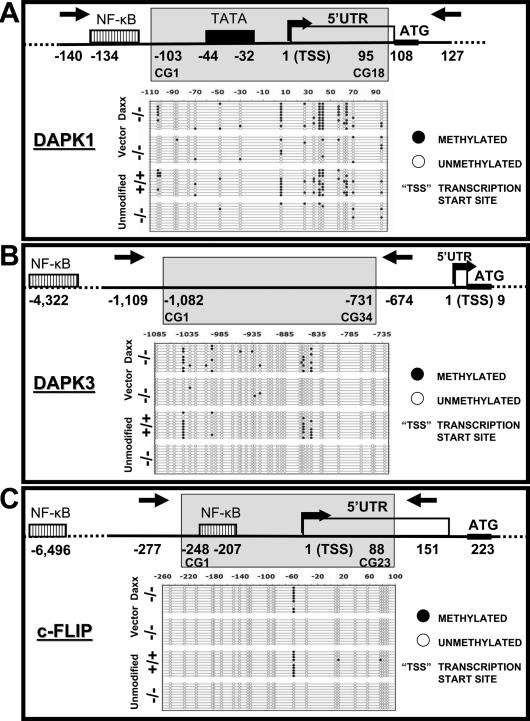
Using retrovirus-mediated stable gene transfer, we reconstituted Daxx protein expression in daxx−/− MEFs and then measured DNA methylation patterns by bisulfite sequencing. An increase in dapk1, dapk3, and c-flip promoter methylation was observed in the Daxx-reconstituted cells, but not in cells infected with control vector (Fig. 5). The DNA methylation patterns of Daxx-reconstituted cells were very similar to wild-type daxx+/+ MEFs in terms of percentage of CpG dinucleotides methylated within the CpG islands examined (cf. Figs. 4 and 5). In contrast, no difference was seen in traf6 control gene promoter methylation between Daxx- and control-infected cells (Fig. 5). Moreover, when the positions of each methylated CpG within the relevant promoters were examined (Fig. 6), an almost identical methylation pattern was observed for wild-type and Daxx-reconstituted knockout cells for all of the RelB target gene promoters examined (Fig. 6). Specifically, positions 64, 57, 43, 40, 6, and −103 relative to the dapk1 gene transcription start site were consistently methylated only in wild-type and Daxx-reconstituted cells (Fig. 6A); positions −844, −856, −997, and −1041 relative to the dapk3 gene transcription start site were similarly methylated only in wild-type and Daxx-reconstituted cells (Fig. 6B); and position −57 relative to the c-flip gene transcription start site was strictly methylated in wild-type and Daxx-reconstituted cells, but not in Daxx-deficient cells (Fig. 6C). In contrast, the methylation pattern of the traf6 gene promoter showed no consistent differences regardless of Daxx status (Supplemental Fig. S3). In conclusion, these gene reconstitution experiments confirm that the presence of Daxx is required for DNA methylation of the examined RelB target gene promoters.
Figure 5.
Daxx protein reconstitution restores normal DNA methylation patterns to RelB target gene promoters. Genomic DNA from mouse daxx−/− + mDaxx-pBabe (circles) and daxx−/− + pBabe (squares) MEFs was treated with bisulfite, amplified, cloned, and sequenced. The percentage of methylated CpG sites are shown for each promoter examined. Promoters are listed in the order of methylation change between daxx−/− + mDaxx-pBabe and daxx−/− + pBabe MEFs from highest to lowest as follows: dapk1, 24.5%; dapk3, 7%; c-flip, 3.5%; and traf6, 0.3%.
Figure 6.
Comparison of methylation status of the genes in daxx+/+ and daxx−/− cells. Genomic DNA from daxx+/+, daxx−/−, daxx−/− + mDaxx-pBabe, and daxx−/− + pBabe MEFs was subjected to sodium bisulphite modification and sequencing, analyzing dapk1 (A), dapk3 (B), and c-flip (C) proximal promoters. Methylated sites are indicated by solid circles and unmethylated sites are indicated by open circles. Predicted or known transcription factor-binding sites and transcription initiation sites are indicated in the depictions of the genes studied. Gray areas indicate the region analyzed.
Daxx targets Dnmt1 to repressed promoters
Since we demonstrated that Dnmt1 interacts with Daxx in MEFs (Fig. 3A,B), we tested whether Dnmt1 could be recruited to prototypical RelB target gene promoters by Daxx by performing ChIP assays using daxx+/+ and daxx−/− cells. In daxx+/+ cells, Dnmt1 bound to dapk1 and c-flip promoters (Fig. 7A, lane 2). In contrast, in daxx−/− cells, Dnmt1 binding to dapk1 and c-flip promoters was undetectable (Fig. 7A, lane 5). These differences in Dnmt1 association with RelB target gene promoters were specific in that no change was observed between daxx+/+ and daxx−/− cells when ChIP analysis was performed for bcl-x, β-actin, and traf6 gene promoters (Fig. 7A, cf. lanes 2 and 5). Specifically, Dnmt1 bound to the bcl-x gene promoter regardless of Daxx status (possibly due to the binding of other Daxx-interacting proteins to this promoter such as Ets and HSF1) (Supplemental Fig. S7; Boellmann et al. 2004), whereas no Dnmt1 binding to the β-actin or traf6 gene promoters was detected in either daxx+/+ or daxx−/− cells. The specificity of these results was also confirmed by the IgG control of ChIP reactions (Fig. 7A, lanes 3,6). Q-PCR analysis of the ChIP reactions showed approximately fivefold to eightfold more Dnmt1 association with the RelB target gene promoters in daxx+/+ compared with daxx−/− cells (dapk1 and c-flip) (Fig. 7B). Collectively, these data show that Daxx is required for recruitment of Dnmt1 to RelB target gene promoters.
Figure 7.
Daxx recruits Dnmt1 to target promoters, resulting in synergistic repression and increased DNA methylation. (A) Comparison of daxx+/+ and daxx−/− MEFs to determine association of Dnmt1 with RelB candidate target gene promoters (dapk1 and c-flip) and control gene promoters (β-actin, bcl-x, and traf6) in vivo, using ChIP assays. Chromatin was immunoprecipitated with anti-Dnmt1 or control IgG antibodies. Target genes were amplified by PCR (36 cycles) using primers that encompassed the NF-κB-binding sites of each promoter (Supplemental Fig. S7), and then were analyzed by gel electrophoresis. Input represents 10% of the chromatin specimen, subjected directly to PCR without IP. (B) Daxx-dependent Dnmt1 recruitment to promoters. Values represent averages (±SEM) of two duplicate Q-PCR reactions from two ChIP experiments. (C) Daxx-mediated repression of target gene promoters is enhanced by Dnmt1. Wild-type MEFs were transiently transfected with various combinations of expression and reporter gene as indicated (total 1.6 μg of DNA per well). 5-azacytidine (2 μM) or TSA (100 ng/mL) were added 4 h after transfections. Luciferase assays were performed at 24 h post-transfection (mean + SD; n = 3). (*) P < 0.05 for dapk1 and c-flip. (D) DNA methylation analysis of transiently transfected promoters. Various promoter–luciferase constructs were transiently transfected for 24 h with or without plasmids encoding Daxx and/or Dnmt1. Low-molecular-weight (LMW) DNA was recovered from cells and incubated with immobilized MeCP2 to capture methylated DNA, followed by PCR analysis using primers targeting promoter regions cloned into the luciferase constructs. PCR products were visualized by ethidium bromide staining after agarose gel electrophoresis. (E) Q-PCR analysis of MeCP2-capatured LMW DNA was performed to quantify relative methylation levels of the transiently transfected promoter constructs.
Daxx and Dnmt1 synergistically repress target promoters via DNA methylation
We examined the functional significance of Dnmt1 recruitment by Daxx to target gene promoters by performing reporter gene assays using RelB target gene promoter–luciferase contructs (Supplemental Fig. S8). While Daxx transfection decreased expression of dapk1 and c-flip reporter gene constructs, Dnmt1 transfection did not (Fig. 7C). Cotransfection of Daxx together with Dnmt1, however, resulted in a cooperative repressive effect on these RelB target gene promoters, significantly reducing expression beyond Daxx alone (P < 0.05 for dapk1 and c-flip). Moreover, this repression was relieved by Dnmt inhibitor 5-azacytidine, restoring gene expression to the levels of control cells. The Hdac inhibitor TSA had much less effect (Fig. 7C), suggesting a less prominent role in the repression compared with Dnmts. In contrast to the RelB target genes, expression of the bax gene promoter was not affected by Daxx, Dnmt1, or Daxx/Dnmt1 transfections (Fig. 7C). Likewise, the bcl-x gene promoter showed no Daxx-dependent repression, although Dnmt1 transfection reduced the activity of this promoter (Fig. 7C, right-most panel). On the basis of this analysis, we conclude that Dnmt1 collaborates with Daxx to repress RelB target genes.
Given that the chemical inhibitor of Dnmtases, 5-azacytadine, restored expression of the RelB target gene promoters (dapk1 and c-flip), we hypothesized that DNA methylation is responsible for the repression of these promoters. To assess the relative levels of methylation of the transfected plasmids, we transiently transfected various promoter–luciferase constructs either alone or in combination with Daxx, Dnmt1, or Daxx and Dnmt1 (Fig. 7D). The isolated transiently transfected plasmids were subjected to DNA methylation analysis using the MeCP2 affinity capture method. Methylation of the dapk1 and c-flip promoter constructs was increased when either Daxx or Daxx/Dnmt1 was cotransfected with the respective reporter constructs, with the combination of Daxx and Dnmt1 inducing greater methylation than Daxx alone (Fig. 7E). Dnmt1 transfection alone had no effect on methylation (Fig. 7D). In contrast, the bax promoter–luciferase construct (used here as a control) showed no change in DNA methylation in response to Daxx or Daxx/Dnmt1 (Fig. 7D,E). Also, the bcl-x promoter construct showed methylation changes that paralleled the promoter activity analysis (Fig. 7C), with increased DNA methylation induced by Dnmt1 but not Daxx (Fig. 7D,E). Taken together, these experiments directly demonstrate that Daxx is capable of promoting the methylation of the RelB target gene promoters to which it binds.
Discussion
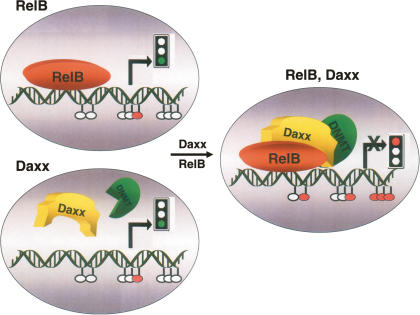
In this report, we demonstrate that Daxx is required for endogenous repression of a variety of RelB target genes, and we link this repressive effect of Daxx to a mechanism involving recruitment of Dnmt1 and DNA hypermethylation. Many studies have shown that Daxx can function as a transcriptional repressor (Salomoni and Khelifi 2006), yet the precise mechanism by which Daxx exerts its suppressive effects on endogenous genes has remained unclear. In this study, we observed that Daxx recruits Dnmt1 onto RelB target gene promoters, resulting in DNA hypermethylation and epigenetic silencing of target regions. Repression of target genes was found to be RelB-dependent (Fig. 2), consistent with data showing that Daxx physically binds RelB without interfering with RelB binding to DNA (Croxton et al. 2006). In this regard, RelB, but not Rel-A or c-Rel, contains a candidate Daxx-interacting domain motif (DID), a motif identified previously in the Ets family of transcription factors (Li et al. 2000), and we reported recently that Daxx physically interacts with RelB but not the other members of the Rel family (Croxton et al. 2006). Thus, the picture emerging (Fig. 8) is that RelB binds to target promoters, resulting in target gene transactivation when Daxx is absent or underexpressed. In contrast, when both RelB and Daxx are present, Daxx interacts with RelB bound to its target sites in the genome, recruiting Dnmtases and resulting in CpG methylation of RelB target gene promoters and gene silencing. Because Daxx lacks domains for sequence-dependent DNA binding (Salomoni and Khelifi 2006), Daxx is unable to suppress RelB target genes when RelB is absent. Thus, Daxx effectively converts RelB from a transactivator to a repressor. Consistent with this hypothesis, we observed reduced expression of the RelB target genes dapk1, c-flip, and birc3 in relB+/+ compared with relB−/− MEFs, implying that, under basal conditions, RelB contributes to the repression of these genes rather than their activation. Upon stimulation with TWEAK, however, expression of these genes increased in relB+/+ but not relB−/− cells, suggesting that, under conditions of cytokine stimulation, repression is reversed.
Figure 8.
Model for Daxx-mediated repression of RelB-binding promoters. A minimalist model is presented, showing Daxx, RelB, and Dnmtases interacting with a RelB target gene promoter. (Top left) In the absence of Daxx, RelB binds to target promoters, resulting in target gene transactivation (green traffic light). Promoters are hypomethylated. (Bottom left) In the absence of RelB, Daxx is unable to suppress RelB target genes because Daxx lacks DNA-binding domains (green traffic light). Promoters remain hypomethylated. (Right) When both RelB and Daxx are present, Daxx interacts with RelB bound to its target sites in the genome, recruits Dnmtases, and induces CpG hypermethylation of RelB target gene promoters, thus resulting in gene silencing (red traffic light).
The RelB/Daxx interaction may represent a novel paradigm for explaining how a single transcription factor—in this case, RelB—can operate as either an activator or repressor of target genes, depending on cellular context. Specifically, RelB has been reported to act as both a transcriptional activator and a repressor of NF-κB-dependent gene expression (Ruben et al. 1992; Ryseck et al. 1992; Bours et al. 1994; Bonizzi and Karin 2004). RelB transactivates through its association with p50 or p52 (Bonizzi and Karin 2004), and it represses when bound to RelA (Marienfeld et al. 2003; Jacque et al. 2005). In addition, a switch from p52/RelB complexes to p50/RelB complexes correlated with repression of skp2 (Schneider et al. 2006). The finding that Daxx converts RelB from activator to repressor thus reveals another mode beyond heterodimerization for flipping the phenotype of this transcriptional regulator. In addition to dual functionality of RelB, likewise RelA can act as both an activator and a repressor of NF-κB target genes, depending on the stimulus for induction (Dong and Goldschmidt-Clermont 2002; Campbell et al. 2004). One class of NF-κB activators (e.g., TNF and etoposide) induces RelA to act as an activator of anti-apoptotic gene expression (XIAP and Bcl-XL) (Campbell et al. 2004), while a second class of activators (e.g., UV-C, doxorubicin, and ARF tumor suppressor) induces RelA to act as a repressor of the same anti-apoptotic genes (Campbell et al. 2004; Perkins and Gilmore 2006). Thus, our findings for the Daxx/RelB interaction suggest that not only RelB but probably also other Rel family members may convert from transactivators to repressors, depending on with what other factors they interact. Interestingly, Daxx is also induced by doxorubicin (Boehrer et al. 2005), UV-C irradiation (Khelifi et al. 2005), and hydrogen peroxide treatment (Khelifi et al. 2005).
Relatively few examples exist thus far that account for selective DNA methylation of specific classes of target genes. Among the known mechanisms in mammalian cells are (1) RP58, a DNA-binding transcriptional repressor that recruits Dnmt3a to silence transcription (Fuks et al. 2001); (2) the PML-RARa fusion protein that results from t(15;17) chromosomal translocations found in certain leukemias, in which the DNA-binding domain of RARα dictates the genomic target sites to which PML recruits Dnmt3a (Di Croce et al. 2002); (3) the Kaposi’s sarcoma-associated herpesvirus oncoprotein LANA, which directly binds to DNA and recruits Dnmt3a to repress specific promoters (Shamay et al. 2006); and (4) Myc transcription factor, which activates genes through direct DNA binding, but represses indirectly through interactions with another transcription factor, inducing silencing via Dnmt3a recruitment (Brenner et al. 2005). Most of these examples differ from the RelB/Daxx interaction in that Dnmtases are directly recruited to sites in the genome by sequence-specific DNA-binding proteins. In contrast, Daxx does not directly bind DNA sequences in the genome (Salomoni and Khelifi 2006), but rather is indirectly brought to specific sites through its ability to bind DNA-binding transcription factor RelB (Croxton et al. 2006). Myc-mediated gene silencing is similar to Daxx in that Myc recruits Dnmtases to genomic target sites when interacting with another DNA-binding protein, Miz1, but not when directly binding its specific target DNA sequences in the genome via its canonical DNA-binding domain (Brenner et al. 2005). However, Daxx differs from Myc in that it lacks a DNA-binding domain (Salomoni and Khelifi 2006). Thus, while sharing similarities with Myc, the mechanism for site-selective DNA methylation described here for Daxx/RelB constitutes a new paradigm for explaining how DNA methylation is achieved in a gene-selective manner in mammals.
Daxx and Dnmt1 synergistically repressed target gene promoters in transient transfection reporter gene assays. The observation that the Dnmt inhibitor 5-azacitydine completely restored gene expression strongly suggests that Dnmts are substantially responsible for the repressive action of Daxx in the context of promoters examined. Plasmid methylation experiments further corroborated these findings, providing direct evidence that Daxx is capable of promoting methylation of the RelB target gene promoters to which it binds, and showing enhancement of DNA methylation by coexpression of Dnmt1 with Daxx but not when expressing Dnmt1 alone. However, using ChIP assays, we found that Daxx also recruits Hdac2 to target promoters (Supplemental Fig. S6). We cannot, therefore, exclude the possibility that Hdacs are also involved as corepressors, although experiments with Hdac inhibitor TSA suggested a more minor role compared with DNA methylation. In this regard, methylated DNA and Hdacs frequently colocate (Gore 2005). To the extent that Hdacs contribute to Daxx-mediated repression, recruitment of Daxx to RelB on genomic sites is reminiscent of Sin3A corepressor complex recruitment to type II nuclear receptors (NRs) in the absence of activating ligands, wherein NRs bound to their target sites in the genome actively repress rather than transactivate genes (Mathur et al. 2001). However, while Sin3A-mediated repression is dependent on Hdacs (Viiri et al. 2006), we found that Daxx-mediated repression involves Dnmts. Also, other studies have shown that DNA methylation is often dominant over Hdac activity in maintaining transcriptional repression (Cameron et al. 1999).
Daxx-dependent changes in the DNA methylation of gene promoters typically involved several CpGs and showed reproducible patterns of CpG methylation (Fig. 6), suggesting that only certain CpG motifs are presented to the DNA methylation machinery in a manner conducive to their modification. In the case of the c-flip gene promoter, a single CpG site (at position −57) was identified that displayed Daxx-dependent methylation, at least within the small region of the promoter that we examined (−248 to 88, relative to the transcription start site). Because only a small portion of the c-flip promoter was analyzed for DNA methylation here, other CpG motifs are likely to be regulated by methylation elsewhere in the c-flip promoter. Also, while we cannot forecast the functional importance of this Daxx-dependent difference in c-flip promoter methylation, it should be noted that a single methylated site per 300 bases within a gene’s promoter has been found, on several occasions, to be sufficient to dramatically repress expression (Hsieh 1994; Robertson et al. 1995; Carvin et al. 2003). It will be interesting to explore how DNA methylation patterns in promoters of the c-flip, dapk1, birc3, and other RelB target genes are impacted by agents that affect the activity of Daxx, such as interferons and arsenical, which induce Daxx recruitment to PML-oncogenic domains within the nucleus.
Daxx reportedly has proapoptotic activity in many contexts but also has been implicated in apoptosis suppression, including in vivo in daxx−/− embryos (Michaelson et al. 1999; Chen and Chen 2003; Salomoni and Khelifi 2006). By analyzing the effects of Daxx on a panel of ∼260 apoptosis-relevant genes, we previously found examples where Daxx represses both anti- and proapoptotic genes (Croxton et al. 2006). For example, Daxx-dependent repression of genes encoding anti-apoptotic proteins Birc3 (cIAP2), Survivin, and c-Flip was observed simultaneously with repression of the gene encoding the proapoptotic protein Dapk3 (Croxton et al. 2006). In the current study, we also included another proapoptotic protein—namely, Dapk1, which is the founding member of the Dapk family of Ser/Thr kinases (Bialik and Kimchi 2006). We observed that Daxx down-regulated expression of both anti-apoptotic genes (c-flip, birc3, and survivin) and proapoptotic genes (dapk1 and dapk3). Thus, perhaps Daxx association with RelB and its regulation of sets of anti- and proapoptotic RelB target genes are at least partly responsible for the reported contradictory roles for Daxx (proapoptotic vs. anti-apoptotic). Our report thus may reconcile longstanding ambiguities about Daxx’s functions, and also points to RelB as a likely link between its transcriptional and apoptotic activities.
Given evidence that Daxx protein levels vary among certain types of tumors, correlating with sensitivity or resistance to chemotherapeutic agents (Lindsay et al. 2007), the role of Daxx as an epigenetic regulator has potential ramifications for cancer biology and cancer therapy. In this regard, drugs that directly inhibit Dnmtases (e.g., 5-azacytidine [Vidaza] and 5-aza-2′-deoxycytidine [Decitabine]) are currently in clinical use for selected types of malignancies, with exploratory clinical trials under way for a wide variety of types of cancers and several oncological contexts. An underlying assumption of these epigenetic therapies is that inhibition of Dnmtases will demethylate genes in dividing cells, restoring the activity of epigenetically silenced tumor suppressor genes (Baylin 2005). Interestingly, however, our study shows that while Daxx induces DNA methylation and inhibits expression of certain genes that operate as tumor suppressors, such as Dapk1 (Bialik and Kimchi 2006; Raval et al. 2007), it also represses expression of several anti-apoptotic genes whose reactivation would presumably be counterproductive in a cancer context (e.g., c-Flip and Survivin), the expression of which has been associated with poor prognostic features of tumors (Valnet-Rabier et al. 2005; Hinnis et al. 2007). Therefore, knowledge of the status of Daxx in tumors may be helpful for identifying those cancer patients for whom Dnmtase inhibitory drugs are most likely to be beneficial (counteracting hypermethylation of tumor suppressors), as well as for possibly avoiding use of these agents in patients for whom Dnmtase inhibition may be counterproductive (counteracting hypermethylation of survival genes).
In addition to RelB, Daxx has been reported to repress a variety of transcription factors including Ets-1 (Li et al. 2000), p73 (Kim et al. 2003), glucocorticoid receptors (GR) (Lin et al. 2003), Tcf4 (Tzeng et al. 2006), Pax-3 (Hollenbach et al. 1999), and Pax-5 (Emelyanov et al. 2002). It remains to be determined whether Daxx recruits Dnmtases to the target genes where these transcription factors bind and induces epigenetic silencing. Thus, although we focused here on apoptosis-regulating RelB target genes, it is likely that the Daxx-dependent mechanism for epigenetic regulation described here applies to other categories of Daxx-regulated genes and other classes of transcription factors that Daxx binds. The cellular consequences of Daxx-mediated gene methylation therefore may be broad, with implications beyond RelB and apoptosis regulation.
Materials and methods
Antibodies and cytokines
The following antibodies were used for immunoblot and immunoprecipitation analyses: rabbit anti-Daxx (Santa Cruz Biotechnology), mouse anti-Dapk1 (Abcam), rabbit anti-ZIPK (Dapk3) (Abcam), goat anti-cIAP2 (Birc3) (Abcam), rat anti-Flip (Alexis Biochemicals), rabbit anti-Survivin (Santa Cruz Biotechnology), rabbit anti-Dnmt1 (Santa Cruz Biotechnology), rabbit anti-Dnmt3a (Santa Cruz Biotechnology), rabbit anti-Dnmt3b (Santa Cruz Biotechnology), rabbit anti-Traf6 (Santa Cruz Biotechnology), mouse anti-β-actin (Sigma), rabbit anti-HA (hemagglutinin epitope tag) (Covance), and mouse anti-Cyclin-D1 (Santa Cruz Biotechnology). The following antibodies were used for ChIP assays: rabbit anti-Daxx (Santa Cruz Biotechnology), mouse anti-Dnmt1 (Imgenex), and rabbit anti-Hdac2 (Abcam). Immunoblotting, following SDS-PAGE, was performed using the enhanced SuperSignal West Pico Chemiluminescent protein detection system (Pierce). Data were quantified by scanning densitometry analysis of scanned gels, using Scion image processing and analysis program. The cytokine TWEAK was purchased from Alexis Biochemicals and used at a concentration of 25 ng/mL to stimulate MEFs for 24 h.
Cell culture conditions and luciferase assays
MEFs, including daxx+/+, daxx−/−, relB+/+, and relB−/−, were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% bovine calf serum (BCS) (Hyclone), and 1% penicillin/streptomycin plus L-glutamine. For luciferase assays, MEFs (85%–90% confluency in 12-well plates) were transiently transfected at 24 h post-plating with various plasmids using LipofectAMINE 2000 reagent (Invitrogen), following the manufacturer’s protocol. The amount of DNA was 1.6 μg per well, the amount of LipofectAMINE was 4 μL per well, and the ratio of experimental vector:coreporter vector was 10:1. Transfection efficiency for MEFs was calculated to be ∼15%, using a GFP plasmid (pGFP2). At 24 h following transfection, cells were lysed following the Dual-Luciferase Reporter Assay System protocol (Promega). Firefly and renilla luciferase activities were measured using a microplate luminometer (BD PharMingen Monolight 3096), normalizing firefly to renilla values. All assays were performed in triplicate. For methylation inhibition studies, 5-azacytidine (Pharmion) or TSA (US Biological) were added 4 h following transfections at a final concentration of 2 μM or 100 ng/mL, respectively.
Plasmid methylation assays
Various promoter–luciferase constructs were transiently transfected for 24 h either alone (control) or in combination with Daxx and/or Dnmt1. Transfection reagents included 4 μg of DNA per well of a six-well plate and LipofectAMINE 2000 reagent (Invitrogen). The transiently transfected plasmids were recovered by isolation of low-molecular-weight (LMW) DNA using a modified protocol available from Qiagen. LMW DNA samples were then subjected to DNA methylation analysis, using the MeCP2 affinity capture method (described below), but without performing restriction digestion, followed by PCR analysis using primers encompassing promoter regions cloned into the luciferase constructs (Supplemental Fig. 8).
ChIP assays
Daxx+/+ and Daxx−/− MEFs were grown to 70%–80% confluency in 15-cm plates. Cells were processed for ChIP assays using the ChIP-IT kit (Active Motif). DNA was enzymatically sheared to a size of 300–900 base pairs (bp) prior to performing the IPs. Sheared chromatin was precleared by incubating with protein G beads (Active Motif) for 2 h at 4°C to reduce nonspecific background. Precleared chromatin was then incubated with various antibodies (3 μg) overnight at 4°C. The kit’s negative control IgG was also used in parallel reactions. Protein G bead incubation, washing of ChIP reactions, DNA elution from protein G, cross-link reversal, RNA removal, and purification of eluted DNA were performed following the kit’s protocol. Isolated DNA was subjected to Q-PCR, using promoter-specific primers as shown in Supplemental Figure 7. Q-PCR products were analyzed using Stratagene’s MxPro software and running agarose gels, stained with ethidium bromide, followed by UV visualization.
Retrovirus transductions
pBABEpuro-mDaxx (HA-tagged) was a generous gift of Dr. Gerd Maul, The Wistar Institute. 293T cells were seeded in 60-mm dishes (50% confluency) and transfected 24 h later with 3 μg of pCL-ECO (retroviral packaging vector) and 9 μg of pBABEpuro-mDaxx-HA (or pBABEpuro-empty as a control) for a total of 12 μg of DNA per 60-mm dish, using a calcium phosphate precipitation method (Promega ProFection Mammalian Tranfection System). Twenty-four hours following calcium phosphate transfection, the supernatant of 293T cells was removed and filtered through a 0.45-μm filter. Filtered supernatants, along with 8 μg/mL polybrene-hexadimethrine bromide (Sigma), were added to Daxx−/− MEFs. MEFs were then grown to confluency before splitting and were cultured for 10 d in media containing 1 μg/mL puromycin.
Plasmids
Birc3 (pGL2-cIAP2-1400-Luc), Bax (BAX-pGL3), and Bcl-x (BCL-X-pGL2) promoter constructs have been described previously (Croxton et al. 2006). The following promoter constructs were kind gifts: mDaxx-pEBB (Dr. David Baltimore, CalTech), mDnmt1-pBluescript-SK (Dr. Tim Bestor, Columbia University), mSurvivin (1430)-pLuc-pGEM3 (Dr. Dario Altieri, University of Massachusetts), mDapk1-pLuc-pGIB (Dr. Chris Marine, University of Ghent, Belgium), and pGL3-FLIP-1500 (Dr. Wafik El-Deiry, University of Pennsylvania School of Medicine).
Q-PCR
Total RNA was isolated using RNeasy mini kit (Qiagen). cDNA was generated using StrataScript First-Strand Synthesis System (Stratagene). RT–PCR (duplicate) was performed using SYBR Green PCR master mix (Qiagen) and Mx3000P Q-PCR system (Stratagene). Values were normalized to the level of CPH or GAPDH mRNA abundance and analyzed using Stratagene’s MxPro software. Thermal DNA melting experiments and gel electrophoresis confirmed generation of a single PCR product of the expected length for each amplification product shown. Graphs were constructed based on ΔΔCt values generated by Q-PCR [(Ctsample − CtCph)Daxx+/+ − (Ctsample − CtCph)Daxx−/−], using the formula 2ΔΔCt.
Bisulfite conversion and genomic sequencing
Methylation status of various promoters was assessed by bisulfite genomic sequencing. Briefly, genomic DNA was isolated from Daxx+/+ and Daxx−/− MEFs using DNeasy kit (Qiagen). Bisulfite reactions were performed using EZ DNA Methylation-Gold Kit (Zymo Research) under conditions that allowed for complete conversion of cytosines, but not 5-methylcytosines, to uracil. The bisulfite-modified DNA was amplified by PCR, using the following conditions: 2 min at 94°C; 40 cycles of 30 sec at 94°C, 30 sec at 50°C, and 1.5 min at 68°C; and finally 10 min at 68°C. Amplified products were subcloned into the pCR2.1 TOPO vector via TA cloning (Invitrogen). Ten to 16 clones were sequenced (per target promoter). The methylation profile of the promoter of interest was determined by comparing the sequence of bisulfite-converted DNA with that of unmodified DNA.
MeCP2 capture
To assess the methylation status of target promoters via an alternative method, an affinity capture method employing an immobilized protein that binds methylated DNA was used (Promoter Methylation PCR, Panomics). Genomic DNA was digested with MseI restriction enzyme (New England Biolabs), then incubated with immobilized MeCP2 (methylation-binding protein), according to the manufacturer’s instructions. Captured methylated DNA fragments were amplified by Q-PCR, comparing Daxx+/+ versus Daxx−/− samples using Stratagene’s MxPro software. The products were visualized using agarose gel electrophoresis.
Acknowledgments
We thank Drs. Manuel Perucho, Sergio Alonso, and Fumi Yamamoto (Burnham Institute for Medical Research) for insightful discussions on DNA methylation studies; Drs. Alexander Hoffmann and Soumen Basak (University of California, San Diego) for helpful advice on retrovirus transduction experiments; Melanie Hanaii and Tessa Siegfried for manuscript submission; and Drs. David Baltimore, Tim Bestor, Dario Altieri, Chris Marine, and Wafik El-Deiry for plasmid reagents. This work was supported by an NIH/National Cancer Institute grant CA69381 to J.C.R., and a dissertation fellowship from California Breast Cancer Research Program (CBCRP) 13GB-0056 to L.A.P.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1632208.
References
- Alcamo E., Hacohen N., Schulte L.C., Rennert P.D., Hynes R.O., Baltimore D. Requirement for the NF-κB family member RelA in the development of secondary lymphoid organs. J. Exp. Med. 2002;195:233–244. doi: 10.1084/jem.20011885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin S.B. DNA methylation and gene silencing in cancer. Nat. Clin. Pract. Oncol. 2005;2 (Suppl. 1):S4–S11. doi: 10.1038/ncponc0354. [DOI] [PubMed] [Google Scholar]
- Baylin S.B., Ohm J.E. Epigenetic gene silencing in cancer—A mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Bialik S., Kimchi A. The death-associated protein kinases: Structure, function, and beyond. Annu. Rev. Biochem. 2006;75:189–210. doi: 10.1146/annurev.biochem.75.103004.142615. [DOI] [PubMed] [Google Scholar]
- Boehrer S., Nowak D., Hochmuth S., Kim S.Z., Trepohl B., Afkir A., Hoelzer D., Mitrou P.S., Weidmann E., Chow K.U. Daxx overexpression in T-lymphoblastic Jurkat cells enhances caspase-dependent death receptor- and drug-induced apoptosis in distinct ways. Cell. Signal. 2005;17:581–595. doi: 10.1016/j.cellsig.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Boellmann F., Guettouche T., Guo Y., Fenna M., Mnayer L., Voellmy R. DAXX interacts with heat shock factor 1 during stress activation and enhances its transcriptional activity. Proc. Natl. Acad. Sci. 2004;101:4100–4105. doi: 10.1073/pnas.0304768101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizzi G., Karin M. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004;25:280–288. doi: 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Bours V., Azarenko V., Dejardin E., Siebenlist U. Human RelB (I-Rel) functions as a κB site-dependent transactivating member of the family of Rel-related proteins. Oncogene. 1994;9:1699–1702. [PubMed] [Google Scholar]
- Brenner C., Deplus R., Didelot C., Loriot A., Viré E., De Smet C., Gutierrez A., Danovi D., Bernard D., Boon T., et al. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 2005;24:336–346. doi: 10.1038/sj.emboj.7600509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkly L., Hession C., Ogata L., Reilly C., Marconi L.A., Olson D., Tizard R., Cate R., Lo D. Expression of relB is required for the development of thymic medulla and dendritic cells. Nature. 1995;373:531–536. doi: 10.1038/373531a0. [DOI] [PubMed] [Google Scholar]
- Cameron E.E., Bachman K.E., Myohanen S., Herman J.G., Baylin S.B. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Campbell K.J., Rocha S., Perkins N.D. Active repression of antiapoptotic gene expression by RelA(p65) NF-κB. Mol. Cell. 2004;13:853–865. doi: 10.1016/s1097-2765(04)00131-5. [DOI] [PubMed] [Google Scholar]
- Carvin C.D., Parr R.D., Kladde M.P. Site-selective in vivo targeting of cytosine-5 DNA methylation by zinc-finger proteins. Nucleic Acids Res. 2003;31:6493–6501. doi: 10.1093/nar/gkg853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.Y., Chen J.D. Daxx silencing sensitizes cells to multiple apoptotic pathways. Mol. Cell. Biol. 2003;23:7108–7121. doi: 10.1128/MCB.23.20.7108-7121.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxton R., Puto L.A., de Belle I., Thomas M., Torii S., Hanaii F., Cuddy M., Reed J.C. Daxx represses expression of a subset of antiapoptotic genes regulated by nuclear factor-κB. Cancer Res. 2006;66:9026–9035. doi: 10.1158/0008-5472.CAN-06-1047. [DOI] [PubMed] [Google Scholar]
- D’Alessio A.C., Szyf M. Epigenetic tete-a-tete: The bilateral relationship between chromatin modifications and DNA methylation. Biochem. Cell Biol. 2006;84:463–476. doi: 10.1139/o06-090. [DOI] [PubMed] [Google Scholar]
- Di Croce L., Raker V.A., Corsaro M., Fazi F., Fanelli M., Faretta M., Fuks F., Lo Coco F., Kouzarides T., Nervi C., et al. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–1082. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- Dong C., Goldschmidt-Clermont P.J. E2F1: A magic bullet for atherosclerosis? Circulation. 2002;106:2640–2641. doi: 10.1161/01.cir.0000043247.87843.aa. [DOI] [PubMed] [Google Scholar]
- Ecsedy J.A., Michaelson J.S., Leder P. Homeodomain-interacting protein kinase 1 modulates Daxx localization, phosphorylation, and transcriptional activity. Mol. Cell. Biol. 2003;23:950–960. doi: 10.1128/MCB.23.3.950-960.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov A.V., Kovac C.R., Sepulveda M.A., Birshtein B.K. The interaction of Pax5 (BSAP) with Daxx can result in transcriptional activation in B cells. J. Biol. Chem. 2002;277:11156–11164. doi: 10.1074/jbc.M111763200. [DOI] [PubMed] [Google Scholar]
- Fraga M.F., Esteller M. Epigenetics and aging: The targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Frigola J., Song J., Stirzaker C., Hinshelwood R.A., Peinado M.A., Clark S.J. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat. Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- Fuks F., Burgers W.A., Godin N., Kasai M., Kouzarides T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore S.D. Combination therapy with DNA methyltransferase inhibitors in hematologic malignancies. Nat. Clin. Pract. Oncol. 2005;2 (Suppl. 1):S30–S35. doi: 10.1038/ncponc0346. [DOI] [PubMed] [Google Scholar]
- Hinnis A.R., Luckett J.C., Walker R.A. Survivin is an independent predictor of short-term survival in poor prognostic breast cancer patients. Br. J. Cancer. 2007;96:639–645. doi: 10.1038/sj.bjc.6603616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Baltimore D. Circuitry of nuclear factor κB signaling. Immunol. Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- Hollenbach A.D., Sublett J.E., McPherson C.J., Grosveld G. The Pax3-FKHR oncoprotein is unresponsive to the pax3-associated repressor hDaxx. EMBO J. 1999;18:3702–3711. doi: 10.1093/emboj/18.13.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbach A.D., McPherson C.J., Mientjes E.J., Iyengar R., Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 2002;115:3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- Hsieh C.L. Dependence of transcriptional repression on CpG methylation density. Mol. Cell. Biol. 1994;14:5487–5494. doi: 10.1128/mcb.14.8.5487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacque E., Tchenio T., Piton G., Romeo P.H., Baud V. RelA repression of RelB activity induces selective gene activation downstream of TNF receptors. Proc. Natl. Acad. Sci. 2005;102:14635–14640. doi: 10.1073/pnas.0507342102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T., Akira S., Reed J.C. ZIP kinase triggers apoptosis from nuclear PML oncogenic domains (PODs) Mol. Cell. Biol. 2003;23:6174–6186. doi: 10.1128/MCB.23.17.6174-6186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khelifi A.F., D’Alcontres M.S., Salomoni P. Daxx is required for stress-induced cell death and JNK activation. Cell Death Differ. 2005;12:724–733. doi: 10.1038/sj.cdd.4401559. [DOI] [PubMed] [Google Scholar]
- Kim E.J., Park J.S., Um S.J. Identification of Daxx interacting with p73, one of the p53 family, and its regulation of p53 activity by competitive interaction with PML. Nucleic Acids Res. 2003;31:5356–5367. doi: 10.1093/nar/gkg741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Pei H., Watson D.K., Papas T.S. EAP1/Daxx interacts with ETS1 and represses transcriptional activation of ETS1 target genes. Oncogene. 2000;19:745–753. doi: 10.1038/sj.onc.1203385. [DOI] [PubMed] [Google Scholar]
- Lin D.Y., Lai M.Z., Ann D.K., Shih H.M. Promyelocytic leukemia protein (PML) functions as a glucocorticoid receptor co-activator by sequestering Daxx to the PML oncogenic domains (PODs) to enhance its transactivation potential. J. Biol. Chem. 2003;278:15958–15965. doi: 10.1074/jbc.M300387200. [DOI] [PubMed] [Google Scholar]
- Lindsay C.R., Scholz A., Morozov V.M., Ishov A.M. Daxx shortens mitotic arrest caused by paclitaxel. Cell Cycle. 2007;6:1200–1204. doi: 10.4161/cc.6.10.4244. [DOI] [PubMed] [Google Scholar]
- Marienfeld R., May M.J., Berberich I., Serfling E., Ghosh S., Neumann M. RelB forms transcriptionally inactive complexes with RelA/p65. J. Biol. Chem. 2003;278:19852–19860. doi: 10.1074/jbc.M301945200. [DOI] [PubMed] [Google Scholar]
- Mathur M., Tucker P.W., Samuels H.H. PSF is a novel corepressor that mediates its effect through Sin3A and the DNA binding domain of nuclear hormone receptors. Mol. Cell. Biol. 2001;21:2298–2311. doi: 10.1128/MCB.21.7.2298-2311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul G.G., Negorev D., Bell P., Ishov A.M. Review: Properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 2000;129:278–287. doi: 10.1006/jsbi.2000.4239. [DOI] [PubMed] [Google Scholar]
- Michaelson J.S. The Daxx enigma. Apoptosis. 2000;5:217–220. doi: 10.1023/a:1009696227420. [DOI] [PubMed] [Google Scholar]
- Michaelson J.S., Bader D., Kuo F., Kozak C., Leder P. Loss of Daxx, a promiscuously interactive protein, results in extensive apoptosis in early mouse development. Genes & Dev. 1999;13:1918–1923. doi: 10.1101/gad.13.15.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muromoto R., Sugiyama K., Takachi A., Imoto S., Sato N., Yamamoto T., Oritani K., Shimoda K., Matsuda T. Physical and functional interactions between Daxx and DNA methyltransferase 1-associated protein, DMAP1. J. Immunol. 2004;172:2985–2993. doi: 10.4049/jimmunol.172.5.2985. [DOI] [PubMed] [Google Scholar]
- Perkins N.D., Gilmore T.D. Good cop, bad cop: The different faces of NF-κB. Cell Death Differ. 2006;13:759–772. doi: 10.1038/sj.cdd.4401838. [DOI] [PubMed] [Google Scholar]
- Raval A., Tanner S.M., Byrd J.C., Angerman E.B., Perko J.D., Chen S.S., Hackanson B., Grever M.R., Lucas D.M., Matkovic J.J., et al. Downregulation of death-associated protein kinase 1 (DAPK1) in chronic lymphocytic leukemia. Cell. 2007;129:879–890. doi: 10.1016/j.cell.2007.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson K.D., Hayward S.D., Ling P.D., Samid D., Ambinder R.F. Transcriptional activation of the Epstein-Barr virus latency C promoter after 5-azacytidine treatment: Evidence that demethylation at a single CpG site is crucial. Mol. Cell. Biol. 1995;15:6150–6159. doi: 10.1128/mcb.15.11.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruben S.M., Klement J.F., Coleman T.A., Maher M., Chen C.H., Rosen C.A. I-Rel: A novel rel-related protein that inhibits NF-κB transcriptional activity. Genes & Dev. 1992;6:745–760. doi: 10.1101/gad.6.5.745. [DOI] [PubMed] [Google Scholar]
- Ryseck R.P., Bull P., Takamiya M., Bours V., Siebenlist U., Dobrzanski P., Bravo R. RelB, a new Rel family transcription activator that can interact with p50–NF-κB. Mol. Cell. Biol. 1992;12:674–684. doi: 10.1128/mcb.12.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomoni P., Khelifi A.F. Daxx: Death or survival protein? Trends Cell Biol. 2006;16:97–104. doi: 10.1016/j.tcb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Schneider G., Saur D., Siveke J.T., Fritsch R., Greten F.R., Schmid R.M. IKKα controls p52/RelB at the skp2 gene promoter to regulate G1- to S-phase progression. EMBO J. 2006;25:3801–3812. doi: 10.1038/sj.emboj.7601259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamay M., Krithivas A., Zhang J., Hayward S.D. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi’s sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. 2006;103:14554–14559. doi: 10.1073/pnas.0604469103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Lallemand-Breitenbach V., Zhu J., De The H. PML nuclear bodies and apoptosis. Oncogene. 2004;23:2819–2824. doi: 10.1038/sj.onc.1207533. [DOI] [PubMed] [Google Scholar]
- Tang J., Wu S., Liu H., Stratt R., Barak O.G., Shiekhattar R., Picketts D.J., Yang X. A novel transcription regulatory complex containing Daxx and the ATR-X syndrome protein. J. Biol. Chem. 2004;279:20369–20377. doi: 10.1074/jbc.M401321200. [DOI] [PubMed] [Google Scholar]
- Torii S., Egan D.A., Evans R.A., Reed J.C. Human Daxx regulates Fas-induced apoptosis from nuclear PML oncogenic domains (PODs) EMBO J. 1999;18:6037–6049. doi: 10.1093/emboj/18.21.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng S.L., Cheng Y.W., Li C.H., Lin Y.S., Hsu H.C., Kang J.J. Physiological and functional interactions between TCF4 and Daxx in colon cancer cells. J. Biol. Chem. 2006;281:15405–15411. doi: 10.1074/jbc.M601807200. [DOI] [PubMed] [Google Scholar]
- Valnet-Rabier M.B., Challier B., Thiebault S., Angonin R., Margueritte G., Mougin C., Kantelip B., Deconinck E., Cahn J.-Y., Fest T. c-Flip protein expression in Burkitt’s lymphomas is associated with a poor clinical outcome. Br. J. Haematol. 2005;128:767–773. doi: 10.1111/j.1365-2141.2005.05378.x. [DOI] [PubMed] [Google Scholar]
- Varfolomeev E., Blankenship J.W., Wayson S.M., Fedorova A.V., Kayagaki N., Garg P., Zobel K., Dynek J.N., Elliott L.O., Wallweber H.J., et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFα-dependent apoptosis. Cell. 2007;131:669–681. doi: 10.1016/j.cell.2007.10.030. [DOI] [PubMed] [Google Scholar]
- Viiri K.M., Korkeamäki H., Kukkonen M.K., Nieminen L.K., Lindfors K., Peterson P., Mäki M., Kainulainen H., Lohi O. SAP30L interacts with members of the Sin3A corepressor complex and targets Sin3A to the nucleolus. Nucleic Acids Res. 2006;34:3288–3298. doi: 10.1093/nar/gkl401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Gibbons R., Yan Z., Yang D., McDowell T.L., Sechi S., Qin J., Zhou S., Higgs D., Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]