Abstract
Particulate matter air pollution (PM) has been linked with chronic respiratory diseases. Real-life exposures are likely to involve a mixture of chemical and microbial stimuli, yet little attention has been paid to the potential interactions between PM components (e.g., Ni) and microbial agents on the development of inflammatory-like conditions in the lung. Using the Toll-like receptor (TLR)-2 agonist MALP-2 as a lipopeptide relevant to microbial colonization, we hypothesized that nickel sensitizes human lung fibroblasts (HLF) for microbial-driven chemokine release through modulation of TLR signaling pathways. NiSO4 (200 μM) synergistically enhanced CXCL8, yet antagonized CXCL10 mRNA expression and protein release from HLF in response to MALP-2. RT2-PCR pathway-focused array results indicated that NiSO4 exposure did not alter the expression of TLRs or their downstream signaling mediators, yet significantly increased the expression of cyclooxygenase 2 (COX-2). Moreover, when NiSO4 was given in combination with MALP-2, there was an amplified induction of COX-2 mRNA and protein along with its metabolic product, PGE2, in HLF. The COX-2 inhibitor, NS-398, attenuated NiSO4 and MALP-2–induced PGE2 and CXCL8 release and partially reversed the NiSO4-dependent inhibition of MALP-2–induced CXCL10 release from HLF. These data indicate that NiSO4 alters the pattern of TLR-2–dependent chemokine release from HLF via a COX-2–mediated pathway. The quantitative and qualitative effects of NiSO4 on microbial-driven chemokine release from HLF shed new light on how PM-derived metals can exacerbate respiratory diseases.
Keywords: COX-2, nickel, inflammation, chemokines, fibroblasts
CLINICAL RELEVANCE
This study examines how metals such as nickel can sensitize cells to microbial-driven inflammation in the lung. These findings will increase our understanding of how environmental exposures interact to modulate pulmonary inflammation.
Particulate matter air pollution (PM) is a contributing risk factor for many adverse health effects, including respiratory diseases (1). Real-life scenarios are likely to involve exposures not only to chemical stresses such as PM, but microbial stimuli as well. While much attention has been paid to microbial-driven inflammation in the lung, limited information is available regarding the potential interactions between PM exposures and microbial stress on respiratory function. There are reports, however, of increased numbers of hospitalizations for respiratory disorders including infections on days of increased levels of PM (2). In support of these epidemiologic findings, several rodent models have shown PM exposure can increase susceptibility to infectious bacteria such as Listeria monocytogenes and Streptococcus pneumoniae (3, 4).
Recent evidence suggests that colonization with species originally considered commensal and not causing overt disease (e.g., Mycoplasma fermentans) also has the potential to interact with other stimuli and/or alter biological responses in infected host cells (5, 6). For example, we have previously shown that exposure of human lung fibroblast cells (HLF) to M. fermentans synergistically enhances the release of IL-6 by PM in the form of residual oil fly ash (ROFA) compared to either stimulus alone (7). It is well recognized that toxicity of PM depends, in part, on the specific chemicals present in PM, and metals are often implicated as causative agents. For example, using data from the Six Cities studies, Laden and coworkers (8) demonstrated that nickel was positively associated with daily deaths. A recent finding by Lippmann and colleagues suggests cardiac dysfunction in response to fine particulate matter is largely attributable to nickel (9), which can also contribute to the onset of asthma (10) and pulmonary fibrosis (11) in the lower airways. In fact, the large synergistic release of IL-6 from HLF seen with ROFA can be recapitulated using NiSO4 (Ni) (but not other metals such as V, Cu, or Fe) and the 2-kD M. fermentans–derived macrophage-activating lipopeptide (MALP-2) (7).
Lung fibroblasts play an active role in the response to tissue injury, contributing to cytokine and chemokine release and the development of fibroproliferative disorders. As such, understanding how HLF respond to chemical and microbial stimuli can contribute to our understanding of the sequela of events leading to pathogenesis of fibrotic lung disease, among others. In order to gain a better understanding of how Ni may influence microbial-driven inflammation in the lung, the current study focuses on changes in CXCL8 and CXCL10 expression in HLF after mixed exposures to Ni and MALP-2. Although any number of chemokines may be modulated during mixed exposures of chemical and microbial stimuli, CXCL8 and CXCL10 were chosen because both of these chemokines can be up-regulated during inflammation (12–14), yet display differential properties that can affect the development of respiratory disorders (e.g., fibrosis). Also included in the study is CCL2, another pro-inflammatory chemokine shown previously to be up-regulated after exposure to M. fermentans and MALP-2 (5, 15). Because MALP-2 stimulates the production of innate immune defense mediators via activation of Toll-like receptor (TLR)-2 (16), we used RT2-PCR pathway-focused arrays to examine the hypothesis that Ni-induced changes in expression of TLR signaling components and/or their downstream targets contribute to alterations in MALP-2–induced release of CXCL8, CXCL10, and CCL2 from HLF. Pathway-focused array results showed that Ni stimulates the expression of cyclooxygenase-2 (COX-2) mRNA. Based on these results, we further addressed the potential role of COX-2 in the Ni-dependent alterations of MALP-2–induced chemokine release from HLF.
MATERIALS AND METHODS
Materials
Minimum essential medium (MEM), penicillin-streptomycin, SYBR GreenER qPCR SuperMix Universal, and fetal bovine serum (FBS) were from Invitrogen (Carlsbad, CA). Low endotoxin bovine serum albumin (BSA) was from Intergen (Purchase, NY). NiSO4 and M. fermentans–derived MALP-2 were from Sigma (St. Louis, MO) and Alexis Biochemicals (San Diego, CA), respectively. The RT2-PCR-Profiler PCR Array Human Toll-Like Receptor Signaling Pathway was from SuperArray Bioscience Corporation (Frederick, MD). Omniscript Reverse Transcriptase, RNAse inhibitor, oligo-dT, and RNAeasy Mini Kit were from Qiagen (Valencia, CA). Bradford protein assay reagent was from Bio-Rad (Hercules, CA). The COX-2 monoclonal antibody, NS-398, PGE2, and PGE2 EIA Kit were from Cayman Chemical (Ann Arbor, MI). The rabbit GAPDH monoclonal antibody, anti-mouse, and anti-rabbit IgG-HRP–linked antibodies were obtained from Cell Signaling Technology, Inc. (Danvers, MA).
Cell Lines and Culture
HLF were isolated as outgrowths from explanted surplus transbronchial biopsy tissues obtained during routine follow-up bronchoscopy of lung transplant recipients as previously described (17) in accordance with a protocol approved by University of Pittsburgh Institutional Review Board. Routine culture was performed in MEM containing 10% FBS, glutamine (2 mM), and 1% penicillin-streptomycin, with weekly passage. All cultures are carried out at 37°C in a humidified atmosphere of 5% CO2/95% air.
RNA Isolation and RT-PCR for Cytokine mRNA Measurement
HLF were treated with 600 pg/ml MALP-2, 200 μM Ni, or the two stimuli in combination in serum-free MEM with 0.1% BSA for 48 hours. Control cells received serum-free MEM with 0.1% BSA alone for 48 hours. The amounts of Ni and MALP-2 used in the current study were chosen on the basis of previous concentration–response analyses that indicate 200 μM NiSO4 and 600 pg/ml MALP-2 are effective at achieving a synergistic induction of IL-6 in HLF (7). These concentrations are also within the range of what others have used in vitro with regard to IL-6 and CXCL8 release in response to MALP-2 (18) or NiSO4 alone (19). After treatment, total RNA was extracted from the cells and cDNA was generated from 1 μg total RNA using Omniscript reverse transcriptase with oligo d(T) as a primer. Reactions were incubated at 37°C for 75 minutes in an Eppendorf Mastercycler Gradient. Real-time RT-PCR analysis was carried out to quantify changes in cytokine mRNA using SYBR GreenER qPCR SuperMixes Universal from Invitrogen. Reactions were carried out for 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds after an initial 10-minutes incubation at 95°C using an Opticon 2 Real-Time PCR Detection System. Relative gene expression was calculated using the 2−ΔΔCt method (20), using RPL13A as the internal control gene to normalize the data for the amount of RNA added to each RT reaction. CXCL8 primers were kindly provided by Dr. Aaron Barchowsky (21). Specific primer pairs for CXCL10, CCL2, COX-2, and RPL13A were designed using Primer3 (22). CXCL10: forward TCCCATCACTTCCCTACATGG, reverse AGATGGGAAAGGTGAGGGAAA; CCL2: forward TGCTCATAGCAGCCACCTTC, reverse CTTGGCCACAATGGTCTTGA; COX-2: forward GCTCAGCCATACAGCAAATCC, reverse CAACGTTCCAAAATCCCTTGA; and RPL13A: forward CGAGGTTGGCTGGAAGTACC, reverse ATTCCAGGGCAACAATGGAG.
Chemokine Production
Cells were seeded in 6-well plates at a density of 4 × 105 cells/well and allowed to attach for 1 day. Complete medium was removed, and serum-free MEM containing 0.1% BSA in the absence (controls) or presence of 200 μM NiSO4 and 600 pg/ml MALP-2 was added for 48 hours. Conditioned medium was then collected and stored at −70°C until use. CXCL8, CXCL10, and CCL2 content in the conditioned media samples were analyzed using specific enzyme-linked immunosorbent assay (ELISA) kits obtained from R&D Systems (Minneapolis, MN) according to the manufacturer's instructions.
PCR Array Analysis of TLR Signaling Pathway
Cells were seeded in 6-well plates at a density of 4 × 105 cells/well and allowed to attach for 1 day. Complete media was then removed and cells were treated with 200 μM NiSO4 in serum-free MEM with 0.1% BSA for 24 hours. Control cells received serum-free MEM with 0.1% BSA alone for 24 hours. After exposure, total RNA was extracted from the cells using the RNAeasy Mini Kit according to manufacturer's instructions. cDNA was generated from 1 μg total RNA using the ReactionReady First Strand cDNA Synthesis kit (SuperArray Bioscience Corporation) according to manufacturer's instructions. The human TLR signaling pathway RT2-PCR-Profiler PCR Array was carried out according to manufacturer's instructions using the Opticon 2 Real-Time PCR Detection System (Bio-Rad).
Immunoblot Detection of COX-2
Cell lysates were prepared using 10 mM Tris, pH 7.4 with 1% SDS containing a cocktail of protease (200 mM PMSF, 1 mg/ml leupeptin, aprotinin, and pepstatin) and phosphatase inhibitors (200 mM each Na3VO4 and NaF). Cell lysates (25 μg protein/lane) were subjected to electrophoresis using NuPAGE 4 to 12% Bis-Tris gels (Invitrogen) under denaturing and reducing conditions using sample buffer containing dithiothreitol (80 mM, final concentration). Proteins were transferred to Polyscreen PVDF Transfer membranes (PerkinElmer, Wellesley, MA). Immunoblots were blocked with 5% nonfat dried milk in Tris-buffered saline containing 1% Tween-20 (TBS-T) for 1 hour, then incubated with an anti–COX-2 monoclonal antibody at a 1:1,000 dilution in TBS-T containing 5% BSA for 2 hours at room temperature. Blots were then rinsed with TBS-T and incubated with an anti-mouse HRP-linked secondary antibody (1:1,000) for 1 hour at room temperature. Immunoblots were developed using chemiluminescence. Membranes were then stripped and reprobed with antibodies to GAPDH (loading control) as described above.
COX-2 Inhibitor and PGE2 Assays
To determine the effects of COX-2 on Ni- and MALP-2–induced PGE2 and chemokine release from HLF, cells were pretreated with 10 μM of the COX-2 inhibitor NS-398 for 30 minutes before the addition of Ni and MALP-2. Dimethyl sulfoxide (DMSO) at a final concentration of 0.1% served as the solvent control. PGE2 and chemokines in conditioned media taken from HLF after exposure were analyzed with enzyme immunoassay (EIA) (Cayman Chemical) or ELISA (R&D Systems), respectively. In subsequent experiments, the effects of exogenously added PGE2 on chemokine release from HLF were examined. Stock solutions of PGE2 were prepared by dissolving the PGE2 in ethanol. On the day of experimental treatment, PGE2 was further diluted in MEM and then added to HLF at a final concentration of 10−7 M alone or in combination with Ni and MALP-2. Levels of CXCL8 and CXCL10 in conditioned medium 48 hours after exposure were measured using ELISA.
Statistical Analysis
Data presented are expressed as mean ± SEM. Ni-induced changes in gene expression using the RT2-PCR-Profiler PCR Array were analyzed using a Student t test. Comparisons between Ni and/or MALP-2 treatment groups were made using a one-way ANOVA with Tukey's multiple comparison tests. Comparisons between treatment groups in the presence or absence of NS-398 and PGE2 were made using two-way ANOVA with Bonferroni post hoc comparison. As needed, data were transformed by the log transformation to correct for the nonnormal distribution of data before ANOVA analysis. Statistical analyses were performed using GraphPad PRISM, version 3.0 (GraphPad Software, San Diego, CA), with P < 0.05 considered significant.
RESULTS
Ni Alters CXCL8 and CXCL10 Chemokine Profiles in HLF in Response to TLR-2 Agonist, MALP-2
HLF were treated with MALP-2 alone, Ni alone, or the two stimuli in combination for 48 hours, and control cells received medium alone for 48 hours. Changes in CXCL8, CXCL10, and CCL2 expression and their release into conditioned medium were analyzed using RT2-PCR and ELISA, respectively (Figure 1). Treatment of HLF with MALP-2 alone for 48 hours induced the expression and release of all three chemokines compared with control-treated cells. Although exposure to Ni alone had no appreciable effects on chemokine expression, the presence of Ni both quantitatively and qualitatively altered CXCL8 and CXCL10 profiles in HLF in response to MALP-2. Figures 1A and 1B show that the stimulation of CXCL8 gene expression was more enhanced when MALP-2 was given in combination with Ni for 48 hours compared with either stimuli alone. Quantitative analysis using real-time PCR indicated a clear, synergistic induction of CXCL8 mRNA in HLF, to levels that were approximately 1,100-fold greater than control-treated cells (Figure 1A; P < 0.001). ELISA analysis of secreted protein in the conditioned media show CXCL8 release from HLF in response to 48 hours of Ni and MALP-2 mixed exposures parallel the changes observed in mRNA (Figure 1B). When cells are co-exposed to Ni and MALP-2 for 48 hours, levels of CXCL8 in conditioned media were synergistically greater than what was observed in media from control-treated cells or cells receiving either stimuli alone (P < 0.001).
Figure 1.
Ni alters the pattern of macrophage-activating lipopeptide (MALP)-2–induced chemokine expression and release in HLF. A, C, and E represent real-time RT-PCR analysis of Ni- and MALP-2–induced changes in chemokine mRNA expression after a 48-hour exposure. Data were normalized to the housekeeping gene RPL13A and are expressed as mean ± SEM fold-increase over control-treated cells (n = 3). B, D, and F represent enzyme-linked immunosorbent assay (ELISA) analysis of Ni- and MALP-2–induced chemokine release in conditioned medium after a 48-hour exposure. Data shown are mean ± SEM (n = 3). (A and B) CXCL8, *different from control, **different from Ni or MALP-2 alone (P < 0.01). (C and D) CXCL10, *different from control, Ni and Ni and MALP-2 co-treated cells (P < 0.05). (E and F). CCL2, *different from control and Ni-treated cells (P < 0.01).
In contrast to CXCL8, the addition of Ni to MALP-2 during co-treatment for 48 hours antagonized MALP-2-induced CXCL10 mRNA expression (Figure 1C). Real-time RT-PCR analysis revealed that MALP-2 alone stimulated a 12-fold increase in CXCL10 mRNA expression over controls (Figure 1C; P < 0.05). In the presence of Ni, however, the increase in CXCL10 transcripts was reduced to levels that were only 3-fold greater than those of control-treated cells. The TLR-2 agonist, MALP-2, also elevated CXCL10 levels in conditioned medium after a 48-hour exposure to levels ranging from 3- to 7-fold greater than what was observed in control-treated cells. Although the extent to which MALP-2 induced CXCL10 release varied across experiments, the response to MALP-2 was always significantly greater when compared with control-treated cells (Figure 1D; P < 0.001). Although Ni exposure alone had no effect, Ni antagonized MALP-2–induced CXCL10 release from HLF. The data in Figure 1D show that in the presence of Ni, MALP-2–induced release of CXCL10 was significantly less than what was seen in cells treated with MALP-2 alone (P < 0.001).
The addition of Ni to HLF did not alter MALP-2–induced CCL2 mRNA expression or protein release into the conditioned medium. The stimulatory effects of MALP-2 on CCL2 mRNA expression were maintained when cells were co-treated with Ni for 48 hours (Figure 1E). Real-time RT-PCR analysis revealed an approximate 7.5-fold increase in CCL2 expression over control-treated cells after treatment with MALP-2 alone or in combination with Ni (Figure 1E, P < 0.01). Figure 1F shows that the stimulatory effect of MALP-2 on release of CCL2 protein from HLF was also unaltered in the presence of Ni.
Ni Exposure Does Not Alter Expression of TLR Signaling Pathway Components
In subsequent experiments it was found that pre-treatment of HLF with Ni for 24 hours synergistically enhanced induction of CXCL8 yet inhibited induction of CXCL10 during a subsequent 24-hour MALP-2 challenge (data not shown), similar to the observations reported above. These data suggest that Ni exposure may modulate components of the TLR signaling pathway, thereby sensitizing HLF cells for an amplified response to MALP-2. In order to assess the ability of Ni to modulate specific components of the TLR signaling pathway, and thereby alter MALP-2–induced release of immune-modulators, an RT2-PCR Profiler Human Toll-Like Receptor Signaling Pathway array was used to screen for Ni-induced changes in 84 genes involved in TLR signaling pathways. Array data indicated that a 24-hour Ni exposure (200 μM) did not alter the mRNA level for TLR receptors 1 through 9, nor did Ni alter the expression of downstream signaling mediators such as MyD88, TRAF6, TBK1, or TICAM1 and 2, among others (Table 1). Three genes contained in the array that were significantly increased in response to Ni exposure are downstream targets of TLR activation: IL-6, CXCL8, and COX-2 (Table 1; P < 0.05). The Ni-induced increase in TLR-10 expression (an orphan member of the TLR family), however, was not consistent in the three replicate experiments that were conducted. Only two of the three array analyses indicated an increase in TLR10 expression after Ni exposure. In addition, the apparent induction could not be repeated in follow-up PCR experiments using independently designed TLR-10 primers. Furthermore, Western blot analysis indicated that a 24-hour exposure to Ni did not alter the expression of TLR-10 protein in HLF compared with control-treated cells (data not shown).
TABLE 1.
NI-INDUCED CHANGES IN TLR SIGNALING PATHWAY COMPONENTS
| Symbol | Fold Change | Symbol | Fold Change | Symbol | Fold Change | Symbol | Fold Change |
|---|---|---|---|---|---|---|---|
| BTK | 1.14 ± 0.29 | IFNG | 1.30 ± 0.15 | MAP3K7 | 1.20 ± 0.08 | SITPEC | 4.88 ± 4.24 |
| CASP8 | 0.37 ± 0.02 | IKBKB | 0.97 ± 0.06 | MAP3K7IP1 | 0.77 ± 0.06 | TBK1 | 0.82 ± 0.10 |
| CCL2 | 0.88 ± 0.09 | IL10 | 2.54 ± 1.72 | MAP4K4 | 1.24 ± 0.14 | TICAM2 | 1.19 ± 0.14 |
| CD14 | 0.71 ± 0.29 | IL12A | 0.93 ± 0.15 | MAPK8 | 1.12 ± 0.01 | TIRAP | 1.24 ± 0.20 |
| CD80 | 1.30 ± 0.15 | IL1A | 1.58 ± 0.56 | MAPK8IP3 | 1.36 ± 0.05 | TLR1 | 1.25 ± 0.27 |
| CD86 | 1.64 ± 0.36 | IL1B | 1.95 ± 0.86 | MYD88 | 0.78 ± 0.09 | TLR10 | 5.03 ± 2.11* |
| CHUK | 0.70 ± 0.23 | IL2 | 1.30 ± 0.15 | NFKB1 | 0.87 ± 0.20 | TLR2 | 2.10 ± 0.76 |
| CLEC4E | 1.18 ± 0.25 | IL6 | 5.66 ± 0.53* | NFKB2 | 1.41 ± 0.14 | TLR3 | 0.64 ± 0.07 |
| CSF2 | 1.71 ± 0.34 | CXCL8 | 6.64 ± 0.57* | NFKBIA | 1.39 ± 0.46 | TLR4 | 1.30 ± 0.19 |
| CSF3 | 1.63 ± 0.47 | IRAK1 | 0.66 ± 0.15 | NFKBIL1 | 1.15 ± 0.11 | TLR5 | 2.51 ± 1.77 |
| CXCL10 | 1.30 ± 0.15 | IRAK2 | 0.70 ± 0.06 | NFRKB | 1.13 ± 0.09 | TLR6 | 1.13 ± 0.16 |
| EIF2AK2 | 1.33 ± 0.56 | IRF1 | 0.64 ± 0.03 | NR2C2 | 1.13 ± 0.14 | TLR7 | 1.27 ± 0.25 |
| ELK1 | 0.91 ± 0.41 | IRF3 | 0.84 ± 0.07 | PELI1 | 1.49 ± 0.20 | TLR8 | 1.30 ± 0.15 |
| FADD | 0.97 ± 0.03 | JUN | 2.71 ± 0.67 | PPARA | 1.42 ± 0.16 | TLR9 | 2.03 ± 1.14 |
| FOS | 1.21 ± 0.29 | LTA | 1.12 ± 0.30 | PRKRA | 0.94 ± 0.12 | TNF | 1.45 ± 0.56 |
| HMGB1 | 0.67 ± 0.06 | CD180 | 2.55 ± 1.69 | COX-2 | 3.91 ± 0.63* | TNFRSF1A | 1.01 ± 0.31 |
| HRAS | 1.28 ± 0.10 | LY86 | 1.68 ± 0.65 | REL | 1.63 ± 0.29 | TOLLIP | 0.83 ± 0.05 |
| HSPA1A | 1.15 ± 0.20 | LY96 | 1.74 ± 0.72 | RELA | 1.18 ± 0.09 | TRAF6 | 1.13 ± 0.12 |
| HSPD1 | 0.89 ± 0.08 | MAP2K3 | 0.91 ± 0.12 | RIPK2 | 0.79 ± 0.17 | TICAM1 | 1.20 ± 0.17 |
| IFNA1 | 2.50 ± 0.34 | MAP2K4 | 0.86 ± 0.10 | SARM1 | 0.90 ± 0.26 | UBE2N | 0.92 ± 0.06 |
| IFNB1 | 2.33 ± 1.53 | MAP3K1 | 0.72 ± 0.09 | SIGIRR | 1.30 ± 0.15 | UBE2V1 | 1.61 ± 0.41 |
Data represent the mean ± SEM (n = 3) of Ni-induced fold-change in gene expression over control-treated cells.
Significantly different from control (P < 0.05).
Ni and MALP-2 Synergistically Induce COX-2 Expression in HLF
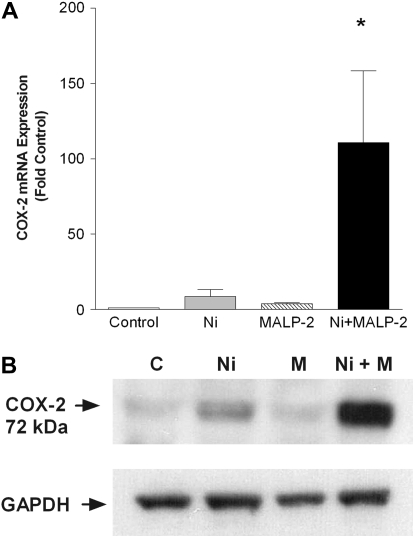
Because COX-2 plays an important contribution to the onset of inflammatory processes, and the Human Toll-Like Receptor Signaling Pathway array revealed Ni treatment alone induced COX-2 expression (Table 1), we hypothesized that this enzyme might contribute to the mechanism whereby Ni modulates MALP-2–induced chemokine production in HLF. Real-time RT-PCR analysis revealed that a 48-hour exposure to Ni alone stimulates an 8-fold induction in COX-2 gene expression over control-treated cells (Figure 2A). Moreover, a synergistic induction in COX-2 mRNA expression was observed in HLF treated with both stimuli for 48 hours, to levels that were 110-fold greater than those of control-treated cells (Figure 2A; P < 0.05). As shown in Figure 2B, this stimulatory effect of Ni and MALP-2 combined exposure on COX-2 is also observed at the protein level.
Figure 2.
Combined exposures to Ni and MALP-2 synergistically induce the expression of cyclooxygenase 2 (COX-2) in HLF. (A) Real-time RT-PCR analysis of Ni- and MALP-2–induced COX-2 expression after a 48-hour exposure. Data were normalized to the housekeeping gene RPL13A and are expressed as mean ± SEM fold-increase over control-treated cells (n = 3). *Different from control, Ni or MALP-2 treatment (P < 0.05). (B) For detection of COX-2 protein, 25 μg of total cellular protein was loaded per lane and subjected to SDS-PAGE and Western blotting with anti–COX-2 antibody. GAPDH is shown as a loading control.
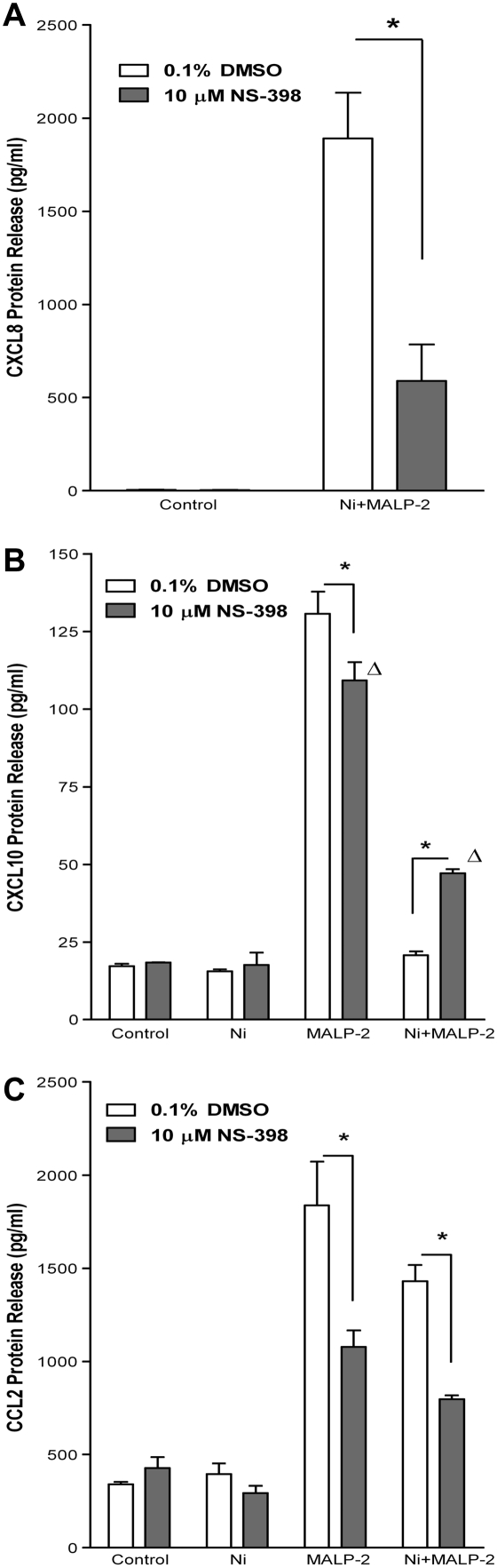
Inhibition of COX-2 Attenuates Ni- and MALP-2–Induced CXCL8 Release and Partially Reverses the Inhibitory Effects of Ni on MALP-2–Induced CXCL10 Release from HLF
Supporting a role for COX-2 in Ni and MALP-2–induced CXCL8 release, the COX-2–specific inhibitor NS-398 attenuated CXCL8 production in HLF after Ni and MALP-2 co-exposure (Figure 3A). Levels of CXCL8 released by Ni and MALP-2 in the presence of the COX-2 inhibitor were reduced by approximately 70%, significantly less than the levels observed in solvent control–treated cells (P < 0.001). Levels of CXCL8 released by HLF in response to Ni or MALP-2 alone were unaffected by the presence NS-398 and were not different from those in control-treated cells (data not shown).
Figure 3.
Inhibition of COX-2 alters Ni-dependent modulation of chemokine release by MALP-2. Cells were pretreated with NS-398 (10 μM) for 30 minutes before the addition of Ni or MALP-2 (48 h). Chemokine levels in conditioned medium were measured using ELISA. Data are shown as mean ± SEM (n = 3); (A) CXCL8, (B) CXCL10, (C) CCL2. *Different from respective treatment in absence of inhibitor (P < 0.01); Δdifferent from control in presence of inhibitor (P < 0.001).
Figure 3B shows that inhibition of COX-2 using NS-398 significantly, albeit not fully, restored the cellular response to MALP-2–induced CXCL10 release in the presence of Ni. In NS-398–treated cells, levels of CXCL10 in conditioned medium from HLF co-treated with Ni and MALP-2 are increased to 47.1 ± 1.4 pg/ml, levels that were significantly greater than cells co-treated with Ni and MALP-2 in the absence of inhibitor, as well as NS-398 control-treated cells (Figure 3B; P < 0.001).
MALP-2–induced stimulation of CCL2 release by human lung fibroblasts was also sensitive to COX-2 inhibition. In the presence of NS-398, levels of CCL2 in conditioned medium from cells treated with MALP-2 were reduced by approximately 41% (Figure 3C; P < 0.01). A similar inhibition of CCL2 release by NS-398 was also observed in cells that were co-exposed to Ni and MALP-2.
Ni and MALP-2 Stimulate PGE2 from HLF
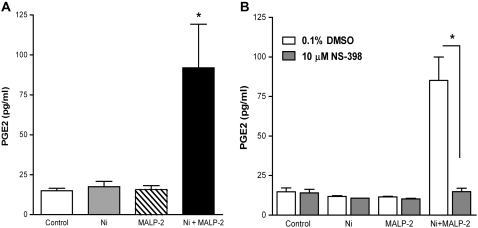
Consistent with the induction of COX-2 mRNA and protein, co-exposure to Ni and MALP-2 for 48 hours stimulated the elaboration of PGE2, a major metabolic product of COX-2, from HLF (Figure 4A). PGE2 in conditioned medium from cells treated with Ni and MALP-2 was stimulated to levels that were 7-fold greater than what was observed in control-treated cells and cells receiving Ni or MALP-2 alone (P < 0.01). The stimulatory effects of co-treatment with Ni and MALP-2 on PGE2 release were likely mediated via COX-2, as opposed to COX-1, as the COX-2–specific inhibitor NS-398 completely attenuated this response (Figure 4B; P < 0.001). Given that MALP-2 alone did not stimulate PGE2 release, the attenuation of CXCL10 release by NS-398 implicates a regulatory role for other COX-2–derived eicosanoids. In addition, the similar levels of CCL2 inhibition by NS-398 observed in cells treated with MALP-2 alone or co-treated with Ni and MALP-2 for 48 h (Figure 3C) suggest a COX-2–mediated pathway in CCL2 release that is PGE2 independent.
Figure 4.
COX-2 mediated PGE2 release from HLF after stimulation with Ni and MALP-2. Data shown are mean ± SEM (n = 3). (A) PGE2 release after a 48-hour exposure to Ni and MALP-2. *Different from control, Ni-, or MALP-2–treated cells (P < 0.01). (B) PGE2 release after a 48-hour exposure to Ni and MALP-2 in the presence of NS-398 (10 μM). *Different from respective treatment in the absence of NS-398 (P < 0.001).
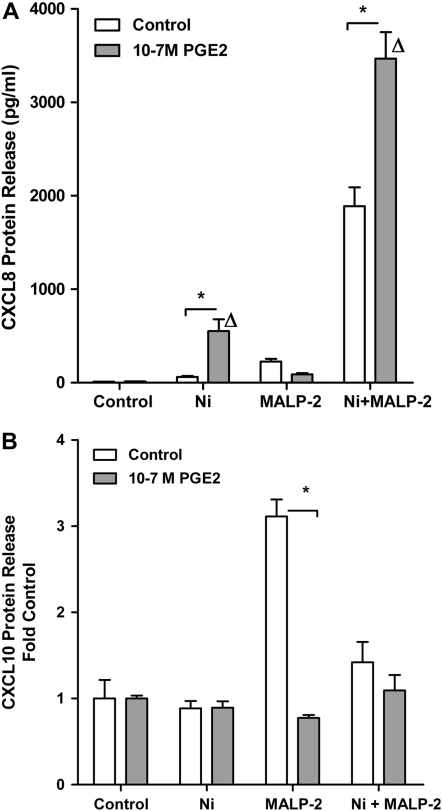
To further investigate the potential role of PGE2 in mediating the Ni-dependent alterations in MALP-2–induced CXCL8 and CXCL10 release, HLF were exposed to Ni and/or MALP-2 alone or in combination with PGE2 (10−7 M) for 48 hours. ELISA analysis revealed that a 48-hour exposure to PGE2 alone had no effect on CXCL8 release from human lung fibroblasts compared to control-treated cells or cells treated with MALP-2 (Figure 5A). However, ELISA analysis on conditioned medium taken from cells co-exposed to Ni and PGE2 showed that PGE2 enhanced Ni-induced CXCL8 release by 9-fold. CXCL8 was stimulated to 552.4 ± 127.8 pg/ml in cells treated with PGE2 and Ni compared to the 62.0 ± 10.1 pg/ml observed in cells treated with Ni alone (Figure 5A; P < 0.05). Furthermore, this stimulation of CXCL8 in response to Ni and PGE2 is greater than the predicted additive response of the two stimuli (552.4 pg/ml vs. 75.1 pg/ml, respectively). Additional studies revealed PGE2 enhanced Ni-induced CXCL8 release from HLF in a dose-dependent manner, beginning at 5 × 10−9 M (data not shown). PGE2 also enhanced the synergistic release of CXCL8 in response to combined Ni and MALP-2 exposure to levels that were significantly greater compared with cells receiving Ni and MALP-2 in the absence of PGE2 (P < 0.05). PGE2 (10−7 M) did not effect CXCL10 release in control, Ni, or combined Ni- and MALP-2–treated cells. However, PGE2 mimicked the effects of Ni in that PGE2 significantly attenuated MALP-2–induced CXCL10 release from human lung fibroblasts (Figure 5B; P < 0.001).
Figure 5.
Effects of PGE2 on Ni- and MALP-2–induced CXCL8 and CXCL10 release from HLF. Cells were treated with Ni and MALP-2 in the presence or absence of PGE2 (10−7 M) for 48 hours. (A) CXCL8. Data are shown as mean ± SEM (n = 3). (B) CXCL10. Data are expressed as fold change in CXCL10 release over control (n = 3).*Different from respective treatment in absence of PGE2 (P < 0.05); Δdifferent from control in presence of PGE2 (P < 0.05).
DISCUSSION
Despite the fact that real-life exposures are likely to involve a mixture of chemical and microbial stimuli, little attention has been paid to the potential interactions between chemical and microbial stress on the evolution of inflammatory-like conditions in the lung. Here we report the novel findings that NiSO4, a metal commonly found in particulate matter air pollution, modulates MALP-2–induced CXCL8 and CXCL10 release from human lung fibroblast cells through a mechanism involving COX-2.
Initially identified as a chemotactic factor for neutrophils released from monocytes (23), CXCL8 has since been shown to be synthesized and released by nonimmune cells such as endothelial cells and fibroblasts and has been implicated in the regulation of angiogenesis (24) and contraction of airway smooth muscle cells (25). Our data showing synergistic induction of CXCL8 in human lung fibroblasts demonstrate the potential impact of combined exposures to chemical and microbial stimuli. PGE2 has been shown to increase CXCL8 gene expression in human colonic epithelial cells through stabilization of CXCL8 mRNA (26). It will be of interest to learn whether the PGE2 released by Ni and MALP-2 behaves similarly to stabilize CXCL8 mRNA in HLF. Alternatively, one cannot overlook interactions at the gene promoter level where Ni, MALP-2, or PGE2 may activate specific sets of transcriptional activators that interact to maximally drive target gene expression. This could explain, in part, the dramatic increases in observed in both mRNA expression and protein release after exposure. The Ni and MALP-2–mediated synergistic induction of CXCL8 in HLF is also reminiscent of our previous observations with IL-6 (7). This is of particular concern in regards to pulmonary health given the correlation of elevated CXCL8 and IL-6 production with numerous respiratory disorders, including asthma (12, 27) and pulmonary fibrosis (13, 28).
In the present study we show Ni not only quantitatively changes chemokine release in response to MALP-2, but also qualitatively alters the chemokine profile in HLF. While Ni promotes the elaboration of the inflammatory mediators CXCL8 and IL-6 (7), Ni antagonizes the release of CXCL10 in response to microbial stimuli. CXCL10 is anti-fibrotic and has been shown to have a protective effect in the bleomycin-induced fibrosis model, limiting drug-induced fibroblast migration (29). The fact that Ni attenuates MALP-2-induced CXCL10 suggests any protective effect offered by chemokines such as CXCL10 may be lost during combined exposures to chemical and microbial stimuli.
Of particular interest are the similarities observed in chemokine profiles after combined Ni and MALP-2 exposures, enhanced elaboration of CXCL8, and decrease in CXCL10, with previous observations made in drug-induced animal models of pulmonary fibrosis and what is seen in human patients with idiopathic pulmonary fibrosis (IPF). Levels of CXCL8 are significantly elevated, whereas levels of CXCL10 are significantly lower in patients with IPF compared with control subjects (30). In the current study, MALP-2–induced CCL2 levels remain elevated in the presence of Ni. CCL-2 mRNA levels in lung epithelial cells from patients with IPF are elevated compared with those from patients without IPF (31), and levels of CCL-2 mRNA are also up-regulated in rat and other rodent models of pulmonary fibrosis (32, 33). The elevated levels of CCL2 in combination with the dichotomous effects of Ni on MALP-2–incuded CXCL8 and CXCL10 release raise the possibility that exposure to Ni and microbial stimuli can set up a pro-fibrotic environment in the lung.
A common thread in the Ni-dependent alterations in MALP-2–induced CXCL8 and CXCL10 release from HLF is the induction of COX-2 and subsequent production of PGE2. COX-2 plays an important role in inflammation. Particulate Ni and air pollution in the form of diesel exhaust particles (DEP) stimulates COX-2 expression in bronchial epithelial BEAS-2B cells (34, 35), which suggests that induction of COX-2 may provide a mechanism whereby air particulate matter pollution stimulates an inflammatory response in pulmonary cells. Microbial stimuli in the form of MALP-2 have also been shown to activate COX-2 (36). It would therefore stand to reason that combined exposures to chemical and microbial stimuli could interact to enhance induction of COX-2 as was observed in the current study. In HLF, Ni and MALP-2 stimulated an approximate 100-fold increase in COX-2 mRNA levels over those of control-treated cells. The human COX-2 promoter contains multiple DNA binding sites including two NF-κB–binding sites, an NF/IL6 site and a CRE/E-box site. It has yet to be determined whether Ni and MALP-2 synergistically induce COX-2 mRNA through transcriptional or post-transcriptional mechanisms. Given that NF-κB and C/EBP are important signaling pathways in inflammatory responses (37, 38), and both MALP-2 and NiSO4 can activate NF-κB (19, 39), one possible explanation is that Ni and MALP-2 activate C/EBP in concert with members of the NF-κB/Rel family to induce COX-2 promoter activity. Alternatively, it is also possible that the dramatic increase in COX-2 may be mediated, in part, through enhanced stabilization of COX-2 mRNA levels. Interestingly, PGE2 has a positive regulatory effect on COX-2 protein expression in mouse lung fibroblasts (40) and can stabilize COX-2 mRNA (41). The possibility that PGE2 stabilizes COX-2 mRNA stability and protein expression through a positive feedback mechanism in HLF after Ni and MALP-2 exposure is of interest and warrants further investigation.
PGE2 has been shown to have regulatory role in both CXCL10 and CXCL8 release. Exogenously added PGE2 (10−7 M) can suppress stimulus-induced release of CXCL10 in A431 cells (42), consistent with current findings that 10−7 M PGE2 inhibited MALP-2–induced CXCL10 release from HLF. It was also observed in the current study that PGE2 levels are increased after combined exposures to Ni and MALP-2, and that inhibition of COX-2 with NS-398 partially restores the cellular response to MALP-2 in the presence of Ni. Although levels of exogenously added PGE2 used in the current study (10−7 M) are higher than what is released from HLF after co-treatment with Ni and MALP-2, the proportion of PGE2 that gets into the cells is likely to be less than 10−7 M, as oxidation of PGE2 after cellular uptake leads to loss of biological activity (43). In addition, there may be higher levels of PGE2 retained in the nucleus of HLF after treatment with Ni and MALP-2 that would go undetected in the ELISA analysis of conditioned medium. Nonetheless, these findings provide evidence that PGE2 contributes, in part, to the mechanism whereby Ni antagonizes MALP-2–induced CXCL10 release from HLF. However, the contribution of COX-2–independent pathways in the Ni-dependent inhibition of MALP-2-induced CXCL10 release cannot be ignored, as NS-398 was unable to completely restore the sensitivity of fibroblast cells to MALP-2 in the presence of Ni.
Although PGE2 is thought to have a suppressive effect on lung fibroblasts, in part through attenuation of proliferation and collagen synthesis, the role of PGE2 in the immune-inflammatory response is more complex. Four transmembrane G protein–coupled PGE2 receptor subtypes (EP1-4) mediate the biological actions of PGE2. Depending on cell-specific expression of the different EP receptor subtypes, PGE2 can either suppress (44) or stimulate CXCL8 production (26, 45). In the current study, PGE2 alone (10−7 M) did not stimulate CXCL8. However, PGE2 significantly enhanced the elaboration of CXCL8 release from HLF in response to Ni alone and combination with MALP-2. Moreover, while the inhibitory effects of PGE2 on MALP-2–induced CXCL10 were only apparent at the 10−7 M concentration, PGE2 could enhance Ni-induced CXCL8 release at much lower concentrations (5 × 10−9 M). This finding suggests that chemical stimuli can alter cellular responsiveness to PGE2, allowing for a stimulatory, as opposed to an inhibitory, effect on pulmonary fibroblasts. In fact, a recent finding by Moore and coworkers (46) show a loss of PGE2 suppression in BLEO-induced pulmonary fibrosis associated with reduced cellular expression of EP2. Furthermore, it was found that PGE2 can stimulate fibroblast proliferation 14 and 21 days after BLEO administration (46). The possibility that Ni can alter EP receptor profiles in human lung fibroblast cells, allowing for a stimulatory effect of PGE2 on CXCL8 release, is currently under investigation.
Epidemiologic studies, although limited, have indicated an increased number of hospitalizations for respiratory disorders on days of increased levels of PM (2). These reports have been supported by studies in rodent models that provide evidence that PM can modulate pulmonary immune responses (47). The observed interactions between Ni and MALP-2 on the elaboration of chemokine release from HLF suggest that pulmonary immune responses in humans may also be altered as a consequence of PM exposure. As always, it is difficult to extrapolate whether the doses of stimuli employed in in vitro studies are relevant to real-world exposures. To our knowledge, MALP-2 has never been measured during an in vitro infection with Mycoplasma; however, we should point out that MALP-2 is at least three orders of magnitude more potent that the classically used microbial stimulus, LPS. We have previously estimated (7) that exposure to 100 ng Ni/m3 would produce approximately 1 μM concentration of Ni in the noncellular volume of the human lung. While this is indeed less than the concentrations chosen here, it should be noted that the synergistic interactions with MALP-2 can be seen with as little as 20 μM (7),and heterogeneous deposition, alterations in breathing patterns, and accumulation over time may produce regional concentrations similar to those used here. Nonetheless, given their important role in the onset of fibroproliferative disorders, understanding how HLF respond to PM and microbial stimuli is an important step in learning how such exposures contribute to disease onset. Epithelial cells and macrophages also play a vital role in inflammation and fibrotic disorders. How chemokine production in these cells may be altered in response to mixed exposures of PM-derived metals and microbial stimuli, and the resultant effects on fibroblast phenotype and behavior, are also of interest and warrant further investigation.
In summary, this is the first study to show that combined exposures to Ni and MALP-2 can synergistically enhance COX-2 expression. This is of importance as (1) real life exposures are more like to involve mixtures of chemical and microbial agents, and (2) induction of COX-2 plays an important role in the production of immune-modulators and is widely recognized as having an important role in the onset of inflammation in the lung (39, 48–50). The findings that COX-2 contributes to the synergistic production of CXCL8 and inhibition of CXCL10 in HLF present a potential mechanism whereby exposure to PM-derived metals can exacerbate cellular responses to microbial stimuli and modulate pulmonary inflammation and its consequences such as fibrosis and angiogenesis.
Acknowledgments
The authors thank Dr. Aaron Barchowsky, Linda Klei, and Antonia Nemec for their valuable input and technical assistance.
This work was supported in part by National Institutes of Health grant RO1-ES-011986 awarded to J.P.F. K.A.B. is supported as a post-doctoral scholar on NIH NRSA 1F32ES015966-01.
Originally Published in Press as DOI: 10.1165/rcmb.2007-0314OC on December 20, 2007
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Davidson CI, Phalen RF, Solomon PA. Airborne particulate matter and human health: a review. Aerosol Sci Technol 2005;39:737–749. [Google Scholar]
- 2.Schwartz J. What are people dying of on high air pollution days? Environ Res 1994;64:26–35. [DOI] [PubMed] [Google Scholar]
- 3.Sigaud S, Goldsmith CA, Zhou H, Yang Z, Fedulov A, Imrich A, Kobzik L. Air pollution particles diminish bacterial clearance in the primed lungs of mice. Toxicol Appl Pharmacol 2007;223:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang HM, Antonini JM, Barger MW, Butterworth L, Roberts BR, Ma JK, Castranova V, Ma JY. Diesel exhaust particles suppress macrophage function and slow the pulmonary clearance of Listeria monocytogenes in rats. Environ Health Perspect 2001;109:515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fabisiak JP, Gao F, Thomson RG, Strieter RM, Watkins SC, Dauber JH. Mycoplasma fermentans and TNF-beta interact to amplify immune-modulating cytokines in human lung fibroblasts. Am J Physiol Lung Cell Mol Physiol 2006;291:L781–L793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyama K, Zhang S, Lo SC. Effects of mycoplasmal LAMPs on receptor responses to steroid hormones in mammalian cells. Curr Microbiol 2001;43:163–169. [DOI] [PubMed] [Google Scholar]
- 7.Gao F, Barchowsky A, Nemec AA, Fabisiak JP. MIcrobial stimulation by Mycoplasma fermentans synergistically amplifies IL-6 release by human lung fibroblasts in response to residual oil fly ash (ROFA) and nickel. Toxicol Sci 2004;81:467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laden F, Neas LM, Dockery DW, Schwartz J. Association of fine particulate matter from different sources with daily mortality in six U.S. cities. Environ Health Perspect 2000;108:941–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lippmann M, Ito K, Hwang JS, Maciejczyk P, Chen LC. Cardiovascular effects of nickel in ambient air. Environ Health Perspect 2006;114:1662–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cruz MJ, Costa R, Marquilles E, Morell F, Munoz X. [Occupational asthma caused by chromium and nickel.] Arch Bronconeumol 2006;42:302–306. (in Spanish) [DOI] [PubMed] [Google Scholar]
- 11.Oller AR, Costa M, Oberdorster G. Carcinogenicity assessment of selected nickel compounds. Toxicol Appl Pharmacol 1997;143:152–166. [DOI] [PubMed] [Google Scholar]
- 12.Ordonez CL, Shaughnessy TE, Matthay MA, Fahy JV. Increased neutrophil numbers and IL-8 levels in airway secretions in acute severe asthma: clinical and biologic significance. Am J Respir Crit Care Med 2000;161:1185–1190. [DOI] [PubMed] [Google Scholar]
- 13.Tsoutsou PG, Gourgoulianis KI, Petinaki E, Germenis A, Tsoutsou AG, Mpaka M, Efremidou S, Molyvdas PA. Cytokine levels in the sera of patients with idiopathic pulmonary fibrosis. Respir Med 2006;100:938–945. [DOI] [PubMed] [Google Scholar]
- 14.Cerri C, Chimenti D, Conti I, Neri T, Paggiaro P, Celi A. Monocyte/macrophage-derived microparticles up-regulate inflammatory mediator synthesis by human airway epithelial cells. J Immunol 2006;177:1975–1980. [DOI] [PubMed] [Google Scholar]
- 15.Luhrmann A, Deiters U, Skokowa J, Hanke M, Gessner JE, Muhlradt PF, Pabst R, Tschernig T. In vivo effects of a synthetic 2-kilodalton macrophage-activating lipopeptide of Mycoplasma fermentans after pulmonary application. Infect Immun 2002;70:3785–3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Means TK, Golenbock DT, Fenton MJ. Structure and function of Toll-like receptor proteins. Life Sci 2000;68:241–258. [DOI] [PubMed] [Google Scholar]
- 17.Fabisiak JP, Weiss RD, Powell GA, Dauber JH. Enhanced secretion of immune-modulating cytokines by human lung fibroblasts during in vitro infection with Mycoplasma fermentans. Am J Respir Cell Mol Biol 1993;8:358–364. [DOI] [PubMed] [Google Scholar]
- 18.Into T, Dohkan J, Inomata M, Nakashima M, Shibata K, Matsushita K. Synthesis and characterization of a dipalmitoylated lipopeptide derived from paralogous lipoproteins of Mycoplasma pneumoniae. Infect Immun 2007;75:2253–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ade N, Antonios D, Kerdine-Romer S, Boisleve F, Rousset F, Pallardy M. NF-{kappa}B plays a major role in the maturation of human dendritic cells induced by NiSO4 but not by DNCB. Toxicol Sci 2007;99:488–501. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- 21.Barchowsky A, Soucy NV, O'Hara KA, Hwa J, Noreault TL, Andrew AS. A novel pathway for nickel-induced interleukin-8 expression. J Biol Chem 2002;2002:24225–24231. [DOI] [PubMed] [Google Scholar]
- 22.Rozen S, Skaletsky H. Primer3 on the WWW for general users and biologist programmers, Bioinformatics methods and protocols: methods in molecular biology. Totowa, NJ: HUmana Press; 2000. [DOI] [PubMed]
- 23.Yoshimura T, Matsushima K, Tanaka S, Robinson EA, Appella E, Oppenheim JJ, Leonard EJ. Purification of a human monocyte-derived neutrophil chemotactic factor that has peptide sequence similarity to other host defense cytokines. Proc Natl Acad Sci USA 1987;84:9233–9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belperio JA, Keane MP, Arenberg DA, Addison CL, Ehlert JE, Burdick MD, Strieter RM. CXC chemokines in angiogenesis. J Leukoc Biol 2000;68:1–8. [PubMed] [Google Scholar]
- 25.Nakamura H, Yoshimura K, Jaffe HA, Crystal RG. Interleukin-8 gene expression in human bronchial epithelial cells. J Biol Chem 1991;266:19611–19617. [PubMed] [Google Scholar]
- 26.Yu Y, Chadee K. Prostaglandin E2 stimulates IL-8 gene expression in human colonic epithelial cells by a posttranscriptional mechanism. J Immunol 1998;161:3746–3752. [PubMed] [Google Scholar]
- 27.Elias JA, Zhu Z, Chupp G, Homer RJ. Airway remodeling in asthma. J Clin Invest 1999;104:1001–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emad A, Emad Y. Levels of cytokine in bronchoalveolar lavage (BAL) fluid in patients with pulmonary fibrosis due to sulfur mustard gas inhalation. J Interferon Cytokine Res 2007;27:38–43. [DOI] [PubMed] [Google Scholar]
- 29.Tager AM, Kradin RL, LaCamera P, Bercury SD, Campanella GS, Leary CP, Polosukhin V, Zhao LH, Sakamoto H, Blackwell TS, et al. Inhibition of pulmonary fibrosis by the chemokine IP-10/CXCL10. Am J Respir Cell Mol Biol 2004;31:395–404. [DOI] [PubMed] [Google Scholar]
- 30.Keane MP, Arenberg DA, Lynch JPR, Whyte RI, Iannettoni MD, Burdick MD, Wilke CA, Morris SB, Glass MC, DiGiovine B, et al. The CXC chemokines, IL-8 and IP-10, regulate angiogenic activity in idiopathic pulmonary fibrosis. J Immunol 1997;159:1437–1443. [PubMed] [Google Scholar]
- 31.Antoniades HN, Neville-Golden J, Galanopoulos T, Kradin RL, Valente AJ, Graves DT. Expression of monocyte chemoattractant protein 1 mRNA in human idiopathic pulmonary fibrosis. Proc Natl Acad Sci USA 1992;89:5371–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang K, Gharaee-Kermani M, Jones ML, Warren JS, Phan SH. Lung monocyte chemoattractant protein-1 gene expression in bleomycin-induced pulmonary fibrosis. J Immunol 1994;153:4733–4741. [PubMed] [Google Scholar]
- 33.Inoshima I, Kuwano K, Hamada N, Hagimoto N, Yoshimi M, Maeyama T, Takeshita A, Kitamoto S, Egashira K, Hara N. Anti-monocyte chemoattractant protein-1 gene therapy attenuates pulmonary fibrosis in mice. Am J Physiol Lung Cell Mol Physiol 2004;286:L1038–L1044. [DOI] [PubMed] [Google Scholar]
- 34.Ding J, Zhang X, Li J, Song L, Ouyang W, Zhang D, Xue C, Costa M, Melendez JA, Huang C. Nickel compounds render anti-apoptotic effect to human bronchial epithelial Beas-2B cells by induction of cyclooxygenase-2 through an IKKbeta/p65-dependent and IKKalpha- and p50-independent pathway. J Biol Chem 2006;281:39022–39032. [DOI] [PubMed] [Google Scholar]
- 35.Cao D, Bromberg PA, Samet JM. COX2 expression induced by diesel particles involves chromatin modification and degradation of HDAC1. Am J Respir Cell Mol Biol 2007;37:232–239. [DOI] [PubMed] [Google Scholar]
- 36.Mitsunari M, Yoshida S, Shoji T, Tsukihara S, Iwabe T, Harada T, Terakawa N. Macrophage-activating lipopeptide-2 induces cyclooxygenase-2 and prostaglandin E(2) via toll-like receptor 2 in human placental trophoblast cells. J Reprod Immunol 2006;72:46–59. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol 1998;16:225–260. [DOI] [PubMed] [Google Scholar]
- 38.Poli V. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J Biol Chem 1998;273:29279–29282. [DOI] [PubMed] [Google Scholar]
- 39.Mitsunari M, Yoshida S, Shoji T, Tsukihara S, Iwabe T, Harada T, Terakawa N. Macrophage-activating lipopeptide-2 induces cyclooxygenase-2 and prostaglandin E(2) via toll-like receptor 2 in human placental trophoblast cells. J Reprod Immunol 2006;72:46–59. [DOI] [PubMed] [Google Scholar]
- 40.Vichai V, Suyarnsesthakorn C, Pittayakhajonwut D, Sriklung K, Kirtikara K. Positive feedback regulation of COX-2 expression by prostaglandin metabolites. Inflamm Res 2005;54:163–172. [DOI] [PubMed] [Google Scholar]
- 41.Faour WH, He Y, He QW, de Ladurantaye M, Quintero M, Mancini A, Di Battista JA. Prostaglandin E(2) regulates the level and stability of cyclooxygenase-2 mRNA through activation of p38 mitogen-activated protein kinase in interleukin-1 beta-treated human synovial fibroblasts. J Biol Chem 2001;276:31720–31731. [DOI] [PubMed] [Google Scholar]
- 42.Kanda N, Watanabe S. Cyclooxygenase-2 inhibitor enhances whereas prostaglandin E2 inhibits the production of interferon-induced protein of 10 kDa in epidermoid carcinoma A431. J Invest Dermatol 2002;119:1080–1089. [DOI] [PubMed] [Google Scholar]
- 43.Piper PJ, Vane JR, Wyllie JH. Inactivation of prostaglandins by the lungs. Nature 1970;225:600–604. [DOI] [PubMed] [Google Scholar]
- 44.Takayama K, Garcia-Cardena G, Sukhova GK, Comander J, Gimbrone MA Jr, Libby P. Prostaglandin E2 suppresses chemokine production in human macrophages through the EP4 receptor. J Biol Chem 2002;277:44147–44154. [DOI] [PubMed] [Google Scholar]
- 45.Caristi S, Piraino G, Cucinotta M, Valenti A, Loddo S, Teti D. Prostaglandin E2 induces interleukin-8 gene transcription by activating C/EBP homologous protein in human T lymphocytes. J Biol Chem 2005;280:14433–14442. [DOI] [PubMed] [Google Scholar]
- 46.Moore BB, Ballinger MN, White ES, Green ME, Herrygers AB, Wilke CA, Toews GB, Peters-Golden M. Bleomycin-induced E prostanoid receptor changes alter fibroblast responses to prostaglandin E2. J Immunol 2005;174:5644–5649. [DOI] [PubMed] [Google Scholar]
- 47.Zelikoff JT, Schermerhorn KR, Fang K, Cohen MD, Schlesinger RB. A role for associated transition metals in the immunotoxicity of inhaled ambient particulate matter. Environ Health Perspect 2002;110:871–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deva R, Shankaranarayanan P, Ciccoli R, Nigam S. Candida albicans induces selectively transcriptional activation of cyclooxygenase-2 in HeLa cells: pivotal roles of Toll-like receptors, p38 mitogen-activated protein kinase, and NF-kappa B. J Immunol 2003;171:3047–3055. [DOI] [PubMed] [Google Scholar]
- 49.Rhee SH, Hwang D. Murine TOLL-like receptor 4 confers lipopolysaccharide responsiveness as determined by activation of NF kappa B and expression of the inducible cyclooxygenase. J Biol Chem 2000;275:34035–34040. [DOI] [PubMed] [Google Scholar]
- 50.Park GY, Christman JW. Involvement of cyclooxygenase-2 and prostaglandins in the molecular pathogenesis of inflammatory lung diseases. Am J Physiol Lung Cell Mol Physiol 2006;290:L797–L805. [DOI] [PMC free article] [PubMed] [Google Scholar]