Abstract
The Glut4 glucose transporter undergoes complex insulin-regulated subcellular trafficking in adipocytes. Much effort has been expended in an attempt to identify targeting motifs within Glut4 that direct its subcellular trafficking, but an amino acid motif responsible for the targeting of the transporter to insulin-responsive intracellular compartments in the basal state or that is directly responsible for its insulin-stimulated redistribution to the plasma membrane has not yet been delineated. In this study we define amino acid residues within the C-terminal cytoplasmic tail of Glut4 that are essential for its insulin-stimulated translocation to the plasma membrane. The residues were identified based on sequence similarity (LXXLXPDEXD) between cytoplasmic domains of Glut4 and the insulin-responsive aminopeptidase (IRAP). Alteration of this putative targeting motif (IRM, insulin-responsive motif) resulted in the targeting of the bulk of the mutant Glut4 molecules to dispersed membrane vesicles that lacked detectable levels of wild-type Glut4 in either the basal or insulin-stimulated states and completely abolished the insulin-stimulated translocation of the mutant Glut4 to the plasma membrane in 3T3L1 adipocytes. The bulk of the dispersed membrane vesicles containing the IRM mutant did not contain detectable levels of any subcellular marker tested. A fraction of the total IRM mutant was also detected in a wild-type Glut4/Syntaxin 6-containing perinuclear compartment. Interestingly, mutation of the IRM sequence did not appreciably alter the subcellular trafficking of IRAP. We conclude that residues within the IRM are critical for the targeting of Glut4, but not of IRAP, to insulin-responsive intracellular membrane compartments in 3T3-L1 adipocytes.
The Glut4 glucose transporter (1-4) is responsible for the insulin-mediated increase in glucose uptake into skeletal muscle, heart, and fat cells that occurs in response to elevated insulin levels in the postprandial state. The transport step is rate limiting for glucose metabolism in these cell types, and therefore the appropriate regulation of Glut4 is critical to the maintenance of normal whole body glucose homeostasis (5). Dys-regulation of insulin-stimulated Glut4 activity appears to be the proximal cause of insulin resistance associated with type 2 diabetes (6, 7), and thus there has been an intensive effort to understand the molecular mechanisms involved in insulin-stimulated Glut4 function.
Under basal conditions, the bulk of Glut4 molecules in adipocytes are present in intracellular membrane compartments (8, 9). Morphologically, these compartments appear as tubular-vesicular elements clustered in the perinuclear region of the cell and as small spherical vesicles dispersed throughout the cytoplasm (10-12). Insulin stimulates the recruitment of Glut4 from intracellular compartments to the plasma membrane (8, 9), and the increased level of the transporter at the cell surface appears to largely account for the concomitant increase in transport activity (12, 13). The precise itinerary of Glut4 trafficking in the basal and insulin-stimulated states is still the subject of controversy. For example, it remains unclear to what extent Glut4 recycles through the plasma membrane in the basal state or whether it is sequestered in one or more “static” intracellular membrane compartments (14-16).
Much effort has been expended on identifying the amino acid motifs in Glut4 that are responsible for its insulin-regulated subcellular trafficking (reviewed in Ref. 17). Early studies often appeared to yield contradictory results, most likely because of differences in the cell systems and/or the techniques employed. A phenylalanine motif (FQQI) within the cytoplasmic N terminus of Glut4 (18, 19) appears to function as an endocytic signal in cultured cell lines (20), primary rat adipocytes (21), and in cultured 3T3L1 adipocytes (22). Interestingly, this motif appears to play little if any role in the steady-state distribution of Glut4 in L6 myocytes (23), consistent with other evidence that Glut4-targeting motifs may be cell-type specific (17). Conversely, a dileucine motif within the C-terminal cytoplasmic tail of Glut4 appears to act as an endocytic motif in cultured cell lines expressing ectopic Glut4 (24, 25) and in L6 myocytes (23), but does not appreciably affect the steady-state distribution of the protein in 3T3L1 adipocytes (22, 26) or in primary rat adipocytes (21).
In L6 myocytes, the C-terminal 25-amino acid residues of Glut4 appear to be both necessary and sufficient for its insulin-responsive subcellular trafficking (23), a result consistent with earlier studies conducted in Chinese hamster ovary cells (27) and in Xenopus oocytes (28) implicating the extreme C terminus of the protein in Glut4 trafficking. Appropriate trafficking in L6 myocytes was shown to be dependent on the dileucine motif as well as additional unidentified residues downstream of this motif. Birnbaum and colleagues (22) reported that the C-terminal 30 residues of Glut4 are both necessary and sufficient for targeting of the protein to an insulin-responsive intracellular compartment in 3T3L1 adipocytes but that the dileucine motif does not play a role in this targeting behavior. Subsequently, James and colleagues (29) identified a sequence motif in the C-terminal tail of Glut4 that is similar to a motif in the N-terminal cytoplasmic tail of the insulin-responsive aminopeptidase (IRAP)2 (30, 31), a protein whose intracellular trafficking closely resembles that of Glut4 (17). This acidic motif (TELEY) was reported to be responsible for the movement of Glut4 from the general endosomal recycling pathway to a Syntaxin 6/16-containing post-endosomal compartment that was proposed to constitute a subcompartment of the trans-Golgi network (16, 32). Mutations in this motif increased the level of Glut4 in a plasma membrane-enriched subcellular fraction in the basal state but did not abrogate insulin-stimulated movement of Glut4 to the plasma membrane fraction. The importance of this motif in the overall steady-state distribution of Glut4 in the basal and insulin-stimulated states is unclear, because data in which wild-type Glut4 was colocalized morphologically with TELEY mutants have not been published. Other residues within the C-terminal tail of Glut4 have been shown to influence the subcellular trafficking of the transporter in various cell types (33, 34), however, a specific motif responsible for the trafficking of Glut4 to an insulin-sensitive membrane compartment or that is directly responsible for insulin-stimulated accumulation of Glut4 in the plasma membrane has not yet been identified. Thus, although the C terminus of Glut4 appears to contain one or more motifs that are necessary for trafficking of the protein to an insulin-sensitive subcellular compartment, the specific amino acids comprising these motifs have not yet been clearly defined.
In the present study, we identify a putative targeting motif in the C-terminal cytoplasmic tail of Glut4. This motif LXXLXP-DEX(D/E), designated IRM (insulin-responsive motif), is shared with a conserved amino acid sequence in the N-terminal cytoplasmic domain of IRAP, but the sequence similarity is based on a different amino acid alignment than that used by James and colleagues (29) to identify the acidic (TELEY) motif. Unlike other putative Glut4-targeting motifs that have been examined in a native, insulin-responsive cell-type, altering the IRM results in a targeting phenotype that radically differs from that of wild-type Glut4 in both the basal and insulin-stimulated states. We demonstrate that mutations in the IRM abolish insulin-stimulated redistribution of Glut4 to the plasma membrane. Additionally, in the basal state, the bulk of the IRM mutant accumulates in subcellular compartments that do not contain detectable levels of wild-type Glut4 or any other subcellular marker tested. Interestingly, mutation of the IRM does not appear to appreciably change the targeting of IRAP. These data indicate that the IRM is critical for the appropriate insulin-sensitive subcellular trafficking of Glut4, but not of IRAP, in 3T3L1 adipocytes.
EXPERIMENTAL PROCEDURES
Materials—DMEM and calf serum were purchased from Invitrogen. Fetal calf serum was obtained from HyClone. BCA protein assay kits were purchased from Pierce. All other chemicals were obtained from Sigma-Aldrich. Polyclonal rabbit antibodies against the hemagglutinin (HA) epitope was purchased from Abcam (Cambridge, MA). Monoclonal HA antibody and Giantin polyclonal antibodies were obtained from Covance (Berkeley, CA). The monoclonal antibody against Syntaxin 6 was from BD Transduction Laboratories. Trapα (translocon-associated protein-α) polyclonal antibody was a gift from Maurine Linder (Washington University School of Medicine, Saint Louis, MO). LysoTracker was obtained from Molecular Probes. The GLUT4 rabbit polyclonal antiserum was raised against a synthetic peptide corresponding to the C-terminal 15 amino acid residues of rat GLUT4 (35). Anti-endosome-associated antigen-1 antibody was a gift from Dr. Marvin J. Fritzer (University of Calgary). Fluorescent secondary antibodies were purchased from Molecular Probes (Eugene, OR). The pcDNA3-GLUT4/eGFP plasmid was a kind gift from Jeffrey E. Pessin (State University of New York, Stoneybrook).
Cell Culture—3T3L1 fibroblasts (from American Tissue Type Culture Collection) were cultured on plastic dishes in DMEM supplemented with 20% calf serum, 100 μg/ml streptomycin, 10 units/ml penicillin, 2 mml-glutamine, in a 5% CO2, 37 °C incubator. 2 days after achieving confluence, cells were incubated in differentiation medium (DMEM containing 10% fetal bovine serum, 2 mml-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin supplemented with 670 nm insulin, 0.5 mm 1-isobutyl 3-methylxanthine, and 25 nm dexamethasone) for 48 h. The cells were then placed in the same medium containing 670 nm insulin but lacking additional supplements for an additional 2 days. Afterward, cells were maintained in the same medium lacking insulin until adenovirus infection 6 days post-differentiation.
Adenovirus Production and Infection—The pShuttle-CMV constructs were linearized with PmeI and cotransformed with pAdeasy-1 into Escherichia coli BJ5183. Recombinants were selected and amplified in E. coli DH5α. The clones were then linearized with PacI and transfected into HEK 293A cells (Q. BIOgene) using Lipofectamine 2000 kits (Invitrogen) for the production of recombinant adenoviruses. The large scale amplification of virus was achieved as described previously (36). 3T3L1 adipocytes were infected with adenovirus in serum-free DMEM containing 0.5% bovine serum albumin at day 6 after differentiation. Following overnight infection, the medium was replaced with DMEM containing 10% fetal bovine serum, and the infected cells were used for experiments 48 h later.
Subcellular Fractionation—Subcellular fractionation was conducted using minor modifications of previously published procedures (37, 38). 3T3-L1 adipocytes on 10-cm dishes were serum-starved overnight, incubated in the absence or presence of 1 μm insulin for 30 min at 37 °C, washed in cold phosphate-buffered saline, pH 7.4 (PBS), and then scraped into HES buffer (50 mm Hepes, pH 7.4, 0.25 m sucrose, 1 mm EDTA) containing a protease inhibitor mixture (1 μg/ml leupeptin, 1 μg/ml benzamidine, 1 μg/ml antipain, 1 μg/ml chymostatin, 1 μg/ml pepstatin A, 5 μg/ml trypsin inhibitor, 0.082 trypsin inhibitory units/ml aprotinin). All of the following procedures were carried out at 4 °C. The cells were passed through a Yamato SC homogenizer 12 times at 1,200 rpm. After removing unbroken cells by centrifugation in a Sorvall H6000A rotor at 2,000 rpm for 5 min, the supernatant was centrifuged for 20 min at 12,500 rpm in a Sorvall SS-34 rotor. The resulting supernatant was then centrifuged at 18,500 rpm in a Sorvall SS-34 rotor for 20 min, and the pellet was designated the high density microsomal fraction. The 18,500 rpm supernatant was further centrifuged at 44,000 rpm in a Beckman SW55.2 rotor for 2 h, the pellet was defined as the low density microsomal fraction, and the supernatant was defined as the cytoplasmic fraction. The 12,500 rpm pellet was resuspended in HES buffer, layered onto a sucrose cushion (1.12 m sucrose, 1 mm EDTA, 50 mm Hepes, pH 7.4), and then centrifuged at 25,000 rpm for 1 h in a Beckman SW41 rotor. The mitochondrial/nuclear fraction corresponded to the resulting pellet. The fluffy band at the sucrose cushion interface was collected and resuspended in HES buffer, and then pelleted by centrifugation at 17,500 rpm in a Sorvall SS-34 rotor for 20 min to obtain the plasma membrane (PM) fraction.
Western Blotting—Infected 3T3L1 adipocytes were rinsed with cold PBS buffer and then lysed in TENET buffer (1% Triton, 0.5 m EDTA, 20 mm M Tris, pH 8.0, 0.15 m NaCl) containing the protease inhibitor mixture described above. The solubilized lysate was cleared by centrifugation at 12,500 rpm for 15 min in an Eppendorf microcentrifuge. Equal amounts of total protein were then subjected to SDS-PAGE, transferred to nitro-cellulose membrane, incubated in 5% milk in TBST buffer (10 mm Tris, pH 7.6, 150 mm NaCl, 0.5% Tween 20) for 1 h at room temperature, and then subsequently probed with primary antibody overnight at 4 °C. After washing with TBST buffer for 30 min, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. The proteins were detected using the enhanced chemiluminescent substrate kit (Amersham Biosciences). The chemiluminescent bands were quantified using NIH Image software version 1.62.
Glycosylation Assays—Cell lysates were solubilized in 1% SDS and incubated at 37 °C for 1 h. For endoglycosidase H digestion (Roche Diagnostics, Germany), 20 μl of cell lysate was digested with 2.5 milliunits enzyme at 37 °C for 1 h after the addition of 0.5 μl3 m sodium acetate, pH 5.5. For N-glycanase digestion (New England Biolabs), 25 μl of the 1% SDS cell lysate was adjusted to a final concentration of 0.25% SDS by the addition of 1.9% Nonidet P-40, 30 mm Tris-HCl, pH 8.0, and then digested with 1.5 μl of enzyme at 37 °C overnight.
Laser Confocal Immunofluorescence Microscopy—Infected 3T3-L1 adipocytes expressing Glut4 constructs were serum-starved at 37 °C for 2 h, treated with or without 1 μm insulin for 30 min, washed with cold PBS three times, fixed in 4% paraformaldehyde in PBS for 15 min at room temperature or in methanol at -20 °C for 10 min, and then permeabilized with 0.5% Triton for 15 min. After blocking with 2.5% bovine serum albumin and 2.5% goat serum in 0.1% Triton for 1 h at room temperature, cells were incubated with primary antibodies for 1 h, washed with PBS three times and then incubated with Alexa-488 or Alexa-594-conjugated secondary antibodies at room temperature for 1 h. After extensive washing with PBS, the coverslips were mounted in Vectashield mounting medium (Vector, Burlingame, CA) on glass slides. The fluorescent staining of the cells was imaged using a 63×/1.4 numerical aperture oil immersion objective, equipped with an MRC 1024 laser confocal image system (Bio-Rad). The quantification of fluorescence intensity was carried out using Zeiss LSM 5 software, version 3.2 SP2 (Carl Zeiss, Thornwood, NY).
Labeling of Cell Surface Glut4 in Living Cells—3T3-L1 adipocytes expressing Glut4/DsRed or IRM/DsRed were serum-starved for 2 h at 37 °C. The cells were then incubated with Alexa 488-conjugated monoclonal HA antibody for 3 h in the presence or absence of 1 μm insulin, rinsed in ice-cold PBS, and then fixed in 4% paraformaldehyde for 20 min. The cells were then processed and imaged by confocal microscopy as described above.
RESULTS
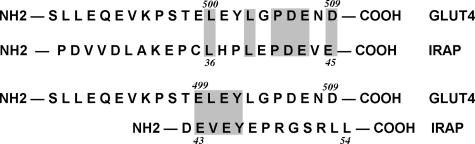
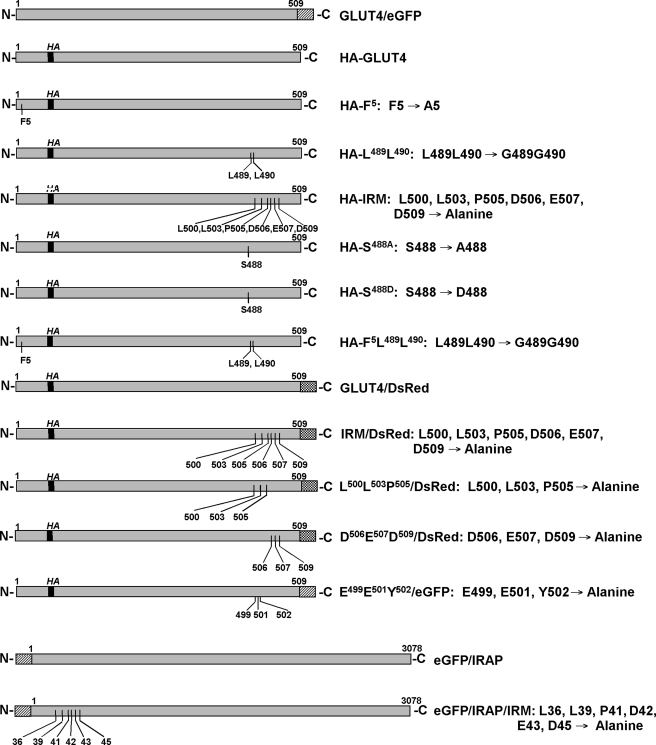
Fig. 1 (upper alignment) shows a sequence alignment between the extreme cytoplasmic C-terminal tail of Glut4 and the cytoplasmic N-terminal domain of IRAP. A conserved motif (IRM) is shared between these two proteins corresponding to the amino acid sequence LXXLXPDEX(D/E), where X is any amino acid. To determine whether residues within this shared sequence are involved in the regulated subcellular trafficking of Glut4, a mutant was constructed in which the six conserved residues were all changed to alanine residues, and the mutant was tagged with a HA epitope in the first exofacial loop (HA-IRM) (Fig. 2). Additionally, we constructed HA-tagged mutants within the previously characterized N-terminal cytoplasmic FQQI motif, the C-terminal dileucine motif, a mutant containing substitutions in both of these motifs, and two mutants at Ser-488, the only known site of phosphorylation in Glut4 (39). The Ser-488 mutants involved changes to either alanine or aspartate (see Fig. 2) to abrogate phosphorylation at this site or to mimic constitutive phosphorylation, respectively.
FIGURE 1.
Sequence alignments between the N-terminal cytoplasmic tail of IRAP and the C-terminal cytoplasmic tail of Glut4. The upper alignment identifies the shared targeting motif characterized in the present study, and the lower alignment identifies the motif characterized by Shewan et al. (29).
FIGURE 2.
Schematic diagrams and nomenclature of the Glut4 mutants examined in this study. Amino acid residues are designated by the single-letter code.
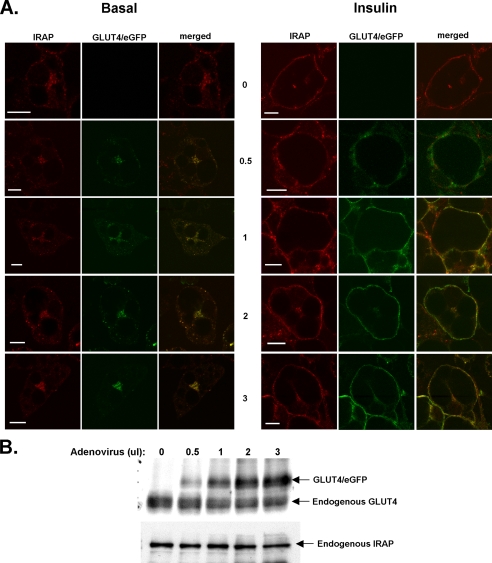
Recombinant adenoviruses were constructed that encode each of the HA-tagged Glut4 mutants illustrated in Fig. 2, and the various constructs were used to infect 3T3L1 adipocytes. In the following experiments, eGFP-tagged wild-type Glut4 (Glut4/eGFP) and the various HA-tagged mutants were coexpressed in the same cell population, so that the extent of colocalization or mistargeting of the mutants compared with wild-type Glut4 could be directly assessed. The coexpression of an internal wild-type control also circumvented the possibility that the differential targeting behavior of a mutant compared with wild-type Glut4 might be caused by the saturation of normal trafficking mechanisms (26). It was not possible to directly compare the targeting of the tagged, ectopically expressed wild-type Glut4 construct with endogenous Glut4, because all available antibodies against native Glut4 also recognize the tagged wild-type controls. However, we did assess the targeting behavior of the ectopically expressed wild-type Glut4/eGFP by comparing its subcellular distribution to that of endogenous IRAP, a protein that appears to target very similarly, if not identically, to Glut4 in the basal and insulin-stimulated states (40). Fig. 3A demonstrates that the targeting of Glut4/eGFP under basal and insulin-stimulated conditions was virtually indistinguishable from that of endogenous IRAP at four different levels of expression that encompass the range of expression obtained throughout the current study. Fig. 3B shows that the expression of Glut4/eGFP did not significantly affect the expression of endogenous Glut4 or of endogenous IRAP. Additionally, the targeting behavior of the tagged wild-type Glut4 constructs in the following experiments strongly suggests that normal Glut4 subcellular trafficking patterns were not disrupted by the levels of ectopic protein expression attained. Specifically, the tagged wild-type Glut4 constructs exhibited the characteristic Glut4 perinuclear and dispersed cytoplasmic distribution in the basal state (41). The wild-type constructs were excluded from the plasma membrane in the basal state and robustly redistributed to the plasma membrane after insulin stimulation. Thus, the subcellular trafficking behavior of the tagged wild-type Glut4 constructs was qualitatively indistinguishable from that of endogenous Glut4 in 3T3L1 adipocytes (41), in agreement with numerous studies demonstrating that wild-type Glut4 constructs containing the HA epitope in the first exofacial loop and/or with GFP protein fused to the C terminus target very similarly, if not identically, to native Glut4 (see a recent careful analysis by McGraw and colleagues (42)).
FIGURE 3.
Adenoviral-expressed Glut4/eGFP colocalizes with endogenous IRAP. 3T3L1 adipocytes were infected with increasing quantities of recombinant adenoviruses expressing Glut4/eGFP reflecting the range of ectopic expression obtained in these studies. Two days after infection, adipocytes were either maintained under basal conditions or treated with insulin for 30 min. A, the cells were then fixed in 4% paraformaldehyde, permeabilized in Triton X-100, incubated with anti-IRAP antibodies, and then imaged by laser confocal microscopy. B, cells were lysed and subjected to SDS-PAGE followed by immunoblotting with antibodies against Glut4 or IRAP. Scale bars represent 10 μm.
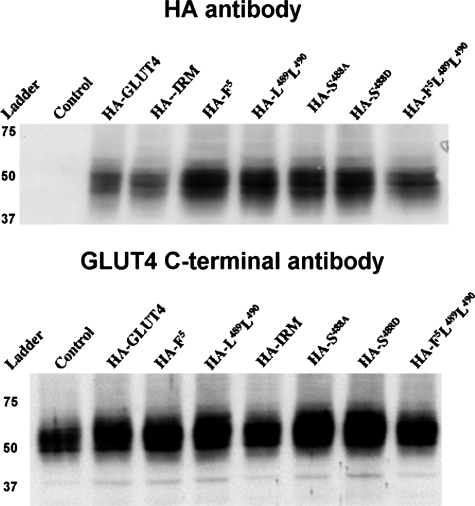
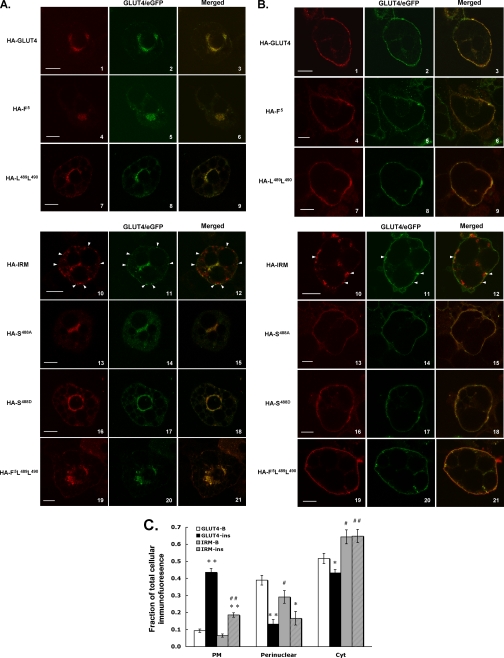
Typical relative expression levels of the various mutant Glut4 proteins, using antibodies against either the HA epitope or the C terminus of Glut4, are shown in Fig. 4 (note that the IRM mutant is not recognized by antibodies directed against the C terminus of Glut4). Similar levels of coexpression were observed for the wild-type control and the mutants, and the colocalization results did not vary with the expression levels attained in numerous independent experiments. The nearly identical subcellular targeting behavior of HA-Glut4 and Glut4/eGFP in the basal and insulin-stimulated states is shown by dual-label immunofluorescence laser confocal microscopy in Fig. 5 (A and B, panels 1-3), demonstrating that the differential tagging did not affect subcellular targeting. In the remaining panels, adipocytes were co-infected with recombinant adenoviruses expressing wild-type Glut4 tagged with GFP as an internal control and with one of the targeting motif mutants tagged with the HA epitope (see Fig. 2). The tagged wild-type Glut4 labeling is shown in green in the center panels, the labeled mutants are shown in red in the left panels, and merged labeling is shown in the far right panels with colocalization between the two ectopically expressed proteins presented in yellow. Mutations in the N-terminal FQQI motif (panels 4-6), the C-terminal di-leucine motif (panels 7-9), in both of these motifs (panels 19-21), or at serine-488 (panels 13-18), resulted in steady-state distributions under basal (Fig. 4A) and insulin-stimulated (Fig. 4B) conditions that were very similar to that of GFP-tagged wild-type Glut4. Notably, none of these mutations discernibly affected translocation of Glut4 to the plasma membrane in response to insulin. Thus, none of the amino acid residues within these putative targeting motifs appear to be critical for the targeting of Glut4 to an insulin-responsive compartment in 3T3L1 adipocytes or for the insulin-stimulated steady-state redistribution of Glut4 to the plasma membrane. Strikingly, however, converting the six residues shared by the cytoplasmic N terminus of IRAP and the C terminus of Glut4 (LXXLXPDXEXD) to alanine residues in the HA-IRM mutant resulted in dramatically different subcellular distributions compared with wild-type Glut4 in both the basal and insulin-stimulated states (panels 10-12). Under basal conditions (Fig. 5A), HA-IRM was largely present in dispersed vesicular structures throughout the cytoplasm that lacked detectable levels of the wild-type Glut4/eGFP control. These dispersed HA-IRM-containing structures also differed qualitatively from the dispersed Glut4/eGFP-containing structures in that the former were much larger than the latter. Extensive examination of Z-sections through many infected cells demonstrated that there was a relatively small degree of colocalization between total HA-IRM and wild-type Glut4 under basal conditions and that this was primarily confined to a perinuclear compartment, although a fraction of small vesicles lying just beneath the plasma membrane appeared to contain both proteins (see arrows in panels 10-12). After insulin stimulation (Fig. 5B), the wild-type Glut4/eGFP control and all of the mutants except for HA-IRM exhibited robust redistribution to the plasma membrane and diminished levels in the cytoplasm. In sharp contrast, HA-IRM did not discernibly translocate to the plasma membrane after insulin stimulation but remained primarily in structures dispersed throughout the cytoplasm. Some colocalization between HA-IRM and Glut4/eGFP was again observed in small vesicles that appeared to lie just beneath the plasma membrane (see arrows in panels 10-12). Quantification of the targeting of HA-IRM and Glut4/eGFP to the perinuclear region, to dispersed cytoplasmic compartments, and to within ∼500 nm of the cell surface (encompassing the plasma membrane) is shown in Fig. 5C. The targeting of HA-IRM to dispersed cytoplasmic compartments was significantly enhanced relative to that of Glut4/eGFP, whereas its targeting to the perinuclear region was reduced. Importantly, the redistribution of HA-IRM to the surface of the cell after insulin treatment was dramatically reduced relative to that of Glut4/eGFP, and most of the labeling of HA-IRM near the cell surface appeared to be in small vesicles lying just beneath the plasma membrane (see Fig. 5B).
FIGURE 4.
Expression levels of tagged Glut4 constructs. 3T3L1 adipocytes were solubilized in TENET buffer 48 h after infection with recombinant adenoviruses expressing the various HA-tagged Glut4 constructs. 10 μg of total protein was then analyzed by SDS-PAGE followed by immunoblotting with primary antibodies against either the HA epitope (upper panel) or against the C terminus of Glut4 (lower panel).
FIGURE 5.
Colocalization of GFP-tagged wild-type Glut4 and HA-tagged targeting mutants by dual-label immunofluorescence laser confocal microscopy. 3T3L1 adipocytes were co-infected with recombinant adenoviruses encoding the GFP-tagged wild-type Glut4 and one of the HA-tagged mutants. 48 h later the cells were serum-starved for 2 h, either exposed to insulin for 30 min or maintained in the basal state, and then prepared for imaging as described under “Experimental Procedures.” Tagged wild-type Glut4 is shown in the middle panels in green, the tagged mutants are shown in the left panels in red, and the merged images are shown in the right panels with colocalization between the two coexpressed proteins presented in yellow. A, localization of ectopically expressed proteins in 3T3L1 adipocytes under basal conditions. B, localization of ectopically expressed proteins in 3T3L1 adipocytes 30 min after insulin stimulation. C, quantification of the localization of Glut4/eGFP and HA-IRM in the perinuclear region, in dispersed cytosolic compartments (Cyt), and within ∼0.5 μm of the outermost periphery of the cell (PM). The PM fractional intensity includes labeling in the plasma membrane as well as in membrane vesicles that may lie just beneath the plasma membrane. Quantification of the fraction in dispersed cytosolic compartments was determined by subtracting the fluorescence intensity in the PM and perinuclear compartments from total cellular fluorescence intensity. The data are expressed as the mean ± S.E. from four independent experiments. **, p < 0.01 compared with basal conditions for the same construct;*,p<0.05 compared with basal conditions for the same construct;##,p<0.01 compared with the same wild-type control fraction;#,p<0.05 compared with the same wild-type control fraction. The scale bars represent 10 μm.
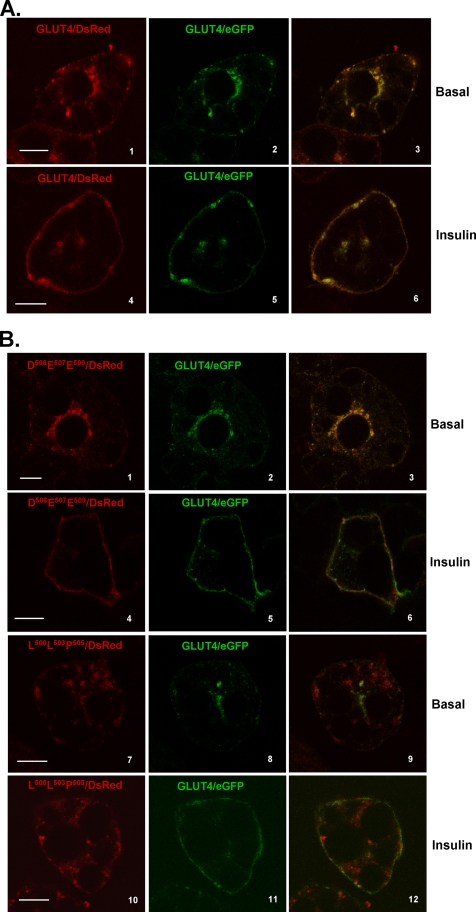
To more precisely define this putative Glut4 targeting motif, two additional mutants were constructed in which either the three N-terminal shared residues (LLP) or the three C-terminal shared residues (DED) of the IRM motif were converted to alanine residues (see Fig. 2). These constructs were tagged within the first exofacial loop with the HA epitope and at their C termini with DsRed fluorescent protein. Fig. 6 (panels 1-3) shows that wild-type Glut4 tagged with both the HA epitope and DsRed exhibited a nearly indistinguishable steady-state distribution compared with Glut4/eGFP in either the basal or insulin-stimulated state (Fig. 6A). The steady-state distributions of HA-Asp506-Glu507-Asp509/DsRed in the basal and insulin-stimulated states also could not be distinguished from the coexpressed wild-type Glut4/eGFP control (Fig. 6B). However, HA-Leu500-Leu503-Pro505/DsRed exhibited subcellular distribution patterns very similar to the parental IRM mutant and displayed much less overlap with wild-type Glut4/eGFP under basal or insulin-stimulated conditions. Like the parental IRM mutant, HA-LLP/DsRed exhibited no discernible translocation to the plasma membrane in response to insulin. We conclude that Leu500, Leu503, and Pro505 comprise three critical amino acid residues within a putative targeting motif that is responsible for the insulin-mediated accumulation of Glut4 in the plasma membrane in 3T3L1 adipocytes.
FIGURE 6.
The N-terminal three residues of the IRAP/Glut 4 shared amino acid sequence motif are critical for the altered targeting behavior of the IRM mutant. 3T3L1 adipocytes were co-infected with recombinant adenoviruses encoding the GFP-tagged wild-type Glut4 and either HA-Glut4/DsRed (A) or the alanine mutants HA-Asp506-Glu507-Asp509/DsRed or HA-Leu500-Leu503-Pro505/DsRed (B). 48 h later the cells were serum-starved for 2 h, either exposed to insulin for 30 min or maintained in the basal state, and then prepared for imaging as described under “Experimental Procedures.” Tagged wild-type Glut4 is shown in the middlepanels in green, the tagged mutants are shown in the left panels in red, and the merged images are shown in the right panels with colocalization between the two coexpressed molecules presented in yellow. The scale bars represent 10 μm.
Next, we attempted to identify the distinct subcellular compartment or compartments to which the IRM mutant was confined in the basal state. In this series of experiments, HA-IRM and HA-Glut4 were expressed in different adipocyte populations, and the infected cells were then stained with an antibody against the HA tag and a series of antibodies against marker proteins for various subcellular membrane compartments. In Fig. 7 the HA-tagged transporters are shown in red and the marker proteins are shown in green (except in the panels where the transporter-expressing cells were stained with LysoTracker to identify acidic membrane compartments where the coloring is reversed). Colocalization in merged images is shown in yellow. Enlargements of regions of the merged images are shown in the extreme right hand panels.
FIGURE 7.
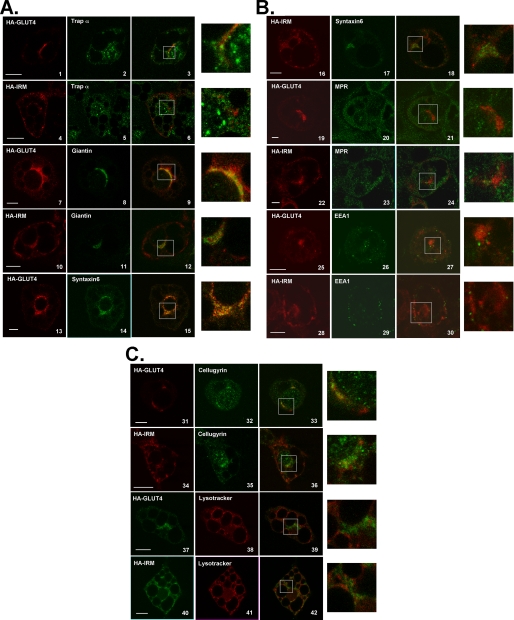
The bulk of HA-IRM does not colocalize with known subcellular markers in 3T3L1 adipocytes. 3T3L1 adipocytes were infected with recombinant adenovirus encoding either HA-Glut4 or HA-IRM. 48 h later the cells were serum-starved for 2 h and then processed for imaging as described under “Experimental Procedures.” The left panels show the localization of either HA-Glut4 or HA-IRM in red, the center panels show the localization of various subcellular marker proteins in green, and the right panels show merged images with colocalization presented in yellow. The color scheme is reversed in cells stained with LysoTracker. Enlarged images of the colocalization panels are shown in the far right panels. Scale bars represent 10 μm.
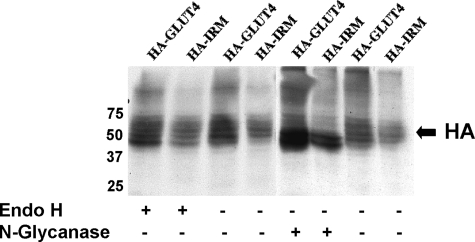
As expected, both wild-type Glut4 and HA-IRM showed a small degree of colocalization with markers for early biosynthetic compartments, including the rough endoplasmic reticulum (trap α, panels 1-6) and the cis and medial Golgi (Giantin, panels 7-12). A smaller fraction of HA-IRM appeared to colocalize with these compartments compared with wild-type Glut4. These data indicate that the IRM mutant was not being trapped in an early biosynthetic compartment due to misfolding. Supporting this conclusion, the IRM mutant was resistant to digestion with endoglycosidase H, but its N-linked oligosaccharides were removed by N-glycanase, indicating that the mutant protein traversed at least the cis-Golgi complex (Fig. 8). A previous study demonstrated that Glut4 partially colocalizes with Syntaxin 6 in the perinuclear region of 3T3L1 adipocytes, which was suggested to represent a subdomain of the trans-Golgi network involved in the intracellular sorting of Glut4 (32). Fig. 7 shows that both the wild-type control and the IRM mutant partially colocalized with Syntaxin 6 (panels 13-18). This probably corresponds to the degree of colocalization of wild-type Glut4 and the IRM mutant in the perinuclear region evident in Fig. 5. However, the major fraction of HA-IRM was present in dispersed vesicles and not in the Syntaxin 6 compartment (Fig. 5C). To determine whether HA-IRM was localized to the general endosomal recycling pathway, we employed antibodies against endosome-associated antigen-1, a standard marker for early endosomes and antibodies against the mannose 6-phosphate receptor, a marker for late endosomes. The tagged wild-type control exhibited a very small degree of colocalization with endosome-associated antigen-1 (panels 25-27) and mannose 6-phosphate receptor (panels 19-21). In contrast, HA-IRM exhibited minimal, if any, colocalization with either mannose 6-phosphate receptor (panels 22-24) or endosome-associated antigen-1 (panels 28-30). This indicates that HA-IRM is largely or completely excluded from the general endosomal recycling pathway, consistent with its complete lack of insulin responsiveness. Wild-type Glut4 partially colocalized with Cellugyrin (panels 31-33), a protein proposed to represent a marker for an intermediate compartment between endosomes and Glut4 storage vesicles (GSVs) (43). However, HA-IRM did not colocalize with Cellugyrin (panels 34-36). Finally, we determined that neither wild-type Glut4 nor HA-IRM noticeably colocalized with LysoTracker (panels 37-42), a marker for acidic compartments including lysosomes.
FIGURE 8.
The bulk of HA-IRM is not trapped in early biosynthetic membrane compartments. 3T3L1 adipocytes were solubilized 48 h after infection with recombinant adenovirus expressing HA-IRM or HA-Glut4. The protein lysate was then either left untreated or digested with endoglycosidase H or with N-glycanase as described under “Experimental Procedures.” 10 μg of total protein was then analyzed by SDS-PAGE followed by immunoblotting with primary antibodies against the HA epitope.
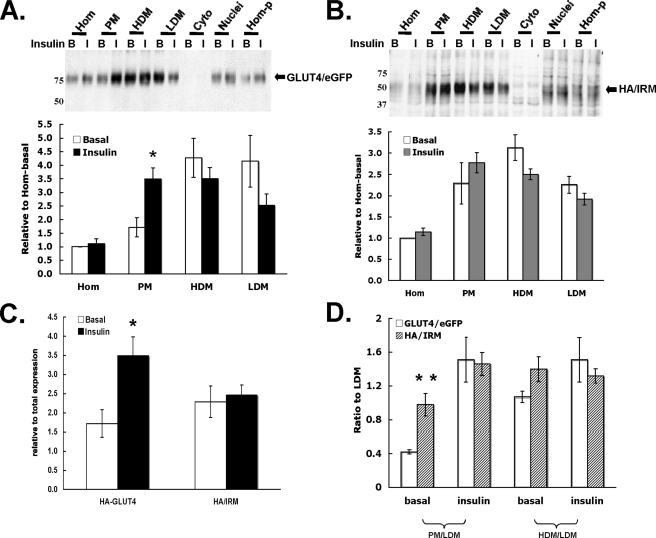
Next, we examined the behavior of coexpressed Glut4/eGFP and the HA-IRM mutant during the traditional subcellular fractionation procedure used for adipocytes. Fig. 9 shows that the relative level of the Glut4/eGFP fusion protein increased in the plasma membrane fraction and decreased in the low density microsomal fraction after insulin stimulation, although the latter change did not quite achieve statistical significance. However, the level of HA-IRM did not increase in the plasma membrane fraction after insulin stimulation. Because the previous morphological analyses clearly demonstrated that HA-IRM is not detectable in the plasma membrane under basal or insulin-stimulated conditions, these data suggest that HA-IRM partially accumulates in a subcellular membrane compartment that cofractionates with plasma membranes. This compartment may represent the fraction of total HA-IRM that was observed in vesicles lying just beneath the plasma membrane (see Fig. 5, A and B, panels 10-12).
FIGURE 9.
Localization of the HA-IRM Glut4 mutant in adipocyte subcellular fractions. 3T3L1 adipocytes were co-infected with recombinant adenoviruses encoding HA-IRM and wild-type Glut4/eGFP. 48 h post-infection the cells were serum-starved overnight and then either maintained in the basal state or treated with insulin for 30 min. The cells were then subjected to subcellular fractionation as described under “Experimental Procedures,” and the fractions were subjected to SDS-PAGE and immunoblotting using primary antibodies against either eGFP (A) or the HA epitope (B). Representative immunoblots are shown, and the quantification of four independent experiments is shown below each immunoblot. Panel C directly compares the relative ratios of the two proteins present in the PM fractions under basal or insulin-stimulated conditions. Panel D shows the ratios of the two proteins present in PM or high density microsomal versus low density microsomal. *, p < 0.05; **, p < 0.01 compared with basal.
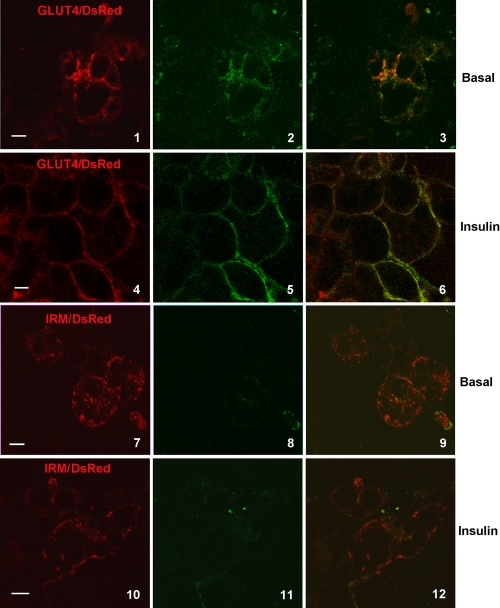
To confirm that HA-IRM does not traffic through the plasma membrane, we studied the exoplasmic accessibility of the HA epitope in a HA-IRM construct tagged at its C terminus with DsRed (IRM/DsRed) and an identically tagged wild-type Glut4 molecule (Glut4/DsRed). 3T3L1 adipocytes expressing the constructs were incubated in the presence or absence of insulin for 3 h in serum-free medium containing Alexa 488-conjugated antibodies against the HA epitope. The red channel (left panels in Fig. 10) shows the steady-state subcellular distribution of the total IRM/DsRed or Glut4/DsRed, and the green channel (center panels) shows the distribution of those molecules that were inserted into the plasma membrane at some point during the 3-h incubation period and reacted with the HA antibody in the medium. The panels on the right show the merged channels with colocalization between the two labels in yellow. Fig. 10 demonstrates that a significant amount of the tagged, wild-type Glut4 reached the plasma membrane under both basal and insulin-stimulated conditions. However, labeling of the HA epitope was virtually undetectable for tagged IRM/DsRed under either condition, indicating that the mutant is largely or completely excluded from the plasma membrane under both basal and insulin-stimulated conditions.
FIGURE 10.
HA/IRM does not recycle through the plasma membrane. 3T3L1 adipocytes were infected with recombinant adenoviruses encoding Glut4/DsRed or IRM/DsRed. 48 h post infection, cells were serum starved for 2 h and then incubated in the presence or absence of insulin and in the presence of Alexa 488-conjugated anti-HA antibody for 3 h. Cells were then imaged for laser confocal microscopy as described under “Experimental Procedures.” The distribution of total DsRed-tagged Glut4 and IRM are shown in the left panels in red, molecules that reached the cell surface and reacted with the HA antibody are shown in green in the center panels, and merged images are shown in the right panels. Scale bars represent 10 μm.
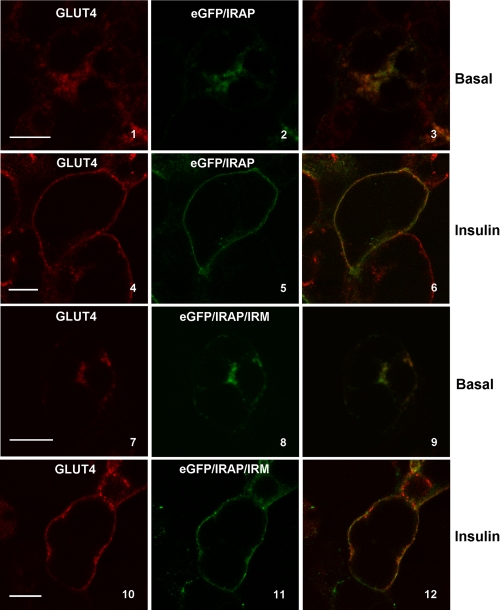
Next, we examined whether the IRM sequence is important for the subcellular trafficking of IRAP, because its initial identification was based on sequence conservation between cytoplasmic domains of Glut4 and IRAP. The subcellular distribution under basal and insulin-stimulated conditions of an IRAP construct tagged at its N terminus with eGFP and containing alanine substitutions at the 6 IRM residues (see Fig. 2) is shown in Fig. 11. The eGFP/IRAP/IRM construct (center panels, green) was colocalized with endogenous Glut4 (left panels, red) with merged images presented in the right panels. Unexpectedly, the data indicate that the IRM mutations had minimal affect on the targeting of IRAP, in sharp contrast to their affect on Glut4. In particular, eGFP/IRAP/IRM showed robust translocation to the plasma membrane in response to insulin (panels 10-12) similar to that of tagged, wild-type IRAP (panels 4-6).
FIGURE 11.
eGFP/IRAP/IRM colocalizes with endogenous Glut4 and does not exhibit aberrant subcellular targeting. 3T3L1 adipocytes were infected with recombinant adenovirus encoding either eGFP/IRAP or eGFP/IRAP/IRM. 48 h post-infection the cells were serum-starved for 2 h and then incubated in the presence or absence of insulin for 30 min. The cells were fixed, permeabilized, stained with Glut4 antibody, and then processed for imaging as described under “Experimental Procedures.” All images represent optical sections approximately midway through the depth of the cell. Endogenous Glut4 is shown in red in the left panels, and eGFP/IRAP and eGFP/IRAP/IRM are shown in green in the middle panels. Merged images are shown in the right panels with colocalization in yellow. Scale bars represent 10 μm.
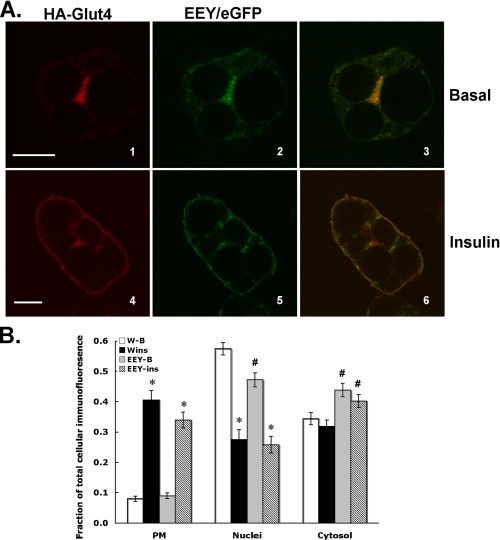
James and colleagues (29, 32) have characterized the subcellular targeting behavior of mutant Glut4 molecules (TELEY or Glu499-Glu501-Tyr502 mutants) containing amino acid substitutions within a region of the C-terminus of Glut4 that partially overlaps with the IRM sequence described in the present study, but is based on a different sequence alignment between Glut4 and IRAP (see Fig. 1, bottom alignment). The TELEY or EEY mutants were reported to be defective in trafficking between the general endosomal recycling pathway and a Syntaxin 6/16-containing intracellular compartment but still exhibited insulin-stimulated redistribution to the plasma membrane. To determine whether the IRM-targeting sequence is functionally distinct from the EEY motif, we compared the subcellular distributions of the EEY mutant and tagged wild-type Glut4 (Fig. 12). These data demonstrate that, under basal conditions (panels 1-3) or after 30 min of insulin stimulation (panels 4-6), the EEY mutant and tagged wild-type Glut4 exhibited substantial colocalization. Most notably, the EEY mutant accumulated in the plasma membrane after insulin stimulation to a similar extent as tagged wild-type Glut4. Thus, the targeting phenotype of the EEY mutant was very distinct from that of the IRM mutant.
FIGURE 12.
Colocalization of HA-Glu499-Glu501-Tyr502/eGFP and HA-Glut4. 3T3L1 adipocytes were co-infected with recombinant adenoviruses encoding HA-Glut4 and Glu499-Glu501-Tyr502/eGFP. 48 h post-infection the cells were serum-starved for 2 h and then either maintained in the basal state or treated with insulin for 30 min. The cells were then processed for laser confocal immunofluorescence microscopy as described under “Experimental Procedures.” A, the localization of HA-Glu499-Glu501-Tyr502/eGFP is shown in green in the middle panels. The localization of HA-Glut4 is shown in red in the left panels. Merged images are shown in the right panels with colocalization between the two different proteins indicated in yellow. Scale bars represent 10 μm. B, quantification of the fluorescence intensity in different regions of the cell as a fraction of total cellular fluorescence intensity. The data represent the mean ± S.E. of 5-7 independent experiments. See the legend to Fig. 5 for details concerning the quantification procedure and the definition of the subcellular regions. *, p < 0.01 relative to the corresponding basal fraction; #, p < 0.01 relative to the corresponding fraction from HA-Glut4.
DISCUSSION
Many studies over the past decade have attempted to identify amino acid motifs in Glut4 that regulate its subcellular trafficking (for a review, see Ref. 17). Because of technical difficulties involved in achieving ectopic expression of proteins in native Glut4-expressing, insulin-responsive cell-types, initial studies were conducted using cultured cell lines that were not derived from fat or muscle cells and do not normally express Glut4. These early studies identified two distinct amino acid motifs that altered the steady-state distribution of ectopically expressed Glut4. The first motif identified was FQQI in the cytosolic N-terminal domain of Glut4 (19). The second motif identified was a dileucine sequence in the cytosolic C-terminal domain of Glut4 (24). Subsequent experiments in which Glut4 proteins containing mutations in these motifs were expressed in 3T3L1 adipocytes demonstrated that neither of these motifs is responsible for the targeting of Glut4 to an insulin-responsive compartment (22, 26). However, both motifs appear to function as cell-surface internalization signals in adipocytes and may act to decrease the steady-state level of Glut4 molecules in the plasma membrane that recycle via the general endosomal pathway in the basal state.
Shortly after these initial studies, two studies were published demonstrating that the extreme C terminus of Glut4 was both necessary and sufficient for its targeting to insulin-responsive intracellular compartments. The results of one study indicate that the C-terminal 25 amino acid residues of Glut4 are necessary and sufficient for the targeting of Glut4 to an insulin-sensitive intracellular compartment in L6 myocytes (23). The data presented in this study included immunofluorescence confocal microscopy, semi-quantitative immunogold-labeling electron microscopy, and functional analysis. Birnbaum and colleagues (22) published a study that demonstrated that the C-terminal 30 residues of Glut4 are necessary and sufficient for the targeting of Glut4 to insulin-sensitive subcellular compartments in 3T3L1 adipocytes. A major difference in the results reported in these two studies was the role of the C-terminal dileucine motif. In L6 myocytes, the dileucine motif appears to be necessary, but not sufficient, for the targeting of Glut4 to an insulin-sensitive subcellular compartment. In contrast, in 3T3L1 adipocytes, the dileucine motif appears to be superfluous to this targeting event. This apparent contradiction may be due to cell-type-specific targeting or to the limitations in the techniques used to assess subcellular distribution. It is notable that the role of Glut4 trafficking signals has never been studied in native skeletal muscle fibers, which possess architecture distinct from any other cell type and is not mimicked by any muscle-derived cell line. A large fraction of Glut4 in skeletal muscle is associated with transverse tubules and with vesicles closely associated with muscle triads (44-46). These structures are not present in muscle-derived cell lines.
James and colleagues (29) reported the identification of an acidic motif (TELEY) in the C-terminal cytoplasmic tail of Glut4 downstream of the dileucine motif that, when mutated to alanine residues, causes accumulation of Glut4 in a plasma membrane-enriched subcellular fraction in the basal state but does not prevent insulin-stimulated movement of Glut4 to a plasma membrane fraction isolated from 3T3L1 adipocytes. The data presented suggested that mutations in this motif cause Glut4 to accumulate in the general endosomal recycling pathway. A subsequent publication from this group identified the EEY residues within this shared motif to be essential and demonstrated that mutation of these three residues delayed newly endocytosed Glut4 from reaching a Syntaxin 6/16-containing perinuclear compartment (32). This acidic motif in Glut4 was originally identified based on sequence similarity with the N-terminal cytoplasmic domain of IRAP, a protein whose intracellular trafficking closely resembles that of Glut4.
A unique aspect of the current study is that mutant Glut4 molecules corresponding to most of the previously proposed subcellular targeting motifs were examined in the same insulin-responsive cell type using the same expression system, and that all of the mutants were directly assessed for colocalization with wild-type Glut4 by coexpression of differentially tagged constructs in the same cells. Based on a different alignment between the sequences of the N terminus of IRAP and the C terminus of Glut4, we identified a different sequence similarity between the two proteins that overlaps with the acidic motif identified by James and colleagues (32). Mutation of three residues (Leu500-Leu503-Pro505) within this putative motif shared between Glut4 and IRAP resulted in a much more dramatic phenotype than that demonstrated by mutations within the acidic motif or any other previously characterized Glut4-trafficking motif. The IRM or LLP mutant protein targeted primarily to relatively large, dispersed membrane compartments that were devoid of wild-type Glut4 in the basal state, and both basal and insulin-stimulated recycling through the plasma membrane appeared to be completely abrogated. These observations suggest that the mutants are excluded from recycling endosomes and from GSVs in 3T3L1 adipocytes, because both of these compartments are insulin-sensitive in this cell line (17).
The major colocalization observed between wild-type Glut4 and the HA-IRM mutant was a fraction of both molecules present in a perinuclear Syntaxin 6-containing compartment, most likely the putative trans-Golgi subcompartment previously identified by James and colleagues (32). The fraction of the total HA-IRM mutant that localized to this compartment, however, was less than the total fraction of HA-tagged wild-type Glut4. Glut4 appears to reach this compartment after movement to the plasma membrane in response to insulin and subsequent endocytosis but may also recycle through this compartment in the basal state (32). One possibility is that Glut4 exits the Syntaxin 6 compartment to an intermediate sorting compartment that subsequently targets a fraction of Glut4 to both the endosomal recycling pathway and to specialized GSVs. Both of these sorting events may be dependent on the IRM motif. The existence of this hypothetical post-Syntaxin 6 compartment is consistent with the observations that the IRM mutant is detectable in the Syntaxin 6 compartment, but that the bulk of the mutant in the steady-state appears to be present in one or more subcellular compartments that do not colocalize with any tested subcellular marker, including wild-type Glut4 or the EEY mutant (32). Additionally, the IRM and LLP mutants appear to be completely unresponsive to insulin with regard to translocation to the plasma membrane, suggesting that this mutant does not reach (or substantially accumulate) in either recycling endosomes or the specialized GSVs. Because wild-type Glut4 appears to be absent from the vast bulk of the IRM mutant-containing dispersed vesicles in the steady state, if wild-type Glut4 normally transits via this compartment to recycling endosomes and/or GSVs, then its transit time through this compartment must be very rapid. Alternatively, the IRM mutant may be targeted to an intracellular membrane compartment that wild-type Glut4 never traverses. If so, then it appears to be an intracellular compartment that has not been previously identified or is poorly characterized, because it does not appear to contain significant quantities of any of the standard subcellular markers that were examined.
Interestingly, the IRM does not appear to be essential for the targeting of IRAP to insulin-sensitive compartments in 3T3L1 adipocytes, despite the fact that the IRM was originally identified due to its conservation in both IRAP and Glut4. There are several possible explanations for this apparent anomaly. IRAP and Glut4 may reach the same insulin-sensitive intracellular compartments via different pathways either during initial partitioning of nascent molecules or after recycling events, and the IRM may play only a subtle role in the trafficking of IRAP. Another possibility is that IRAP has redundant motifs that are involved in its targeting to insulin-sensitive compartments. A third possibility is that the IRM is not required for IRAP trafficking in adipocytes but may play an essential role in other cell types, such as skeletal muscle, heart, or neurons. In this latter case it is conceivable that in adipocytes IRAP forms oligomers with another membrane protein on which it “piggy-backs” to reach insulin-sensitive Glut4 compartments. The initial partitioning of nascent IRAP may be defective in this hypothetical case, but it may eventually reach subcellular compartments containing the interacting protein and may subsequently be redirected to Glut4-containing insulin-sensitive vesicles via interaction with another protein. The observations, that 1) the IRM represents the most highly conserved sequence conservation between the cytoplasmic 25-30 residues of Glut4 that are known to be critical for its insulin sensitive targeting and the cytoplasmic N-terminal domain of IRAP; 2) residues within the IRM are clearly critical in the targeting of Glut4; and 3) the only unique characteristic shared between Glut4 and IRAP is their highly similar subcellular targeting, strongly suggest that the IRM is likely to play an essential role in the subcellular trafficking of IRAP, even though it may be in a cell type other than adipocytes.
Two recent studies employing total internal reflection microscopy have examined the insulin-regulated behavior of Glut4-containing membrane vesicles within ∼100-250 nm of the plasma membrane in 3T3L1 adipocytes (47, 48). The data presented by Bai et al. (47) suggest that insulin has a minimal affect on the rate of the initial docking of Glut4 storage vesicles with the cytoplasmic face of the plasma membrane and that the major insulin-regulated step may be the fusion of docked vesicles with the plasma membrane. Our data indicate that mutants within the IRM motif colocalize with wild-type Glut4 in small vesicles just beneath the plasma membrane, but the mutants do not discernibly accumulate in the plasma membrane after insulin stimulation. Either these shared vesicles are not capable of docking or fusion, or the amount of the IRM mutant that reaches the shared vesicles is insufficient to detect accumulation of the mutant protein in the plasma membrane.
The data presented in this study, combined with previously published data, suggest that there may be multiple overlapping but functionally distinct targeting motifs within the C-terminal cytoplasmic tail of Glut4. The IRM mutant exhibits the most dramatic targeting phenotype in an insulin-sensitive cell-type of any putative trafficking motif thus far reported for Glut4. Our data suggest that the IRM mutant accumulates in a previously unrecognized or at least poorly characterized intracellular membrane compartment. Further experiments are clearly required to identify the precise subcellular compartment(s) to which the IRM Glut4 mutant is targeted.
Acknowledgments
We thank Matthew Storck for construction of several of the mutants examined in this study and Dr. Susanna Keller (University of Virginia, Charlottesville) for kind gifts of IRAP cDNA and IRAP antibodies.
This work was supported by the American Diabetes Association and by National Institutes of Health Grants R01 DK067229 and R01 DK043695. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IRAP, insulin-responsive aminopeptidase; IRM, insulin-responsive motif; PM, plasma membrane; MPR, mannose 6-phosphate receptor; GSV, Glut4 storage vesicle; PBS, phosphate-buffered saline.
References
- 1.James, D. E., Brown, R., Navarro, J., and Pilch, P. F. (1988) Nature 333 183-185 [DOI] [PubMed] [Google Scholar]
- 2.James, D. E., Strube, M., and Mueckler, M. (1989) Nature 338 83-87 [DOI] [PubMed] [Google Scholar]
- 3.Birnbaum, M. J. (1989) Cell 57 305-315 [DOI] [PubMed] [Google Scholar]
- 4.Charron, M. J., Brosius, F. D., Alper, S. L., and Lodish, H. F. (1989) Proc. Natl. Acad. Sci. U. S. A. 86 2535-2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueckler, M. (1995) News Physiol. Sci. 10 22-29 [Google Scholar]
- 6.Cline, G. W., Petersen, K. F., Krssak, M., Shen, J., Hundal, R. S., Trajanoski, Z., Inzucchi, S., Dresner, A., Rothman, D. L., and Shulman, G. I. (1999) N. Engl. J. Med. 341 240-246 [DOI] [PubMed] [Google Scholar]
- 7.Rothman, D. L., Shulman, R. G., and Shulman, G. I. (1992) J. Clin. Invest. 89 1069-1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cushman, S. W., and Wardzala, L. J. (1980) J. Biol. Chem. 255 4758-4762 [PubMed] [Google Scholar]
- 9.Suzuki, I., and Kono, T. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 2542-2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slot, J. W., Geuze, H. J., Gigengack, S., Lienhard, G. E., and James, D. E. (1991) J. Cell Biol. 113 123-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slot, J. W., Geuze, H. J., Gigengack, S., James, D. E., and Lienhard, G. E. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 7815-7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malide, D., Ramm, G., Cushman, S. W., and Slot, J. W. (2000) J. Cell Sci. 113 4203-4210 [DOI] [PubMed] [Google Scholar]
- 13.Holman, G. D., Kozka, I. J., Clark, A. E., Flower, C. J., Saltis, J., Habberfield, A. D., Simpson, I. A., and Cushman, S. W. (1990) J. Biol. Chem. 265 18172-18179 [PubMed] [Google Scholar]
- 14.Zeigerer, A., Lampson, M. A., Karylowski, O., Sabatini, D. D., Adesnik, M., Ren, M., and McGraw, T. E. (2002) Mol. Biol. Cell. 13 2421-2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karylowski, O., Zeigerer, A., Cohen, A., and McGraw, T. E. (2004) Mol. Biol. Cell 15 870-882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Govers, R., Coster, A. C., and James, D. E. (2004) Mol. Cell. Biol. 24 6456-6466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holman, G. D., and Sandoval, I. V. (2001) Trends Cell Biol. 11 173-179 [DOI] [PubMed] [Google Scholar]
- 18.Piper, R. C., Tai, C., Slot, J. W., Hahn, C. S., Rice, C. M., Huang, H., and James, D. E. (1992) J. Cell Biol. 117 729-743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piper, R. C., Tai, C., Kulesza, P., Pang, S., Warnock, D., Baenziger, J., Slot, J. W., Geuze, H. J., Puri, C., and James, D. E. (1993) J. Cell Biol. 121 1221-1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garippa, R. J., Judge, T. W., James, D. E., and McGraw, T. E. (1994) J. Cell Biol. 124 705-715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Al-Hasani, H., Kunamneni, R. K., Dawson, K., Hinck, C. S., Muller-Wieland, D., and Cushman, S. W. (2002) J. Cell Sci. 115 131-140 [DOI] [PubMed] [Google Scholar]
- 22.Verhey, K. J., Yeh, J. I., and Birnbaum, M. J. (1995) J. Cell Biol. 130 1071-1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haney, P. M., Levy, M. A., Strube, M. S., and Mueckler, M. (1995) J. Cell Biol. 129 641-658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verhey, K. J., and Birnbaum, M. J. (1994) J. Biol. Chem. 269 2353-2356 [PubMed] [Google Scholar]
- 25.Corvera, S., Chawla, A., Chakrabarti, R., Joly, M., Buxton, J., and Czech, M. P. (1994) J. Cell Biol. 126 979-989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsh, B. J., Alm, R. A., McIntosh, S. R., and James, D. E. (1995) J. Cell Biol. 130 1081-1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czech, M. P., Chawla, A., Woon, C. W., Buxton, J., Armoni, M., Tang, W., Joly, M., and Corvera, S. (1993) J. Cell Biol. 123 127-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall, B. A., Murata, H., Hresko, R. C., and Mueckler, M. (1993) J. Biol. Chem. 268 26193-26199 [PubMed] [Google Scholar]
- 29.Shewan, A. M., Marsh, B. J., Melvin, D. R., Martin, S., Gould, G. W., and James, D. E. (2000) Biochem. J. 350 99-107 [PMC free article] [PubMed] [Google Scholar]
- 30.Keller, S. R., Scott, H. M., Mastick, C. C., Aebersold, R., and Lienhard, G. E. (1995) J. Biol. Chem. 270 23612-23618 [DOI] [PubMed] [Google Scholar]
- 31.Kandror, K. V., Yu, L., and Pilch, P. F. (1994) J. Biol. Chem. 269 30777-30780 [PubMed] [Google Scholar]
- 32.Shewan, A. M., Van Dam, E. M., Martin, S., Luen, T. B., Hong, W., Bryant, N. J., and James, D. E. (2003) 14 973-986 [DOI] [PMC free article] [PubMed]
- 33.Cope, D. L., Lee, S., Melvin, D. R., and Gould, G. W. (2000) FEBS Lett. 481 261-265 [DOI] [PubMed] [Google Scholar]
- 34.Martinez-Arca, S., Lalioti, V. S., and Sandoval, I. V. (2000) J. Cell Sci. 113 1705-1715 [DOI] [PubMed] [Google Scholar]
- 35.Haney, P. M., Slot, J. W., Piper, R. C., James, D. E., and Mueckler, M. (1991) J. Cell Biol. 114 689-699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He, T. C., Zhou, S., da Costa, L. T., Yu, J., Kinzler, K. W., and Vogelstein, B. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 2509-2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson, I. A., Yver, D. R., Hissin, P. J., Wardzala, L. J., Karnieli, E., Salans, L. B., and Cushman, S. W. (1983) Biochim. Biophys. Acta 763 393-407 [DOI] [PubMed] [Google Scholar]
- 38.Piper, R. C., Hess, L. J., and James, D. E. (1991) Am. J. Physiol. 260 C570-C580 [DOI] [PubMed] [Google Scholar]
- 39.Lawrence, J. C., Jr., Hiken, J. F., and James, D. E. (1990) J. Biol. Chem. 265 2324-2332 [PubMed] [Google Scholar]
- 40.Keller, S. R. (2003) Front. Biosci. 8 s410-s420 [DOI] [PubMed] [Google Scholar]
- 41.Bryant, N. J., Govers, R., and James, D. E. (2002) Nat. Rev. Mol. Cell Biol. 3 267-277 [DOI] [PubMed] [Google Scholar]
- 42.Martin, O. J., Lee, A., and McGraw, T. E. (2006) J. Biol. Chem. 281 484-490 [DOI] [PubMed] [Google Scholar]
- 43.Xu, Z., Huang, G., and Kandror, K. V. (2006) Mol. Endocrinol. 20 2890-2897 [DOI] [PubMed] [Google Scholar]
- 44.Wang, W., Hansen, P. A., Marshall, B. A., Holloszy, J. O., and Mueckler, M. (1996) J. Cell Biol. 135 415-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marette, A., Burdett, E., Douen, A., Vranic, M., and Klip, A. (1992) Diabetes 41 1562-1569 [DOI] [PubMed] [Google Scholar]
- 46.Dudek, R. W., Dohm, G. L., Holman, G. D., Cushman, S. W., and Wilson, C. M. (1994) FEBS Lett. 339 205-208 [DOI] [PubMed] [Google Scholar]
- 47.Bai, L., Wang, Y., Fan, J., Chen, Y., Ji, W., Qu, A., Xu, P., James, D. E., and Xu, T. (2007) Cell Metabolism 5 47-57 [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez, E., and McGraw, T. E. (2006) Mol. Biol. Cell 17 4484-4493 [DOI] [PMC free article] [PubMed] [Google Scholar]