Abstract
The RNA-dependent RNA polymerases (RdRPs) of Cystoviridae bacteriophages, like those of eukaryotic viruses of the Reoviridae, function inside the inner capsid shell in both replication and transcription. In bacteriophage Φ6, this inner shell is first assembled as an icosahedral procapsid with recessed 5-fold vertices that subsequently undergoes major structural changes during maturation. The tripartite genome is packaged as single-stranded RNA molecules via channels on the 5-fold vertices, and transcripts probably exit the mature capsid by the same route. The RdRP (protein P2) is assembled within the procapsid, and it was thought that it should be located on the 5-fold axes near the RNA entry and exit channels. To determine the initial location of the RdRP inside the procapsid of bacteriophage Φ6, we performed cryo-electron microscopy of wild type and mutant procapsids and complemented these data with biochemical determinations of copy numbers. We observe ring-like densities on the 3-fold axes that are strong in a mutant that has ∼10 copies of P2 per particle; faint in wild type, reflecting the lower copy number of ∼3; and completely absent in a P2-null mutant. The dimensions and shapes of these densities match those of the known crystal structure of the P2 monomer. We propose that, during maturation, the P2 molecules rotate to occupy positions closer to adjacent 5-fold vertices where they conduct replication and transcription.
Viruses of the Cystoviridae and the Reoviridae families encapsidate RdRPs, virally encoded enzymes that have the dual functions of replicating the genome and synthesizing transcripts (1, 2). The reoviruses have a complex assembly pathway whereby genome replication and packaging proceed concurrently (2). Cystoviridae, on the other hand, first assemble a closed procapsid (3), into which the three plus-sense single-stranded RNA segments of the viral genome are packaged sequentially via conduits on one or more of the twelve 5-fold vertices.
Bacteriophage Φ6, the type species of the Cystoviridae, is a spherical, triple-layered, double-stranded RNA virus (diameter, 860 Å) of the bacterium Pseudomonas syringae (3). In Φ6 assembly, the first particle produced is the procapsid or polymerase complex, composed of the proteins P1, P2, P4, and P7. Its shell, containing 120 copies of P1, has a distinctive dodecahedral shape with deeply recessed vertices (4). P4 is a hexameric NTPase that is mounted in a symmetry-mismatched location above the 5-fold vertices (5, 6) on the outer surface of the procapsid where it translocates single-stranded RNA segments into the P1 shell. The locations of P2, the RdRP, and P7, a protein that appears to aid in packaging, neither of which is required for procapsid assembly (7, 8), have been less clear. P2 is inside the P1 shell and has been estimated to be present at ∼14 molecules/particle (9). This copy number being close to 12 and the observation that the reovirus polymerase, λ3, is located close to its 5-fold vertices (10) suggested that P2 should also be located on or near the 5-fold axes, an assumption used in interpreting neutron diffraction data on polymerase complexes (11).
In this study, we purified procapsids from several different expression vectors and determined their stoichiometries by biochemical quantitation and their structures by cryo-electron microscopy (cryo-EM)3 and image reconstruction. The procapsids turned out to have differing amounts of P2; one, a P2-null mutant, lacked this protein entirely, allowing us to correlate certain internal densities with the presence of P2. The size and shape of these densities are compatible with the known crystal structure (12) of the P2 monomer, confirming this identification.
EXPERIMENTAL PROCEDURES
Bacteriophage Φ6 Strains—Wild type procapsids, designated P1247, were produced, using the Escherichia coli JM105 strain LM4213 with the plasmid, pLM450. The procapsids do not contain either cellular or viral RNA because specific pac sequences are required for genomic packaging and maturation (13). The cell line LM3000 with the plasmid pLM2541 produced a procapsid with a mutation in P1 (E390A) that is able to package the M and L segments without the S segment, and this is called P1′247 (14). A mutant procapsid lacking P2, P147, was produced with cell line LM4419 and plasmid pLM1906, which is derived from plasmid pLM687 by deleting the AflII-SphI fragment (15). P124 procapsids lacking P7 were produced with cell line LM1115 and plasmid pLM574 (16).
Procapsid Isolation—Cell cultures were grown overnight in minimal medium and induced with 1 mm isopropyl β-d-thiogalactopyranoside. The cells were lysed with a French press, and the particles were purified by zone and equilibrium centrifugation in sucrose gradients. Zone gradients contained Buffer A (10 mm potassium phosphate, 1 mm magnesium chloride pH 7.5) and a linear gradient of 10 to 30% sucrose (w/v). Equilibrium gradients contained Buffer A and linear gradients of 25 to 50% sucrose (w/v). Sucrose was removed from bands collected from the equilibrium gradients by passage through Sephadex G-50 spin columns equilibrated with Buffer A.
Quantitation of P2 Content—For labeling experiments to quantitate P2, cell cultures were grown in minimal medium with 5 or 10 μg/ml l-methionine plus 10 μCi/ml [35S]methionine. After lysis with a French Press, the samples were mixed with lysates of unlabeled, procapsid-producing, cells and further purified as described above. The purified particles were dissolved and heated at 100 °C in cracking buffer (1% SDS, 1% 2-mercaptoethanol, 10% glycerol, 2 mm EDTA, 50 mm Tris-HCl, pH 6.8, bromphenol blue) and applied to 8 or 13% polyacrylamide gels. The gels were dried and incubated in phosphorimaging cassettes and processed (Molecular Dynamics Storm 860). The counts in the bands were measured, and the ratios of counts in the bands for P2 and P1 were tabulated. The number of P2 molecules was calculated on the basis that there are 120 copies of P1/procapsid and that the number of methionine residues is 19 and 24 for P1 and P2, respectively. In addition, because the band of P2 is very close to that of P1, the amount of spillover was estimated using procapsids produced from the plasmid, pLM3548, a construct not coding for P2. This correction amounts to a subtraction equivalent to 1.6 P2 molecules/procapsid.
Cryo-electron Microscopy—Samples (3 μl) at 1–5 mg/ml protein were applied to grids with holey carbon films, blotted, and plunge-frozen in liquid ethane at liquid nitrogen temperature. The vitrified specimens were imaged at 50,000× magnification with a CM200 FEG microscope (FEI, Eindhoven, The Netherlands) fitted with a Gatan 626 cryoholder and operating at an accelerating voltage of 120 keV. Focal pairs of micrographs were recorded at ∼12–15 e-/Å2 per exposure at 0.8–1.5 and 1.5–2.6 μm underfocus for the first and second images, respectively (see Table 1).
TABLE 1.
Reconstruction statistics for maps of the Φ6 procapsids and the occupancy of the RNA-dependent RNA polymerase, P2, based on integrated density or comparative 35S labeling
| Reconstructiona | P1247 | P1′247 | P147 | P124 |
|---|---|---|---|---|
| Micrograph pairs | 25 | 23 | 18 | 17 |
| Pixel size (Å/pixel) | 2.54 | 2.54 | 2.54 | 2.54 |
| Defocus (μm) | 0.9-1.7 | 0.9-1.7 | 1.0-2.3 | 1.3-2.6 |
| Particles used | 3150 | 902 | 1072 | 1238 |
| Resolution (Å)b | 14 | 19 | 11 | 16 |
| P2 occupancyc | ||||
| Density | 1-2 | 10 | 0 | 3-4 |
| 35S labeling | 3-4 | 7-8 | 0 | 3 |
The digits indicate the proteins present. P1'247 is a mutant in P1.
Based on Fourier shell correlation at a cut-off of 0.3.
The values indicate the corresponding number of P2 monomers/procapsid.
Image Processing—Micrographs were digitized using a Nikon Super Coolscan 9000 ED at 4000 dpi (1.27 Å/pixel). Image processing was done with Bsoft and the programs PFT2 and EM3DR2 (17). For each micrograph, the contrast transfer function parameters were estimated in bshow. The particles were picked in the further-from-focus micrographs of focal pairs in bshow, followed by alignment of the focal pairs with bmgalign to find the particles in the closer-to-focus micrographs. The particle images were extracted with bpick and contrast transfer function-corrected by phase-flipping with bctf. All of the micrographs and the picked particles were binned by a factor of 2 to accelerate the image processing, giving 2.54 Å/pixel. As an initial reference, a synthetic model based on the procapsid as rendered in Ref. 4 was constructed with the program beditimg. borient and PFT2 were used to determine the origins and orientations of the particles starting with the synthetic map, using an angular step size of 1° and a resolution range of 10–200 Å for the final runs. The reconstructions were done with breconstruct and EM3DR2 to a resolution limit of 10 Å. The resolution was estimated using bresolve, based on Fourier shell correlation and using a 0.3 cut-off (see Table 1).
Occupancy of P2 Molecules—The occupancy of densities identified as P2 in the procapsid reconstructions was estimated by integration of small spherical volumes (∼22 Å in diameter and ∼5600 Å3 in volume) centered on each density and compared with the same size volumes of background and the most intense part of the P1 shell as the zero and full stoichiometric references, respectively.
Fitting the P2 Structure into the Ring-like Densities—P2 is a monomer of approximately the same size as the observed ring-like densities. Because these densities are located on the 3-fold axes, they represent 3-fold symmetrized versions of the P2 molecule. One of these densities was excised from the P1′247 map with bshow, rotated to align its 3-fold axis with the z-axis and symmetrized again with bsym to remove any interpolation artifacts. The P2 structure, 1UVJ (12), was obtained from the Protein Data Base and centered, and a density map was generated using bsf at the same pixel size as the procapsid maps to a resolution limit of 12 Å. To best align this representation of P2 with a ring-like density from the cryo-EM3 map, a correlationbased search of starting orientations was conducted with a 4° step size and shifts with 1 pixel step size ± 2 pixels in all directions, using the program bsymcomp. For each shift, a full grid search was conducted over the three Euler angles, and the maximum correlation coefficient was selected as the best fit (mean coefficient over all views, 0.82, with a standard deviation of 0.03). A map of these correlation coefficient maxima shows three symmetry-related peaks (with values of 0.913–0.915) within a background consistently decreasing with distance from the peaks.
RESULTS AND DISCUSSION
Procapsids were prepared from four different combinations of expression plasmids and host cell strains. Following earlier practice (5), we use the term P1247 to denote procapsids assembled with the wild type versions of all four proteins: P1, P2, P4, and P7; P147 for procapsids with P1, P4, and P7 but lacking P2, etc. P1′247 are procapsids with all four proteins but with a point mutation in P1.
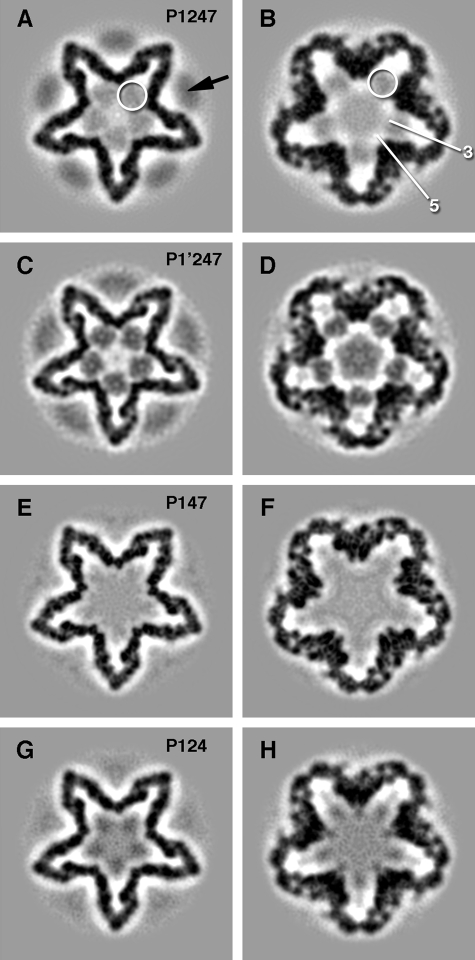
Reconstructions of Φ6 Procapsids—Cryo-electron micrographs were recorded under standardized conditions and used to calculate density maps (Fig. 1 and Table 1). In each case, the P1 surface shell was similar both to those reported in previous studies (4, 5) and to each other, apart from differences in resolution. The highest resolution, 11 Å, was achieved with the P147 capsid (Figs. 1, E and F, and 2, B and D). The P1 shell of this map was essentially indistinguishable from that in the other maps (such as P1′247; Figs. 1, C and D, and 2, A and C) when band-limited to the same resolution (data not shown).
FIGURE 1.
Sections through the bacteriophage Φ6 procapsid reconstructions at 70 Å (A, C, E, and G) and 30 Å (B, D, F, and H) from the procapsid center. A and B, the wild type procapsid showing faint ring-like densities (white circles) on the 3-fold axes between the recessed 5-fold vertices, as well as the hexameric P4 protein (black arrow). The approximate locations of the 3- and 5-fold axes are shown in B. C and D, the procapsid of the P1 mutant with prominent ring-like densities and some density in the center. E and F, the procapsid lacking the P2 molecule has no ring-like densities between the 5-fold vertices. G and H, the procapsid lacking the P7 protein has density between the 5-fold vertices.
FIGURE 2.
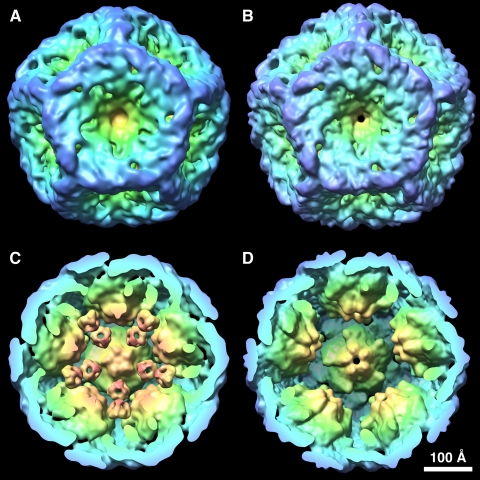
Comparison of the procapsid reconstructions of P1′247 (A and C) and P147 (B and D) showing the ring-like densities on the 3-fold axes between the 5-fold vertices in the former and their complete absence in the latter. The P4 protein densities and the central density were excised from the P1′247 map for clarity in the comparison. The isosurfaces were generated at a level of 1.5 σ.
It has been demonstrated that hexamers of P4 overlie the 5-fold vertices of the Φ6 procapsid and that the amount of P4 retained per particle is dependent on preparation conditions (18). Such was also our experience in the present studies. The strongest P4-related densities were observed with P1247 and P1′247 procapsids (Fig. 1, A and C). This density is somewhat blurred compared with the sharply defined density in the P1 shell, which is a result of the 5:6 symmetry mismatch and possibly also of some misalignment of the 6-fold axis of the P4 hexamer with the 5-fold axis of the capsid (5).
P2 has not been unambiguously localized in previous studies but is thought to be internal (5, 11). Examining wild type P1247 procapsids, we observed ring-like densities on the 3-fold axes and located between the inverted 5-fold vertices (white circles in Fig. 1, A and B). The corresponding densities in the P1′247 mutant were much more pronounced as seen in grayscale sections of the procapsid (Fig. 1, C and D). The identity of these densities was considered to be either P2 or P7, and procapsid maps were reconstructed for both P147 and P124. P147 completely lacks the ring-like densities, and the map inside the P1 shell is featureless (Fig. 1, E and F). Similar, albeit faint, densities were discernible for the P124 procapsids (Fig. 1, G and H). Surface renderings of the interiors of the P1′247 and P147 procapsids confirm that ring-like densities are present in the former (Fig. 2C) and absent in the latter (Fig. 2D). These observations suggested that the densities in question represent encapsidated P2 molecules and that the P1247 and P124 procapsids have unexpectedly low copy numbers of this protein.
Quantitation of P2 Contents—To test this hypothesis, we performed biochemical quantitations of the amount of P2 present in radiolabeled preparations of all four kinds of procapsids (Table 1). These copy numbers may be compared with estimates made from the cryo-EM density maps where the occupancy of the inferred site may be taken from the ratio between its peak density above background and the peak density in the P1 shell (representing 100% occupancy), and the copy numbers estimated by multiplying this fraction by the number of symmetry-related sites per capsid, in this case, 20. There is excellent agreement between the biochemical quantitation and the cryo-EM based quantitation, providing strong support for the inferred localization of P2.
In principle, alternative interpretations of this density are that it might represent P7 or some host protein that fortuitously binds to the procapsid inner surface. The presence of significant density at this site in P124 procapsids that lack P7 argues against the first possibility. The complete absence of density at this site in P147 capsids rules out the host protein hypothesis.
The P2-related density, like the P4-related density, varies in intensity among these reconstructions, but the origins of the observed variability differ. It appears that the P1247 procapsid that is produced from a very low copy plasmid has a lower content of P2 than particles encoded by high copy plasmids. For a wild type procapsid encoded by a plasmid, pLM1413, of the same copy number as that producing P1′247, we measured a P2 content of ∼6 molecules/particle. Thus, the higher content of P2 in P1′247 procapsids appears to reflect a fortuitously higher expression level in this system and/or a change in a P1 residue at the P2-binding site that results in a higher level of incorporation.
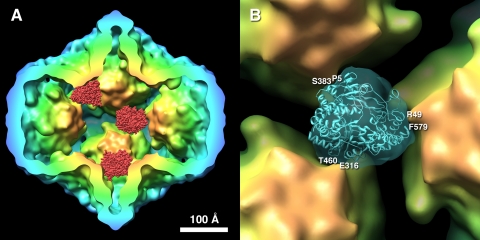
P2 Structure Matches the Ring-like Densities—P2 is a monomer whose structure is known to high resolution (12). However, its location on the icosahedral 3-fold axes means that in our maps, it appears as 3-fold symmetrized. To confirm that the observed densities are consistent with the P2 structure, it was subjected to 3-fold symmetrization and band-limited to a resolution consistent with the cryo-EM density map. Densities were thus generated for all possible starting orientations of P2 and compared to a ring-like density extracted from the P1′247 cryo-EM map. The search yielded three symmetry-related correlation coefficient maxima, corresponding to three equivalent orientations of the molecule fitted within one ring-like density. To illustrate the location of P2, three P2 molecules were placed in the best fitted orientation at three arbitrarily chosen sites of this kind, shown in Fig. 3A. The best match shows good consistency by visual criteria (Fig. 3B). A more quantitative comparison, in terms of the Fourier shell correlation, showed them to be consistent to a resolution of 17 Å, close to the resolution of the complete P1′247 procapsid map (Table 1). Interestingly, the P2 molecule has a somewhat triangular appearance in this orientation. This experiment provided our best estimate of how P2 is oriented relative to the P1 shell.
FIGURE 3.
A, three P2 molecules (red) placed in three of the 20 possible positions in the P1′247 procapsid based on their fit to the ring-like densities. B, the fit of the P2 structure into the ring-like density from the P1′247 procapsid suggests an orientation and interactions with the three neighboring 5-fold vertices. The main contacts are with the palm (Glu316 and Thr460), the thumb (Arg49 and Phe579), and finger (Pro5 and Ser383) domains of the P2 molecule. The substrate pore is directed away from the viewer and toward the 3-fold vertex of the P1 shell. The isosurface was generated at a level of 1 σ.
Location of P2 in the Procapsid—In an attempt to localize P2 by small angle neutron scattering with contrast matching, Ikonen et al. (11) concluded that P2 is distributed with a radius of gyration of about 110 Å from the procapsid center. Our analysis places the P2 center of mass at a similar distance of 104 Å from the procapsid center. Because the neutron diffraction data were one-dimensional, it could not be used to assign azimuthal coordinates for P2, and these authors suggested on the basis of symmetry arguments and stoichiometry that P2 is on the 5-fold axes. According to our localization, it is offset by approximately one molecular diameter from the 5-fold axes and centered on the icosahedral 3-fold axes, although at a ∼60 Å lower radius than the point at which that axis passes through the surface shell.
Any given P2 molecule has three potential contact sites with the surrounding P1 shell, at positions distributed around its 3-fold axis and with the adjacent 5-fold vertices (Fig. 3B). These contacts are, in fact, distinct because P2 is a monomer; thus one of them should dominate. As the shell expands during packaging, only one of these links can be maintained. Therefore, the P1 shell may carry P2 with it during expansion, possibly bringing it closer to the channel on the 5-fold. Such positioning would emulate that of the reovirus polymerase (10).
The P2 molecule is oriented with the substrate pore pointing outwards toward the 3-fold vertex, whereas the template tunnel is directed sideways (Fig. 3B). The C-terminal domain that presumably covers the product exit (19) is positioned toward the center of the procapsid.
Polymerase Stoichiometry—For reoviruses, a widely entertained but unproven notion is that each RdRP in a virion is associated with one of the 10–12 genomic segments. Because of its location at the 5-fold vertex (10), the maximum number of RdRPs is 12/virion. A persuasive circumstantial argument in favor of this scenario is that no reoviruses with more than 12 segments have been observed. The tenet that any given RdRP processes only a single segment implies that, at least for reoviruses, the full complement of RdRPs is required for functional virions. Applying this scenario to Φ6 implies that three copies of P2 would suffice, and this number is, in fact, what we observe in P1247 and P124 procapsids, although quantitation of native procapsids suggests that the P2 copy number is higher at 8–10 and the lower number of ∼3 is a consequence of the plasmid expression system. In this case, if each RNA segment is replicated by a different P2 molecule, there is an ∼3-fold redundancy in available polymerases.
Although Φ6 normally contains three genomic segments, the capsid is capable of replicating a larger number of shortened segments (20). This is true of the procapsids produced by pLM450 (20) that we find to have, on average, only one to three P2 molecules (Table 1). It appears then, that under these conditions, several RNA molecules can be replicated, presumably sequentially, by a single P2 molecule.
Control of Packaging—The P1′247 procapsid produced by pLM2541 has a mutant protein P1 (E390A) that enables this particle to alter its program of genomic packaging (14). Normally, Φ6 packages the plus strand of segment S before those of M and L. This mutant procapsid is able to package M, and subsequently L, without packaging segment S. According to the hypothesis that the order of packaging is controlled by sequential conformational changes in the maturing capsid (3), one might expect the P1′247 procapsid to be structurally altered, as if segment S were already packaged. Our observation is, however, that this particle is not any more expanded than the wild type P1247 procapsid (Fig. 1). This suggests that the mutations in protein P1 are changing the binding site for segment M to make it available even though the procapsid has not expanded. The P1′247 particle also shows competition between the binding of the S and M transcripts (14), indicating that the binding site for S is changed as well. These findings support the possibility that the binding sites for S and M on the procapsid are the same or overlapping and undergo subtle modifications in conformation during the packaging program.
This work was supported by the Intramural Research Program of NIAMS, National Institutes of Health and by Grant GM34352 (to L. M.) from the National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviation used is: cryo-EM, cryo-electron microscopy.
References
- 1.Poranen, M. M., and Tuma, R. (2004) Virus Res. 101 93–100 [DOI] [PubMed] [Google Scholar]
- 2.Taraporewala, Z. F., and Patton, J. T. (2004) Virus Res. 101 57–66 [DOI] [PubMed] [Google Scholar]
- 3.Mindich, L. (2004) Virus Res. 101 83–92 [DOI] [PubMed] [Google Scholar]
- 4.Butcher, S. J., Dokland, T., Ojala, P. M., Bamford, D. H., and Fuller, S. D. (1997) EMBO J. 16 4477–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Haas, F., Paatero, A. O., Mindich, L., Bamford, D. H., and Fuller, S. D. (1999) J. Mol. Biol. 294 357–372 [DOI] [PubMed] [Google Scholar]
- 6.Huiskonen, J. T., Jaalinoja, H. T., Briggs, J. A., Fuller, S. D., and Butcher, S. J. (2007) J. Struct. Biol. 158 156–164 [DOI] [PubMed] [Google Scholar]
- 7.Juuti, J. T., and Bamford, D. H. (1995) J. Mol. Biol. 249 545–554 [DOI] [PubMed] [Google Scholar]
- 8.Juuti, J. T., and Bamford, D. H. (1997) J. Mol. Biol. 266 891–900 [DOI] [PubMed] [Google Scholar]
- 9.Day, L. A., and Mindich, L. (1980) Virology 103 376–385 [DOI] [PubMed] [Google Scholar]
- 10.Zhang, X., Walker, S. B., Chipman, P. R., Nibert, M. L., and Baker, T. S. (2003) Nat. Struct. Biol. 10 1011–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikonen, T., Kainov, D., Timmins, P., Serimaa, R., and Tuma, R. (2003) J. Appl. Crystallogr. 36 525–529 [Google Scholar]
- 12.Salgado, P. S., Makeyev, E. V., Butcher, S. J., Bamford, D. H., Stuart, D. I., and Grimes, J. M. (2004) Structure 12 307–316 [DOI] [PubMed] [Google Scholar]
- 13.Gottlieb, P., Qiao, X., Strassman, J., Frilander, M., and Mindich, L. (1994) Virology 200 42–47 [DOI] [PubMed] [Google Scholar]
- 14.Qiao, J., Qiao, X., Sun, Y., and Mindich, L. (2003) J. Bacteriol. 185 4572–4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mindich, L., Qiao, X., Onodera, S., Gottlieb, P., and Frilander, M. (1994) Virology 202 258–263 [DOI] [PubMed] [Google Scholar]
- 16.Gottlieb, P., Strassman, J., Qiao, X. Y., Frucht, A., and Mindich, L. (1990) J. Bacteriol. 172 5774–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heymann, J. B., and Belnap, D. M. (2007) J. Struct. Biol. 157 3–18 [DOI] [PubMed] [Google Scholar]
- 18.Olkkonen, V. M., and Bamford, D. H. (1987) J. Virol. 61 2362–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butcher, S. J., Grimes, J. M., Makeyev, E. V., Bamford, D. H., and Stuart, D. I. (2001) Nature 410 235–240 [DOI] [PubMed] [Google Scholar]
- 20.Mindich, L., Qiao, X., and Qiao, J. (1995) Virology 212 213–217 [DOI] [PubMed] [Google Scholar]