Abstract
We have previously shown that ASK1-interacting protein 1 (AIP1) transduces tumor necrosis factor-induced ASK1-JNK signaling. Because endoplasmic reticulum (ER) stress activates ASK1-JNK signaling cascade, we investigated the role of AIP1 in ER stress-induced signaling. We created AIP1-deficient mice (AIP1-KO) from which mouse embryonic fibroblasts and vascular endothelial cells were isolated. AIP1-KO cells show dramatic reductions in ER stress-induced, but not oxidative stress-induced, ASK1-JNK activation and cell apoptosis. The ER stress-induced IRE1-JNK/XBP-1 axis, but not the PERK-CHOP1 axis, is blunted in AIP1-KO cells. ER stress induced formation of an AIP1-IRE1 complex, and the PH domain of AIP1 is critical for the IRE1 interaction. Furthermore, reconstitution of AIP1-KO cells with AIP1 wild type, not an AIP1 mutant with a deletion of the PH domain (AIP1-ΔPH), restores ER stress-induced IRE1-JNK/XBP-1 signaling. AIP1-IRE1 association facilitates IRE1 dimerization, a critical step for activation of IRE1 signaling. More importantly, AIP1-KO mice show impaired ER stress-induced IRE1-dependent signaling in vivo. We conclude that AIP1 is essential for transducing the IRE1-mediated ER stress response.
Proteins synthesized in the endoplasmic reticulum (ER)3 are properly folded with the assistance of ER chaperones. Misfolded proteins are disposed of by ER-associated protein degradation. Accumulation of misfolded protein in the ER triggers an adaptive ER stress response termed the unfolded protein response (1, 2). For experimental purposes, chemicals such as thapsigargin (which depletes Ca2+ from ER), tunicamycin (which inhibits protein N-linked glycosylation), and dithiothreitol (which disrupts protein disulfide bonds) are used to induce ER stress in cultured cells or animals. There are three distinct signaling pathways that are triggered in response to ER stress, initiated by protein kinase activated by ds RNA (PKR)-like ER kinase (PERK), activating transcription factor 6 (ATF6), and inositol-requiring enzyme-1 (IRE1). PERK is an ER-resident serine/threonine protein kinase that phosphorylates eIF2α (the subunit of translational initiation factor 2), attenuating new protein synthesis to prevent protein loading into the ER. Interestingly, eIF2α phosphorylation induces translation of ATF4 and its target genes such as CHOP, contributing to apoptosis. ATF6 is cleaved to release a cytosolic fragment that translocates to the nucleus to induce gene expression of chaperones that promote correct protein folding. IRE1 is a serine-threonine protein kinase/endonuclease that contains an ER luminal domain, a transmembrane domain, a cytoplasmic kinase, and an endonuclease domain. IRE1 has two isoforms; IRE1α is ubiquitously expressed whereas IREβ is primarily expressed in intestine tissues (we will use the term IRE1 to refer to IRE1α unless specified otherwise). Upon activation in response to ER stress, IRE1 has at least two different actions. First, the endoribonuclease activity of IRE1 cleaves XBP-1 mRNA, converting it into a potent transcriptional activator that, in turn, induces gene expression of proteins involved in protein degradation (3, 4). Second, recent studies from several laboratories have shown that IRE1 links ER stress to the activation of JNK signaling pathways. Specifically, IRE1 binds to TRAF2 and through its kinase activity couples ER stress to activation of JNK (5). The activation of JNK by ER stress requires the presence of ASK1 (6, 7). It has therefore been proposed that ER stress induces the formation of IRE1-TRAF2 complex that leads to ASK1-JNK activation (7).
Among MAP3Ks, apoptosis signal-regulating kinase-1 (ASK1) is an enzyme that specifically activates a cascade ending with JNK/p38 mitogen-activated protein kinase (MAPK) (but not NF-κB) activation. Structurally, ASK1 is a 170-kDa protein containing an inhibitory N-terminal domain, an internal kinase domain, and the C-terminal domain (8, 9). ASK1 via its C-terminal domain binds to TNF receptor-associated factors (TRAFs) and this association is required for ASK1 activation (9). The current model for ASK1 activation by TNF involves several critical steps, including release of inhibitors (thioredoxin and 14-3-3) (10, 11), TRAF-dependent homodimerization/polymerization (12), and ASK1 autophosphorylation at Thr-845 (13). We have recently shown that AIP1 (ASK1-interacting protein-1), by interacting with TRAF2-ASK1, functions as a transducer in TNF-induced ASK1-JNK apoptotic signaling (14–16). Specifically, AIP1 preferentially binds to dephosphorylated ASK1, facilitating release of the inhibitor 14-3-3 from ASK1 (14). More recently, we have shown that TNF-activated RIP1 mediates AIP1 phosphorylation at the 14-3-3-binding site Ser-604 and this phosphorylation of AIP1 by RIP1 is critical for TNF-induced AIP1-TRAF2-ASK1 complex formation and JNK signaling (16).
Given the critical role of TRAF2-ASK1 in mediating ER stress-induced JNK signaling, we wished to determine whether AIP1 plays a similar role in this process. To this end, we generated AIP1-deficient mice and isolated the mouse embryonic fibroblasts (MEF) and vascular endothelial cells (EC) to determine the role of AIP1 in ER stress responses. Our data suggest that AIP1 deficiency specifically impairs ER stress-induced IRE1-JNK/XBP-1 axis, but not the PERK-CHOP1 axis. Structural analyses suggest that AIP1 uses its PH domain to associate with IRE1, facilitating IRE1 dimerization and activation. Consistent with the cell culture findings, AIP1-deficient mice show impaired IRE1-JNK/XBP-1 signaling. We conclude that AIP1 plays an essential role in transducing IRE1-mediated ER stress response, analogous to its role in the TNF signaling cascade.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatment—Human umbilical vein EC were purchased from the Yale Vascular Biology and Therapeutic Endothelial Cell Culture Core Facility (Yale University). Bovine aortic EC (BAEC) were purchased from Clonetics (San Diego, CA) and cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Mouse EC isolation from lung tissues was performed as we described (17–19) followed by immuno-selection and immortalization modified from the protocol described by Lim et al. (20). For immunoselection, 10 μl of beads (per T-75 of mouse lung cells) were washed with 1 ml of buffer A (phosphate-buffered saline + 2% fetal bovine serum) three times and resuspended in 100 μl of buffer A. 10 μl (10 μg) of anti-mouse ICAM-2 or 10 μl (10 μg) of PECAM-1 were added and rocked at 4 °C for 2 h. Beads were washed three times and resuspended in 160 μl of buffer A. Confluent mouse lung cells cultured in a T-75 flask were placed at 4 °C for 5 min and incubated with the beads at 4 °C for 1 h. Cells were then washed with warm phosphate-buffered saline and treated with 3 ml of warm trypsin/EDTA. When cells were detached, 7 ml of growth medium was added. An empty 15-ml tube in the magnetic was placed on the holder, and the cell suspension (∼10 ml) was added slowly by placing the pipette on the wall of the tube so that the cells passed through the magnetic field. Cells were incubated for 5 min, and the medium was carefully aspirated. The 15-ml tube was removed from the magnetic holder and the beads/cells were resuspended in 10 ml of medium. The selected cells were plated on 0.2% gelatin-coated flasks and cultured for 3–7 days. When the cells were confluent, another round of immunoselection was repeated. Human recombinant TNF was from R&D Systems Inc. (Minneapolis, MN) and used at 10 ng/ml. Cells were also treated with H2O2 (1 mm), t-butyl hydroperoxide (BSO, 0.5 mm), paraquat (1 mm), thapsigargin (5 μm), or tunicamycin (10 μg/ml) as indicated.
Plasmid Construction and Transfection—Expression plasmids for AIP1 have been described previously (14, 15). Expression plasmids for IRE1α (WT and KA) were constructed in Dr. Urano's laboratory (4). Myc-tagged IRE1β constructs were from Dr. David Ron (Skirball Institute of Biomolecular Medicine, New York University School of Medicine). BAEC were transfected by Lipofectamine 2000 (Invitrogen). Mouse lung EC (MLEC) were transfected by HMVEC-L Nucleofector kit (Amaxa) according to the protocol provided by the manufacturer.
ASK1 Kinase Assay—ASK1 in vitro kinase assays were performed as described previously using glutathione S-transferase-MKK4 fusion protein as a substrate (14, 15). Briefly, total 400 μg of cell lysates were immunoprecipitated with 5 μgof antibody against ASK1 (Santa Cruz Biotechnology). The immunoprecipitates were mixed with 10 μg of glutathione S-transferase-MKK4 suspended in the kinase buffer (20 mm Hepes, pH 7.6, 20 mm MgCl2, 25 mm β-glycerophosphate, 100 μm sodium orthovanadate, 2 mm dithiothreitol, 20 μm ATP) containing 1 μl (10 μCi) of [γ-32P]ATP. The kinase assay was performed at 25 °C for 30 min. The reaction was terminated by the addition of Laemmli sample buffer, the products were resolved by SDS-PAGE (12%), and the phosphorylated KST-MKK4 was visualized by autoradiography. ASK1 protein was determined by Western blot with anti-ASK1.
Immunoprecipitation and Immunoblotting—BAEC after various treatments were washed twice with cold phosphate-buffered saline and lysed in 1.5 ml of cold lysis buffer (50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 0.1% Triton X-100, 0.75% Brij 96, 1 mm sodium orthovanadate, 1 mm sodium fluoride, 1 mm sodium pyrophosphate, 10 mg/ml aprotinin, 10 mg/ml leupeptin, 2 mm phenylmethylsulfonyl fluoride, 1 mm EDTA) for 20 min on ice. Immunoprecipitation and immunoblotting were performed as described previously (14, 15). Antibodies against AIP1 and phospho-AIP1 (pS604) were described previously (14–16). Anti-IRE1α and anti-p-IRE1α were generated from Dr. Fumihiko Urano or purchased from Novus Biologicals. Antibodies against ASK1, TRAF2, pPERK, CHOP, XBP1, JNK, and p38 were from Santa Cruz Biotechnology. Antibodies against p-eIF2α, eIF2α, pASK(T845), p-JNK, p-P38, and BiP were from Cell Signaling. Anti-Myc monoclonal antibody was purchased from Roche Applied Science, and anti-FLAG monoclonal antibody (M2) antibody was from Sigma. Anti-β-tubulin was from BD Biosciences.
Quantification of Apoptotic Cells—A cell-killing assay was performed as described previously with a modification (21). The propidium iodide (PI) exclusion method for loss of integrity of cell membranes was used to assess viability. In brief, cells were suspended in phosphate-buffered saline containing 25 μg/ml PI for 5 min at 37 °C and then subjected to analytic flow cytometry on a FACSort (BD Biosciences) immediately after labeling. A light scatter gate was set up to eliminate cell debris from the analysis. The PI fluorescence signal was recorded on the FL3 channel and analyzed by using CellQuest software. Phosphatidylserine translocation, which precedes loss of PI exclusion in apoptotic cell death, was assessed by an annexin V- fluorescein isothiocyanate/PI staining kit (Roche Applied Science) following the manufacturer's protocol. For nuclear morphology, cells were stained with 4′,6-diamidino-2-phenylindole and apoptotic cells (nuclei condensation) were visualized under UV microscope.
Targeted Inactivation of AIP1 Gene by Homologous Recombination—The AIP1 targeting arms were isolated from BAC clone identified by screening a BAC library from Research Genetics (Invitrogen) using AIP1 cDNA as a probe. The targeting vector was constructed in pEASYflox backbone to contain a loxP site inserted upstream of AIP1 exon 5 and a neomycin cassette (Neo) flanked by two loxP sites downstream of exon 6 using standard molecular procedures. The linearized targeting construct was electroporated into 129/C57B/6 embryonic stem cells, and the targeted clones were selected with G418 and ganciclovir. Resistant clones were screened for homologous recombination by PCR and confirmed by Southern blot analysis. Two independent AIP1lox embryonic stem cell clones were injected into WT blastocytes. Chimeras were further bred with WT females for germ line transmission. AIP1lox/lox mice were mated with β-actin-Cre mice to mediate a recombination in vivo, resulting in a complete deletion of AIP1 exon 5 and exon 6 and a frameshift of the downstream GAP domain as well the Neo gene to generate the heterozygous mice (AIP+/–). AIP1-KO mice were obtained by breeding heterozygous mice and verified by genotyping using specific primers. A deletion of AIP1 protein in AIP1-KO mouse tissues was also confirmed by Western blotting and immunohistochemistry with an anti-AIP1 antibody.
Animal Protocol—All animal studies were approved by the institutional animal care and use committees of Yale University. AIP1-WT and AIP1-KO mice were confirmed by genotyping with specific primers. Mice were injected intraperitoneally with tunicamycin at 1 μg/g body weight.
Gene Expression in Tissues—Total RNA of lung, liver, or aorta was isolated by using phenol/chloroform and isolated using RNeasy kit with DNase I digestion (Qiagen, Valencia, CA). Reverse transcription was done by standard procedure (Super Script First-Strand Synthesis System; Qiagen) using 1 μg of total RNA. Quantitative real-time PCR was performed by using iQ SYBR Green Supermix on the iCycler Real-Time Detection System (Bio-Rad Laboratories) as we described recently (18, 22). Specific primers for mouse ero1-L, PDI-P5, BiP, and 18 S ribosomal RNA as an internal control were used as follows: 18 S RNA, 5′-TTCCGATAACGAACGAGACTCT-3′ and 5′-TGGCTGAACGCCACTTGTC-3′; ero1L, 5′-TCAGTGGACCAAGCATGATGA-3′ and 5′-TCCACATACTCAGCATCGGG-3′; PDI-P5, 5′-AGCTCGTCAAGGATCGCCT-3′ and 5′-TATCACCTCTGCCCTGCTTTC-3′; CHOP 5′-CCACCACACCTGAAAGCAGAA-3′ and 5′-AGGTGAAAGGCAGGGACTCA; and grp78/bip, 5′-TCATCGGACGCACTTGGAA-3′ and 5′-CAACCACCTTGAATGGCAAGA-3′. Relative amount of mRNA in untreated and tunicamycin (Tm)-injected mouse tissues was quantified.
Statistical Analysis—All data are expressed as means ± S.E. Statistical differences were measured by the nonparametric Mann-Whitney test. A value of p < 0.05 was considered statistically significant.
RESULTS
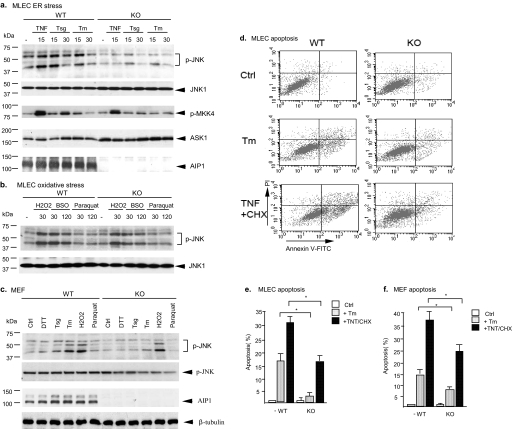
ER Stress-induced Activation of ASK1-JNK and Apoptosis Is Blunted in AIP1-KO Cells—We created AIP1-deficient mice based on a homologous recombination (see “Experimental Procedures” for details). To determine the role of AIP1 in ER stress signaling, we isolated MLEC and MEF. AIP1-WT and AIP1-KO MLEC were treated with an ER stress stimulus, thapsigargin (Tsg, 5 μm) or Tm (1 μm) for the indicated times. TNF treatment (10 ng/ml) was used as a control. ASK1 activity was determined by an in vitro kinase assay using glutathione S-transferase-MKK4 as a substrate. Activation of JNK was detected by Western blotting with a phospho-specific antibody. TNF-induced ASK1-JNK activation was reduced in AIP1-KO EC, consistent with our previous reports that AIP1 is critical for TNF-induced ASK1-JNK signaling (Fig. 1a). Interestingly, AIP1-KO EC also showed a dramatic reduction in ER stress-induced ASK1-JNK signaling (Fig. 1a). EC were also treated with oxidative stress stimuli (H2O2, t-butyl hydroperoxide, or paraquat). In this case AIP1 deficiency had no effects on oxidative stress-induced JNK activation (Fig. 1b). Activation of ASK1-JNK has been linked to EC apoptosis, and we determined ER stress-induced EC apoptosis by a double staining with propidium iodide and annexin V followed by FACS analysis. Consistent with an impaired ASK1-JNK signaling, AIP1-KO EC showed a dramatic reduction in ER stress-induced apoptosis (Fig. 1, c and d). Similarly, reduced ER stress-induced ASK1-JNK activation and apoptosis were observed in AIP1-KO MEF compared with AIP1-WT MEF (Fig. 1, e and f).
FIGURE 1.
ER stress-induced activation of ASK1-JNK and apoptosis are blunted in AIP1-KO cells. Mouse lung microvascular EC (MLEC) and MEFs were isolated from AIP1-WT and AIP1-KO. a, MLEC were treated with TNF (10 ng/ml), thapsigargin (Tsg, 5 μm), or tunicamycin (Tm, 1 μm) for the indicated times. ASK1 activity was determined by an in vitro kinase assay using GST-MKK4 as a substrate. Activation of JNK was detected by Western blotting with a phospho-specific antibody against p-JNK1/2. Total JNK was determined by Western blot with anti-JNK1. b, MLEC were treated with oxidative stress stimuli H2O2 (1 mm), buthionine-sulfoximine (BSO, 0.5 mm), or paraquat (1 mm). Activation of JNK was detected as described in a. c, MEFs were treated with ER stress and oxidative stress as indicated for 30 min. Activation of JNK was detected as described in a. AIP1 and β-tubulin proteins were determined by Western blot with respective antibodies. d and e, MLEC were treated with Tm (1 μm) or TNF (10 ng/ml) plus cycloheximide (CHX, 10 μg/ml) for 6 h. EC apoptosis was also determined with annexin-V/PI staining followed by FACS analysis. Representative FACS is shown in d, and the % of apoptotic cells (both upper and lower right quadruplets in FACS) are quantified in e. f, MEFs were treated with Tm (1 μm) or TNF (10 ng/ml) plus cycloheximide (CHX, 10 μg/ml) for 6 h. EC apoptosis was determined as in d and e. Data are presented in e and f as mean of duplicates from two independent experiments. * indicates a statistical significance between WT and AIP1-KO cells; p < 0.05. Error bars show the calculated S.D.
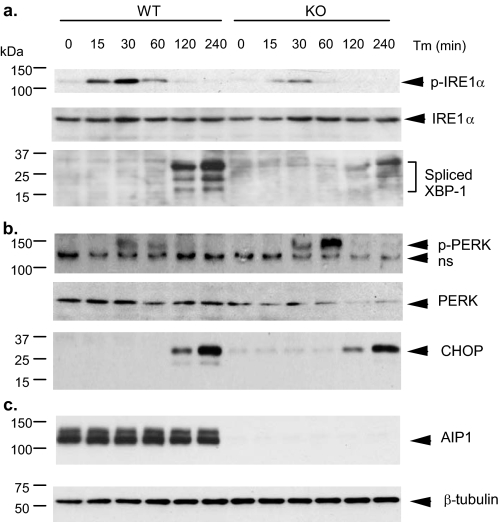
ER Stress-induced Activation of IRE1-JNK/XBP-1 Axis Is Blunted in AIP1-KO EC—We then determined whether AIP1 regulates IRE1, an upstream activator of ASK1-JNK in ER stress signaling. Activation of IRE1 also leads to induction of spliced forms of XBP-1. AIP1-WT and AIP1-KO EC were treated with tunicamycin (which induces ER stress by inhibiting protein N-linked glycosylation) for the indicated times (0–240 min). Phosphorylation of IRE1 and induction of XBP-1 expression were determined by Western blotting with respective antibodies. As shown in Fig. 2a, Tm strongly induced activation of IRE1 at 15–30 min and induction of spliced forms of XBP-1 at 120–240 min. Tm-induced activation of IRE1 and synthesis of XBP-1 were blunted in AIP1-KO EC (Fig. 2a). Activation of the PERK-CHOP axis, a parallel pathway activated by ER stress, was also examined by measuring phosphorylation of PERK and expression of CHOP. In contrast to IRE1-JNK/XBP-1 axis, Tm-induced phosphorylation of PERK and induction of CHOP expression were still activated in AIP1-KO MLEC. AIP1 deletion augmented Tm-induced phosphorylation of PERK, suggesting a possible negative role of AIP1 in PERK signaling (Fig. 2, b and c). Taken together, these observations suggest that AIP1 is specifically involved in ER stress-activated IRE1-JNK/XBP-1 axis but not the PERK-CHOP axis.
FIGURE 2.
ER stress-induced activation of IRE1-JNK/XBP-1 is blunted in AIP1-KO EC. AIP1-WT and AIP1-KO MLEC were treated with Tm (1 μm) for the indicated times (0, 15, 30, 60, 120, and 240 min). Phosphorylation of IRE1 and JNK and induction of the spliced XBP-1 (a) and phosphorylation of PERK and expression of CHOP (b) as well as total AIP1 andβ-tubulin (c) were determined by Western blot with respective antibodies. ns in b, nonspecific band.
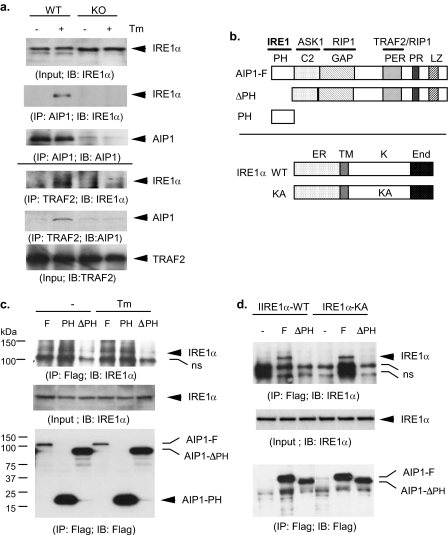
The PH Domain of AIP1 Is Critical for IRE1 Binding—To determine the mechanism by which AIP1 regulates IRE1 activity, we examined whether ER stress induces association of AIP1 with IRE1. MLEC were treated with Tm (1 μm for 15 min), and endogenous complexes of AIP1-IRE1 in EC were determined by co-immunoprecipitation assays. AIP1-IRE1 complex was not detected in resting EC. However, ER stress strongly induced an association of AIP1 with IRE1 (Fig. 3a, top three panels). ER stress also induced an association of TRAF2 with IRE1 and AIP1 as determined by immunoprecipitation assay with anti-TRAF2 (Fig. 3a, bottom three panels). These data suggest that IRE1, AIP1, and TRAF2 form complexes in response to ER stress. Interestingly, IRE1-TRAF2 complex was not detected in AIP1-KO cells, suggesting that the formation of IRE1-AIP1 complex is required for subsequent recruitment of TRAF2 to IRE1.
FIGURE 3.
ER stress induces formation of AIP1-IRE1 complex, and the PH domain of AIP1 is critical for the RE1 binding. a, ER stress-induced formation of endogenous AIP1-IRE1-TRAF2 complexes in EC. MLEC were treated with Tm (1 μm for 15 min). Association of AIP1 with IRE1 was determined by immunoprecipitation with AIP1 followed by Western blot with anti-IRE1 or anti-AIP1 (top three panels). Association of TRAF2 with IRE1 and AIP1 was determined by immunoprecipitation assay with anti-TRAF2 followed by Western blot with anti-IRE1 or anti-AIP1 (bottom three panels). b, schematic diagram for AIP1 and IRE1α structure domains and expression constructs. AIP1 contains a PH, a C2, and a GAP domain at the N-terminal half and a Period-like (PER) domain, a proline-rich (PR) region, and leucine-zipper (LZ) motif. The specific domains responsible for protein interactions with IRE1, ASK1, TRAF2, and RIP1 are indicated. IRE1α WT contains the ER lumen domain (ER), the transmembrane domain (TM), the kinase domain (K), and the endonuclease domain (End). KA, K599A. c, BAEC were transfected with FLAG-tagged AIP1-F, AIP1-ΔPH, or AIP1-PH in the presence of IRE1-WT. Cells were treated with Tm (1 μm for 15 min), and association of AIP1 with IRE1 was determined by immunoprecipitation with anti-FLAG followed by Western blot with anti-IRE1. d, BAEC were transfected with FLAG-tagged AIP1-F or AIP1-PH in the presence of IRE1-WT or IRE1-KA. Association of AIP1 with IRE1 was determined as in c. ns in c and d, nonspecific band.
To map the critical domain of AIP1 for IRE1 binding, BAEC were transfected with FLAG-tagged AIP1-F, AIP1-ΔPH (with a deletion of the PH domain), or AIP1-PH (containing the PH domain only) in the presence of IRE1 (Fig. 3b). Association of AIP1-F and IRE1 was determined by co-immunoprecipitation assays. AIP1-IRE1 complex was weakly detected in resting EC and was enhanced in response to Tm treatment (Fig. 3c). AIP1-PH strongly bound to IRE1 in an ER stress-independent manner. However, AIP1-ΔPH failed to bind to IRE1, suggesting that the PH domain on AIP1 is necessary and sufficient for the binding to IRE1. The kinase activity of IRE1 has been shown to be critical for IRE1-TRAF2 interaction (5). To determine whether the kinase activity of IRE1 is necessary for AIP1 binding, BAEC were transfected with FLAG-tagged AIP1-F and AIP1-ΔPH in the presence of IRE1-WT or IRE1-KA (a kinase-dead form) (Fig. 3b). AIP1-F (but not AIP1-ΔPH) binds to both IRE1-WT and IRE1-KA, indicating that AIP1-IRE1 interaction was independent of the kinase activity of IRE1 (Fig. 3d).
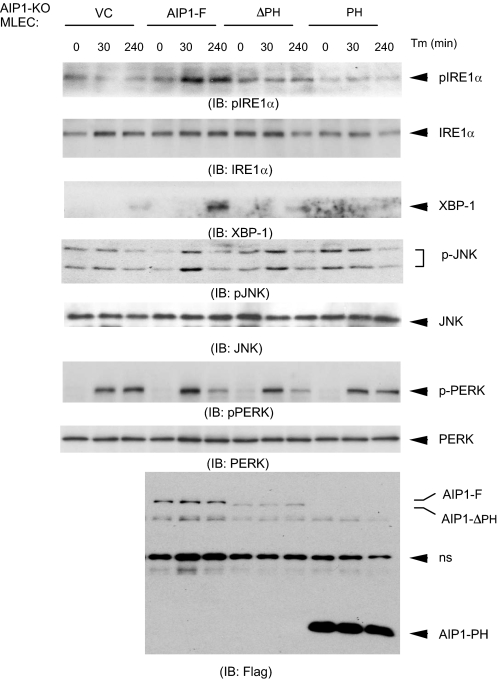
The PH Domain of AIP1 Is Necessary but Not Sufficient to Activate IRE1-JNK/XBP-1 Signaling—The critical role of the AIP1 PH domain in IRE1 binding prompted us to determine its role in AIP1-mediated IRE1 signaling. To eliminate the effect of endogenous AIP1, we reconstituted AIP1-KO MLEC with AIP1-F, AIP1-ΔPH, or AIP1-PH. Reconstituted MLEC were treated with Tm for the indicated times (0, 30, and 240 min), and phosphorylation of IRE1, JNK, and PERK was determined. Reconstitution of AIP1-KO MLEC with AIP1-F restored the ER stress-induced activation of IRE1-JNK/XBP-1 signaling (Fig. 4). In contrast, reconstitution of AIP1-ΔPH or AIP1-PH, like vector control (VC), failed to restore the ER stress response. As a control, expression of different forms of AIP1 had no effects on Tm-induced phosphorylation of PERK. These results suggest that the PH domain of AIP1 is necessary but not sufficient to activate IRE1-JNK/XBP-1 signaling.
FIGURE 4.
The PH domain of AIP1 is necessary but not sufficient to activate IRE1-JNK/XBP-1 signaling. AIP1-KO MLEC were reconstituted with AIP1-F, AIP1-ΔPH, or AIP1-PH by electroporation transfection. Reconstituted MLEC were treated with Tm for the indicated times (0, 30, and 240 min); phosphorylation of IRE1, JNK, and PERK as well as expression of the spliced XBP-1 were determined by Western blot with respective antibodies. Expression of AIP1 proteins was determined by Western blot with anti-FLAG. ns, nonspecific band.
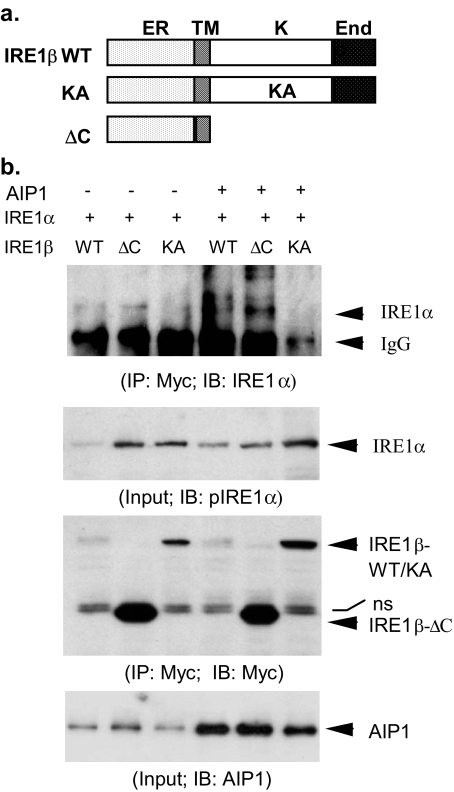
AIP1 Enhances Dimerization of IRE1—Dimerization of IRE1 is a prerequisite step for its activation and the subsequent downstream events (23, 24). We determined whether AIP1 induces IRE1 dimerization. To this end, we determined the effects of AIP1 on the dimerization between IRE1α and IRE1β. Similar to IRE1α (Fig. 3b), IRE1β contains an ER lumen domain, a transmembrane domain, a kinase domain, and an endonuclease domain (Fig. 5a). Myc-tagged IRE1β-WT, -KA, and -ΔC were co-expressed into BAEC with IRE1α in the presence or absence of AIP1. Association of IRE1α and IRE1β was determined by a co-immunoprecipitation assay with anti-Myc followed by Western blot with anti-IRE1α. Association of IRE1α with IRE1β-WT and IRE1β-ΔC was strongly increased by AIP1 co-expression, indicating that IRE1 proteins dimerize via their luminal/transmembrane domain. (Fig. 5b). In contrast, IRE1β-KA failed to associate with IRE1α even in the presence of AIP1, suggesting that the kinase activity of IRE1 is required for their dimerization and IRE1β-KA may function as a dominant negative mutant. These data indicate that AIP1 facilitate IRE1 protein dimerization.
FIGURE 5.
AIP1 enhances dimerization of IRE1 proteins. a, schematic diagram for IRE1β structure domains and expression constructs. IRE1β WT contains similar domains as IRE1α (ER, ER lumen domain; TM, transmembrane domain; K, kinase domain; End, endonuclease domain). KA, K536A; ΔC, deletion of the C-terminal kinase and End domains. b, AIP1 enhances dimerization of IRE1α with IRE1β. Myc-tagged IRE1β WT, -KA, and -ΔC were co-transfected with IRE1α into BAEC in the absence or presence of AIP1 for 24 h. Heterodimerization of IRE1α and IRE1β was determined by immunoprecipitation with anti-Myc followed by Western blot with anti-IRE1α. Total IRE1α, IRE1β, and AIP1 were determined by Western blot with anti-IRE1α, anti-Myc, and anti-AIP1, respectively. ns, nonspecific band.
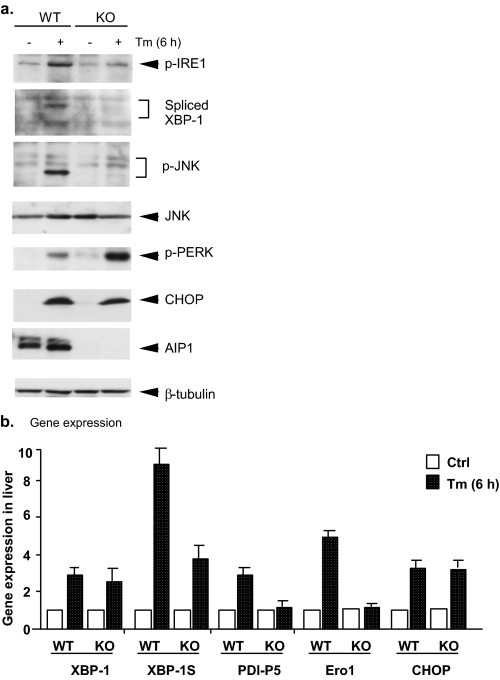
ER Stress Responses Are Blunted in AIP1-KO Mice—Finally, we examined the role of AIP1 in ER stress responses in vivo. AIP1-WT and AIP1-KO mice were administered intraperitoneally with tunicamycin at 1 μg/g body weight, and lung tissue was harvested at 6, 12, and 24 h after injection. ER stress responses were determined by Western blot for signaling. Six hours after Tm injection, phosphorylation of IRE1 and IRE1 downstream target JNK as well as IRE1-dependent expression of XBP-1 were dramatically induced in WT mouse tissues. Activation of IRE1-JNK and expression of XBP-1 in AIP1-KO mice were significantly reduced compared with WT mice. Expression of XBP-1 target genes including the oxidoreductase endoplasmic reticulum oxidoreductin-1 (Ero1-L) and protein disulfide isomerase was determined by quantitative reverse transcription PCR (Fig. 6b). Similarly, activation of PERK and its downstream CHOP1 was detected at 6 h post-injection. In contrast to the IRE1 axis, AIP1 deficiency had no effects on Tm-induced activation of PERK and induction of CHOP (Fig. 6a). These data suggest that AIP1 may specifically involve in ER stress-induced IRE1-JNK/XBP-1 axis without effects on the PERK-CHOP pathway.
FIGURE 6.
ER stress responses are blunted in AIP1-KO mice. AIP1-WT and AIP1-KO mice were administered intraperitoneally with tunicamycin (Tm) at 1 μg/g body weight, and lung tissues were harvested at 6 h after injection. a, activation of the IRE1-JNK/XBP-1 axis and activation of the PERK-CHOP axis was determined by Western blot with respective antibodies. Expression of AIP1 and β-tubulin was also determined with respective antibodies. b, characterization of gene expression in response to ER stress. Expression of ER stress-responsive genes was determined by quantitative reverse transcription PCR with specific primers (see “Experimental Procedures”). Data represent the -fold increases of each gene by taking untreated AIP1-WT as 1.0. n = 3 for each strain/time point. -Fold induction of ER stress-responsive genes is shown. Error bars show the calculated S.D.
DISCUSSION
In the present study, we have defined a critical function of AIP1 in ER stress-mediated signaling and responses in vitro and in vivo. AIP1-KO fibroblasts (MEF) and vascular EC show dramatic reduction in ER stress-induced ASK1-JNK activation and cell apoptosis. We demonstrated that ER stress-induced IRE1-JNK/XBP-1 axis, but not the PERK-CHOP1 axis, is specifically blunted in AIP1-KO cells. AIP1 via its PH domain associates with IRE1. Moreover, the PH domain of AIP1 is critical for AIP1-mediated IRE1 activation as demonstrated by reconstitution of AIP1-KO cells with AIP1 or AIP1 mutant with a deletion of the PH domain. Finally, we showed that AIP1 facilitates IRE1 dimerization, a critical step for activation of IRE1 signaling. Our study demonstrates an essential role of AIP1 in transducing IRE1-mediated ER stress responses.
Recent studies suggest that TNFR1 signaling components, including TNFR1, RIP1, TRAF2, and ASK1, are involved in the activation of JNK as ER stress-induced JNK activation is reduced or diminished in MEFs with deficiency of each individual component (6, 7, 25–27). The current model suggests that TNFR1 signaling components function downstream of IRE1. ER stress could induce association of IRE1 with TNFR1 at 1 h after ER stress stimulation (25). It is not known whether this resulted from an autocrine effect of TNF that has been shown to be induced by ER stress in MEFs (27). Similarly, IRE1 associates with TRAF2 in an IRE1 kinase-dependent manner (5), suggesting that IRE1 recruits TRAF2 after IRE1 activation in response to ER stress. In contrast, we showed that association of AIP1 with IRE1 is not dependent on the kinase activity of IRE1. Furthermore, AIP1 binds to IRE1 in response to ER stress, facilitating IRE1 dimerization. More importantly, ER stress-induced IRE1 activation and its downstream ASK1-JNK signaling are blunted in AIP1-KO EC. So far, no studies have determined the effects of TNF signaling components on IRE1 activity/activation, and our study provides the first evidence that a TNF signaling component, AIP1, may directly regulate IRE1 activity.
One current model of IRE1 activation (so-called “Competitive Deprivation” model of BiP dissociation) (23, 24) proposes that the abundant luminal ER chaperon BiP (also known as glucose-regulated protein 78, GRP78) maintains IRE1 in an inactive state. Upon ER stress, BiP preferentially binds to unfolded proteins, thereby releasing IRE1 to undergo activation by homodimerization and autophosphorylation. Alternative mechanisms, IRE1 activation without release of BiP, have been also proposed (4, 28, 29). In response to ER stress stimulation, AIP1 associates with IRE1 and enhances IRE1 dimerization upon binding to IRE1, suggesting a direct role of AIP1 in regulating IRE1 activity. Similarly, it has been shown that Bcl-2 family proteins BAX and BAK can directly bind to IRE1 and regulate IRE1 activation (30). It needs to be further determined whether AIP1 facilitates the release of IRE1 from BiP, contributing to AIP1-mediated IRE1 dimerization. The PH domain of AIP1 is critical for IRE1 binding. However, reconstitutions of AIP1-KO EC with AIP1-F, AIP1-ΔPH, and AIP1-PH indicate that the PH domain of AIP1 is necessary but not sufficient to mediate ER stress-induced activation of IRE1, suggesting other domains of AIP1 or/and its associated proteins are required in this process. Thus, the exact mechanism by which AIP1 mediates IRE1 activation needs be investigated.
It has been shown that IRE1 recruits TRAF2, which in turn activates ASK1-JNK cascade in response to ER stress (5). Our data show that ER stress induces association of AIP1 with both IRE1 and TRAF2, suggesting formation of a putative IRE1-AIP1-TRAF2 complex. We have previously defined the critical domain of AIP1 in binding with TRAF2. AIP1 utilizes a C-terminal PER (Period-like) domain that binds to the N-terminal RING finger of TRAF2, transducing TNF-induced ASK1-JNK signaling (15). It is conceivable that AIP1-TRAF2 form a similar complex, mediating ER stress-induced activation of the ASK1-JNK signaling pathway. More importantly, our data demonstrate that AIP1 is crucial for ER stress-induced activation of IRE1 and formation of the IRE1-TRAF2 complex. Our results are consistent with a previous report that only an active form of IRE1 associates with TRAF2 (5). Several remaining questions are raised from our current study. The localization of the IRE1-AIP1 complex has not been determined; the role of RIP1, a critical kinase involved in AIP1-mediated ASK1-JNK activation in TNF signaling, is not known. We will address these questions in future studies.
A malfunction of the ER stress response has been associated with a variety of human diseases such as atherosclerosis, diabetes, and neurodegenerative disorders. Given the critical role of AIP1 in ER stress response, the availability of AIP1-flox and AIP1-deficient mice warrants further investigation on AIP1 function in the progression of these diseases.
Acknowledgments
We thank Dr. Jordan S. Pober for discussion.
This work was supported by National Institutes of Health Grants R01 HL65978-5, R01 HL077357-1, and P01 HL070295-6, an Established Investigator Award from the American Heart Association (0440172N) (to W. M.), and National Natural Science Foundation of China Grant 30772392 (to S. T.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ER, endoplasmic reticulum; PERK, PKR-like ER kinase; JNK, c-Jun N-terminal kinase; TNF, tumor necrosis factor; MEF, mouse embryo fibroblast; EC, endothelial cell; BAEC, bovine aortic EC; WT, wild type; MLEC, mouse lung EC; PI, propidium iodide; Tm, tunicamycin; FACS, fluorescence-activated cell sorter; PH, pleckstrin homology; MKK4, mitogen-activated protein kinase kinase 4; KO, knock-out.
References
- 1.Marciniak, S. J., and Ron, D. (2006) Physiol. Rev. 86 1133–1149 [DOI] [PubMed] [Google Scholar]
- 2.Yoshida, H. (2007) FEBS J. 274 630–658 [DOI] [PubMed] [Google Scholar]
- 3.Calfon, M., Zeng, H., Urano, F., Till, J. H., Hubbard, S. R., Harding, H. P., Clark, S. G., and Ron, D. (2002) Nature 415 92–96 [DOI] [PubMed] [Google Scholar]
- 4.Lipson, K. L., Fonseca, S. G., Ishigaki, S., Nguyen, L. X., Foss, E., Bortell, R., Rossini, A. A., and Urano, F. (2006) Cell Metab. 4 245–254 [DOI] [PubMed] [Google Scholar]
- 5.Urano, F., Wang, X., Bertolotti, A., Zhang, Y., Chung, P., Harding, H. P., and Ron, D. (2000) Science 287 664–666 [DOI] [PubMed] [Google Scholar]
- 6.Matsuzawa, A., Nishitoh, H., Tobiume, K., Takeda, K., and Ichijo, H. (2002) Antioxid. Redox Signal. 4 415–425 [DOI] [PubMed] [Google Scholar]
- 7.Nishitoh, H., Matsuzawa, A., Tobiume, K., Saegusa, K., Takeda, K., Inoue, K., Hori, S., Kakizuka, A., and Ichijo, H. (2002) Genes Dev. 16 1345–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichijo, H., Nishida, E., Irie, K., ten Dijke, P., Saitoh, M., Moriguchi, T., Takagi, M., Matsumoto, K., Miyazono, K., and Gotoh, Y. (1997) Science 275 90–94 [DOI] [PubMed] [Google Scholar]
- 9.Nishitoh, H., Saitoh, M., Mochida, Y., Takeda, K., Nakano, H., Rothe, M., Miyazono, K., and Ichijo, H. (1998) Mol. Cell 2 389–395 [DOI] [PubMed] [Google Scholar]
- 10.Saitoh, M., Nishitoh, H., Fujii, M., Takeda, K., Tobiume, K., Sawada, Y., Kawabata, M., Miyazono, K., and Ichijo, H. (1998) EMBO J. 17 2596–2606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, L., Chen, J., and Fu, H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 8511–8515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotoh, Y., and Cooper, J. A. (1998) J. Biol. Chem. 273 17477–17482 [DOI] [PubMed] [Google Scholar]
- 13.Tobiume, K., Saitoh, M., and Ichijo, H. (2002) J. Cell. Physiol. 191 95–104 [DOI] [PubMed] [Google Scholar]
- 14.Zhang, R., He, X., Liu, W., Lu, M., Hsieh, J. T., and Min, W. (2003) J. Clin. Investig. 111 1933–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang, H., Zhang, R., Luo, Y., D'Alessio, A., Pober, J. S., and Min, W. (2004) J. Biol. Chem. 279 44955–44965 [DOI] [PubMed] [Google Scholar]
- 16.Zhang, R., Zhang, H., Lin, Y., Li, J., Pober, J. S., and Min, W. (2007) J. Biol. Chem. 282 14788–14796 [DOI] [PubMed] [Google Scholar]
- 17.Pan, S., An, P., Zhang, R., He, X., Yin, G., and Min, W. (2002) Mol. Cell. Biol. 22 7512–7523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo, D., Luo, Y., He, Y., Zhang, H., Zhang, R., Li, X., Dobrucki, W. L., Sinusas, A. J., Sessa, W. C., and Min, W. (2006) Am. J. Pathol. 169 1886–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang, H., Luo, Y., Zhang, W., He, Y., Dai, S., Zhang, R., Huang, Y., Bernatchez, P., Giordano, F. J., Shadel, G., Sessa, W. C., and Min, W. (2007) Am. J. Pathol. 170 1108–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim, Y. C., Garcia-Cardena, G., Allport, J. R., Zervoglos, M., Connolly, A. J., Gimbrone, M. A., Jr., and Luscinskas, F. W. (2003) Am. J. Pathol. 162 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, Y., Zhang, W., Zhang, R., Zhang, H., and Min, W. (2006) J. Biol. Chem. 281 5559–5566 [DOI] [PubMed] [Google Scholar]
- 22.He, Y., Luo, Y., Tang, S., Rajantie, I., Salven, P., Heil, M., Zhang, R., Luo, D., Li, X., Chi, H., Yu, J., Carmeliet, P., Schaper, W., Sinusas, A. J., Sessa, W. C., Alitalo, K., and Min, W. (2006) J. Clin. Investig. 116 2344–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, C. Y., Schroder, M., and Kaufman, R. J. (2000) J. Biol. Chem. 275 24881–24885 [DOI] [PubMed] [Google Scholar]
- 24.Bertolotti, A., Zhang, Y., Hendershot, L. M., Harding, H. P., and Ron, D. (2000) Nat. Cell Biol. 2 326–332 [DOI] [PubMed] [Google Scholar]
- 25.Yang, Q., Kim, Y. S., Lin, Y., Lewis, J., Neckers, L., and Liu, Z. G. (2006) EMBO Rep. 7 622–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mauro, C., Crescenzi, E., De Mattia, R., Pacifico, F., Mellone, S., Salzano, S., de Luca, C., D'Adamio, L., Palumbo, G., Formisano, S., Vito, P., and Leonardi, A. (2006) J. Biol. Chem. 281 2631–2638 [DOI] [PubMed] [Google Scholar]
- 27.Hu, P., Han, Z., Couvillon, A. D., Kaufman, R. J., and Exton, J. H. (2006) Mol. Cell. Biol. 26 3071–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimata, Y., Oikawa, D., Shimizu, Y., Ishiwata-Kimata, Y., and Kohno, K. (2004) J. Cell Biol. 167 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Credle, J. J., Finer-Moore, J. S., Papa, F. R., Stroud, R. M., and Walter, P. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18773–18784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hetz, C., Bernasconi, P., Fisher, J., Lee, A. H., Bassik, M. C., Antonsson, B., Brandt, G. S., Iwakoshi, N. N., Schinzel, A., Glimcher, L. H., and Korsmeyer, S. J. (2006) Science 312 572–576 [DOI] [PubMed] [Google Scholar]