Abstract
A helical filament of Rad51 on single-strand DNA (ssDNA), called the presynaptic filament, catalyzes DNA joint formation during homologous recombination. Rad52 facilitates presynaptic filament assembly, and this recombination mediator activity is thought to rely on the interactions of Rad52 with Rad51, the ssDNA-binding protein RPA, and ssDNA. The N-terminal region of Rad52, which has DNA binding activity and an oligomeric structure, is thought to be crucial for mediator activity and recombination. Unexpectedly, we find that the C-terminal region of Rad52 also harbors a DNA binding function. Importantly, the Rad52 C-terminal portion alone can promote Rad51 presynaptic filament assembly. The middle portion of Rad52 associates with DNA-bound RPA and contributes to the recombination mediator activity. Accordingly, expression of a protein species that harbors the middle and C-terminal regions of Rad52 in the rad52 Δ327 background enhances the association of Rad51 protein with a HO-made DNA double-strand break and partially complements the methylmethane sulfonate sensitivity of the mutant cells. Our results provide a mechanistic framework for rationalizing the multi-faceted role of Rad52 in recombination and DNA repair.
In eukaryotes, homologous recombination (HR)5 is mediated by genes of the RAD52 epistasis group. HR plays a prominent role in chromosome damage repair and helps restart stalled DNA replication forks (1–3). In addition, meiotic recombination mediated by the RAD52 group genes ensures the proper segregation of homologous chromosomes at meiosis I. Accordingly, mutants of the RAD52 group are sensitive to genotoxic agents, bear a strong mutator phenotype, and exhibit severe meiotic abnormalities (2). In mammals, a deficiency in HR causes cell inviability and can lead to the cancer phenotype (4, 5).
The mechanistic aspects of HR are best understood in the yeast Saccharomyces cerevisiae, within the context of DNA double-strand break (DSB) repair (6, 7). In S. cerevisiae, the RAD51, RAD52, RAD54, RAD55, RAD57, RAD59, and RDH54 genes are the core members of the RAD52 epistasis group. The RAD51-encoded product, an orthologue of the Escherichia coli RecA recombinase (8), mediates the formation of DNA joints that link the recombining DNA molecules (8, 9). In the presence of ATP, Rad51 polymerizes onto ssDNA to form a righthanded filament called the presynaptic filament. The presynaptic filament also harbors a binding site for duplex DNA (8, 10). Once engaged, the duplex DNA is sampled for homology, leading to DNA joint formation with the ssDNA (8, 10). The nascent DNA joint is extended by DNA strand exchange (10, 11). Thus, assembly of the presynaptic filament and its maintenance represent a most critical event in HR (10); see below).
Because nucleation of Rad51 onto ssDNA is slow, presynaptic filament assembly is prone to interference by RPA, the single-strand DNA binding protein in eukaryotes. RPA was first recognized as an accessory factor in the assembly of the presynaptic filament (9) via DNA secondary structure removal (12). Later, it was shown that RPA also possesses a post-synaptic function through sequestering the ssDNA generated during DNA strand exchange (13, 14). However, because RPA has high affinity for ssDNA, it can prevent Rad51 from binding ssDNA and thus strongly inhibit presynaptic filament assembly (12, 15). Inclusion of Rad52 efficiently overcomes this inhibitory effect of RPA (15–17). This recombination mediator function of Rad52 stems from its ability to nucleate Rad51 onto RPA-coated ssDNA to seed the assembly of the presynaptic filament (7, 11).
Cell-based studies have validated the significance of the biochemical data as summarized above. Specifically, in chromatin immunoprecipitation and cytological experiments designed to examine the association of RPA and HR factors with DSBs, loading of RPA onto the DNA substrate occurs first, and then it is gradually replaced by Rad51 (18–22). This replacement of RPA by Rad51 is dependent on Rad52. Aside from its recombination mediator role, Rad52 also functions in a pathway of HR between direct DNA repeats known as single-strand annealing. Herein, Rad52 is thought to anneal RPA-coated complementary single DNA strands (23–25) to form deletion recombinants (7).
Understanding the mechanistic basis of the Rad52 recombination mediator activity is of great importance, because an inability to shepherd Rad51 to DNA damage is a hallmark of cells deficient in the tumor suppressor BRCA2 (4). Several properties of Rad52 are believed to be germane for its recombination mediator function (2, 11). As first recognized by Mortensen et al. (23) and subsequently confirmed by others (16, 17, 26), Rad52 binds both ssDNA and dsDNA, showing a preference for the former. In addition, Rad52 exists as homooligomer (24) of seven or more subunits (27). The N-terminal third of Rad52 harbors the oligomerization domain and a Rad59 interaction domain and also exhibits DNA binding activity (23, 28–30). Rad52 also physically associates with Rad51 through its C terminus (31, 32) and is thought to bind RPA as well (16, 33, 34).
Disruption of the Rad51-binding domain in Rad52, as in the rad52 Δ327 and rad52 Δ409–412 alleles (16, 35), compromises its recombination mediator function. Although not yet formally proven, it seems reasonable to propose that Rad52 DNA binding function is important for the loading of Rad51 onto RPA-coated ssDNA. In this regard, it has been generally assumed that the N-terminal third of Rad52 is critical. In addition, the oligomeric structure of Rad52 may promote cooperative DNA binding that could be relevant for the recombination mediator activity (11).
In our continuing effort to dissect the Rad52 recombination mediator function, we unexpectedly find a robust DNA binding activity within the C-terminal third of the protein. We demonstrate that the Rad52 C terminus alone has recombination mediator activity. Importantly, we show that the middle portion of Rad52 interacts with DNA-bound RPA and makes a contribution to the recombination mediator function. Accordingly, a polypeptide harboring the middle and C-terminal portions of Rad52 partially complements the DNA damage sensitivity of the rad52 Δ327 mutant strain and enhances the ability of these cells to target Rad51 to a DSB site. These results provide new molecular details into the role of Rad52 in HR.
EXPERIMENTAL PROCEDURES
Yeast Strains—The yeast strains are derivatives of JKM179 (MATα Δho Δhml::ADE1 Δhmr::ADE1 ade1-110 leu2,3-112 lys5 trp1::hisG ura3-52 ade3::GAL10:HO) (36). The rad52-Δ327 strain was generated by inserting a stop codon and KanMX right after codon 327 of the chromosomal RAD52 gene, and the rad52Δ strain was made by replacing the chromosomal RAD52 gene by NatMX.
Construction of Plasmids—His6-RAD52 harboring codons 34–504 of RAD52 was ligated into the NcoI site of the pET11d vector. His6-RAD52-C was ligated into BamHI or EcoRI sites of the pRSETc vector. His6-RAD52-N/C was generated by inserting the C-terminal fragment of RAD52 (codons 327–504) into the BamHI site of His6-RAD52-N::pRSETc. His6-RAD52-M/C was constructed by ligating the BamHI/BglII fragment of GST-RAD52::pGEX (35) into the BamHI site of pRSETc. For the expression of the Rad52 species in yeast cells, the various alleles were inserted into the SmaI site of the 2μ vector pRS426 ADH (37) to place them under the control of the ADH promoter. The nuclear localization signal GGPKKKRKVG, derived from the SV40 large T antigen, was inserted at the end of the opening reading frame of the RAD52-M/C and RAD52-C genes.
Purification of Rad52 Species—The His6-tagged Rad52 species were expressed in E. coli strain BL21 (DE3). Overnight cultures grown in Luria broth at 37 °C were diluted 1000-fold with fresh medium and incubated at 37 °C until the A600 reached 1.2. At that time, isopropyl-d-thiogalactopyranoside was added to 0.1 mm, and the culture was incubated at 16 °C for 24 h. The cells were harvested by centrifugation and stored at –80 °C. All of the protein purification steps were carried out at 4 °C. Lysate was prepared from 20 g of E. coli cell paste using a French press in 100 ml of buffer G (50 mm Tris-HCl, pH 7.4, 0.5 mm EDTA, 0.01% Igepal, 1 mm 2-mercaptoethanol, 10% sucrose, and 100 mm KCl that also contained aprotinin, chymostatin, leupeptin, and pepstatin A at 5 μg/ml each). The crude lysate was clarified by centrifugation (100,000 × g, 90 min).
For His6-tagged Rad52-N/C—The clarified lysate was applied onto a SP Sepharose column (20 ml), which was developed with a 160-ml gradient from 100 to 400 mm KCl in K buffer (20 mm K2HPO4, pH7.4,1mm 2-mercaptoethanol, 0.01% Igepal, and 10% glycerol). The fractions containing the His6-tagged protein (12 ml total; 220 mm KCl) were mixed with 1 ml of nickel-nitrilotriacetic acid-agarose (Qiagen) for 3 h at 4°C. The beads were washed three times with 20 ml of buffer K containing 400 mm KCl, and the bound His6-Rad52-N/C was eluted with 5 ml of 10, 50, 100, and 200 mm imidazole in buffer K, respectively. The 100 and 200 mm imidazole eluates were combined, diluted with 10 ml 10% glycerol, and applied onto a 1-ml Macro-HAP (Bio-Rad) column, which was eluted with a 20-ml gradient from 0 to 300 mm KH2PO4 in buffer K. The Macro-HAP fractions (3 ml total; 120 mm KH2PO4) that contained the peak of Rad52-N/C were dialyzed, concentrated to 10 mg/ml, and stored at –80° C. The overall yield of highly purified Rad52-N/C was ∼2 mg.
For His6-tagged Rad52-C and Rad52-M/C—The clarified lysate was fractionated in a SP Sepharose column (20 ml) as above. The fractions containing the His6-tagged proteins (12 ml total; 250 mm KCl) were subjected to affinity purification in nickel-nitrilotriacetic acid-agarose and chromatographic fractionation in Macro-HAP as above. The Macro-HAP fractions (3 ml total; 140 mm KH2PO4) that contained the peak of the Rad52 species were dialyzed, concentrated to 10 mg/ml, and stored as above. We could obtain ∼5 mg of Rad52-C and ∼2.5 mg of Rad52-M/C.
Purification of Other Proteins—The procedures for the expression and purification of the various GST-Rad52 species have been described elsewhere (35). Rad51 and RPA were expressed and purified as described previously (9).
DNA Substrates—The φX 174 viral (+) strand and replicative form I DNA were purchased from New England Biolabs and Invitrogen, respectively. The sequences of the oligonucleotides (oligonucleotides 1 and 2) used in ssDNA annealing and DNA binding experiments and their 32P labeling have been described (35).
DNA Binding Assay—Varying amounts of the Rad52 species were incubated with 32P-labeled Oligo-1 (1.36 μm nucleotides) or dsDNA (1.36 μm base pairs; obtained by hybridizing oligo-nucleotide 1 to oligonucleotide 2) at 37 °C in 10 μl of buffer D (40 mm Tris-HCl, pH 7.8, 50 mm KCl, 1 mm dithiothreitol, and 100 μg/ml bovine serum albumin) for 10 min. After the addition of gel loading buffer (50% glycerol, 20 mm Tris-HCl, pH 7.4, 0.5 mm EDTA, 0.05% orange G), the reaction mixtures were resolved in 12% native polyacrylamide gels in TAE buffer (40 mm Tris-HCl, pH 7.4, 0.5 mm EDTA) at 4 °C. The gels were dried, and the DNA species were quantified using the Quantity One software in the Personal Molecular Imager FX (Bio-Rad). To show that the DNA substrate remained intact, we treated a reaction mixture containing the highest concentration of the Rad52 species with 0.5% SDS and 0.5 mg/ml proteinase K at 37 °C for 5 min before the analysis.
DNA Strand Exchange Assay—The DNA strand exchange reaction was performed according to our published procedure (26). Rad51 (10 μm) was incubated with ssDNA (30 μm nucleotides) in 10 μl of buffer R (35 mm 3-morpholinopropanesulfonic acid potassium salt, pH 7.2, 1 mm dithiothreitol, 50 mm KCl, 2.5 mm ATP, and 3 mm MgCl2) for 5 min. After the addition of RPA (1.2 μm) in 0.5 μl, the reaction mixture was incubated for another 5 min, before the incorporation of dsDNA (25 μm nucleotides added in 1 μl) and 1 μl of 50 mm spermidine hydrochloride (final concentration, 4 mm). At the indicated times, a 4.5-μl portion of the reaction mixture was withdrawn, processed, and analyzed by agarose gel electrophoresis with ethidium bromide staining (9). Gel images were recorded in a NucleoTech gel documentation system and analyzed with the GelExpert software. To examine the Rad52 recombination mediator function, reaction mixtures (12.5 μl final volume) containing Rad51 (10 μm), RPA (2 μm), and the indicated amounts of the various Rad52 species were incubated on ice for 10 min in 9.5 μl of buffer R, followed by the addition of ssDNA in 1 μl and a 10-min incubation. After the incorporation of linear duplex and spermidine, the completed reactions were incubated and analyzed as before. For the time course experiments, the reactions were scaled up four times to 50 μl but were otherwise assembled in the same fashion.
GST Pulldown Assay—RPA or E. coli SSB (2 μm) was incubated with or without either φX174 ssDNA or dsDNA (30 μm nucleotides or base pairs) in 29 μl of buffer T (20 mm Tris-HCl, pH 7.5, 150 mm KCl, 1 mm dithiothreitol, 0.5 mm EDTA, and 0.01% Igepal) at 37 °C for 10 min before GST-Rad52-M (6 μm) was added in 1 μl. Following a 30-min incubation at 4 °C, the reactions were mixed with 10 μl of glutathione-Sepharose-4B beads at 4 °C for 30 min. After washing the beads twice with 150 μl of buffer T, the bound proteins were eluted with 30 μl of 3% SDS. The supernatants and SDS eluates, 10 μl each, were subjected to SDS-PAGE analysis.
Magnetic Bead-based Pulldown Assay—Purified RPA (2 μm) was incubated with 15 μl of magnetic beads containing φX174 ssDNA (30 μm nucleotides) immobilized on the beads through an annealed biotinylated oligonucleotide (38) in 43.5 μl of buffer T at 37 °C for 10 min. The magnetic beads in one-third of the slurry were captured with a magnetic particle separator and treated with 15μl 1% SDS to elute the bound RPA, and 10μl of the eluate (beads) was analyzed by SDS-PAGE. GST-Rad52-M (6 μm) was added to two-thirds of the slurry in 1μl, followed by mixing for 30 min at 4 °C. The beads were captured with the magnetic particle separator and washed with 30μl of the same buffer, and the bound proteins were eluted with 30 μl 1% SDS. The supernatant and SDS eluate, 10 μl each, were analyzed by SDS-PAGE.
Chromatin Immunoprecipitation—The yeast strains harbor a plasmid of the HO endonuclease gene under the control of the GAL10 promoter, which permits the galactose induction of HO expression. Chromatin immunoprecipitation (ChIP) was carried out essentially as described (21). The cells were grown in YEP supplemented with 3% glycerol for 6 h until the mid-log phase, and 2% galactose was added to induce HO expression. At the designated times, the cells were harvested from 45 ml of the cultures, cross-linked with 1% formaldehyde for 20 min, and then quenched with 125 mm glycine for 5 min. After lysing the cells with glass beads and sonication to shear the chromatin, the cell lysates were incubated with anti-Rad51 antibodies (our own stock) or anti-Rad52 antibodies raised against the C terminus of the protein (Santa Cruz Biotechnology) and Dynabeads Protein G (Invitrogen). After a series of washes, the immunoprecipitates were incubated at 65 °C to reverse protein-DNA cross-link. Radioactive semi-quantitative PCR was performed to amplify the precipitated DNA. The primers used for amplifying PHO5 and MAT Z were as described (21). The PCR products were resolved on 6% native polyacrylamide gels and quantified in a phosphorimaging device (Bio-Rad) using the Quantity One software (Bio-Rad).
Immunoblot Analysis—Cells grown in SC-Ura media at 30 °C were lysed with glass beads in buffer (50 mm HEPES-KOH, pH7.5, 500 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, and the following protease inhibitors: aprotinin, chymostatin, leupeptin, and pepstatin A at 5 μg/ml each) and cleared by centrifugation at 15,000 × g and 4 °C for 15 min. Clarified lysates (10 μg of total protein) were subjected to immunoblot analysis with affinity-purified anti-Rad52 antibodies raised against amino acid residues 168–456 of Rad52 (15) and anti-rabbit secondary antibodies conjugated with horseradish peroxidase. The blots were developed with the Super-Signal Substrate (Pierce).
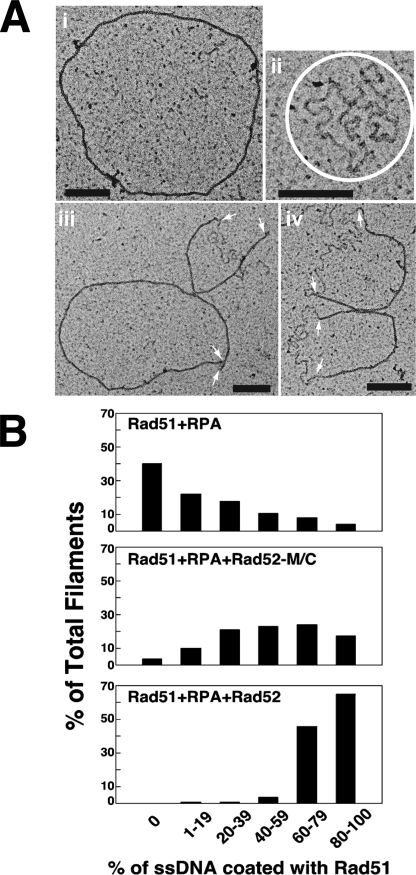
Electron Microscopy—Reactions containing Rad51 (10 μm), RPA (2 μm), and ssDNA (30 μm nucleotides) were assembled in the buffer used for DNA strand exchange and incubated for 15 min either in the absence or presence of Rad52 (1.2 μm) or Rad52-M/C (3 μm). After dilution, the samples were applied onto freshly glow discharged carbon-coated Mesh 400 copper grids, stained with uranyl acetate, and examined with a Technai-12 electron microscope (39).
MMS Survival Assay—rad52Δ and rad52-Δ327 cells harboring plasmids that express Rad52, Rad52-C, and Rad52-M/C tagged with the SV40 nuclear localization signal were grown to the stationary phase in SC-Ura media at 30 °C and diluted 20-fold with fresh medium. When the cultures reached a density of ∼1 × 108 cells/ml, they were serially 10-fold diluted and then spotted on SC-Ura plates containing 0.005% MMS. The plates were incubated at 30 °C for 4 days and then photographed.
RESULTS
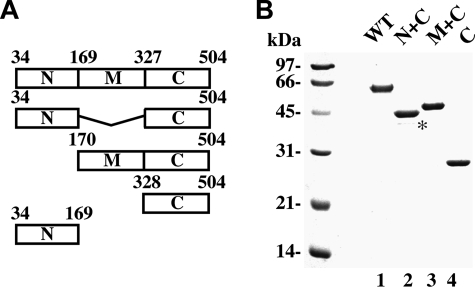
Purification of Rad52 Domains—We previously expressed Rad52 and also the N-terminal third (designated as N), middle third (designated as M), and C-terminal third (designated as C) of this protein as GST fusions (35). We have since expressed and purified His6-tagged Rad52, Rad52-C, and fusions of the M and C regions (Rad52-M/C) and the N and C regions (Rad52-N/C) (Fig. 1). The Rad52 species that contain the N-terminal region of this protein were made from the third ATG codon (corresponding to residue 34) in the RAD52 protein coding frame, because this represents the first start codon that is used in Rad52 protein synthesis in yeast cells (40). At least two independent preparations of each of the Rad52 species were used in our study, and they all gave the same results.
FIGURE 1.
Purification of Rad52 species. A, schematic representation of the Rad52 species used. B, purified His6-tagged proteins: Rad52, Rad52-N/C, Rad52-M/C, and Rad52-C (1 μg each) were run on a 10% denaturing SDS-PAGE and stained with Coomassie Blue. The asterisk designates a proteolytic fragment of Rad52-N/C.
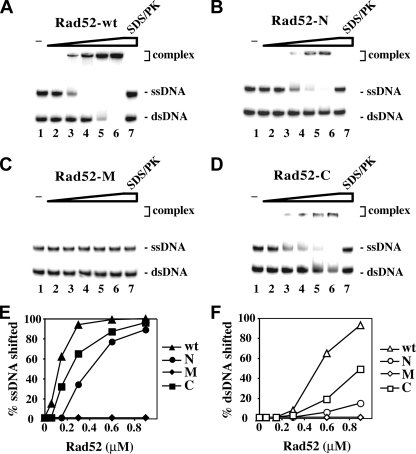
Rad52-C Harbors DNA Binding Activity—Rad52 binds ssDNA and shows a weaker affinity for dsDNA (Ref. 23 and Fig. 2A), and the DNA binding activity is thought to reside within the N terminus (23). This region of Rad52 forms a ring-shaped oligomer that possesses an outer groove (24, 27, 29, 30) lined with basic and aromatic residues that are involved in binding DNA (29).
FIGURE 2.
The C-terminal region of Rad52 harbors a DNA binding activity. A–D, Radiolabeled ssDNA and dsDNA were incubated without protein (lane 1) and with 60, 135, 270, and 400 nm of Rad52 (A), Rad52-N (B), Rad52-M (C), and Rad52-C (D). In lane 7, a reaction mixture containing the highest amount of each of the Rad52 species was treated with SDS and proteinase K before analysis. The results from A–D were plotted in E and F. Note that the ssDNA migrated more slowly than the dsDNA. wt, wild type.
Even though GST-Rad52-N bound 32P-labeled ssDNA and dsDNA, it was not nearly as proficient as GST-tagged Rad52 (Fig. 2B). This prompted us to investigate whether other parts of Rad52 could also be involved in DNA binding. GST-Rad52-M is devoid of any ability to bind DNA (Fig. 2C), but we were surprised to find a DNA binding activity in GST-Rad52-C (Fig. 2D). In fact, Rad52-C appears to be more adept at binding both ssDNA and dsDNA than Rad52-N (Fig. 2, E and F). The same conclusions were reached when the oligonucleotide substrates were replaced with φX174 ssDNA and dsDNA molecules (data not shown). Taken together, the results confirmed that the N terminus of Rad52 harbors a DNA binding function and, rather unexpectedly, revealed a DNA binding activity in the C-terminal third of the protein.
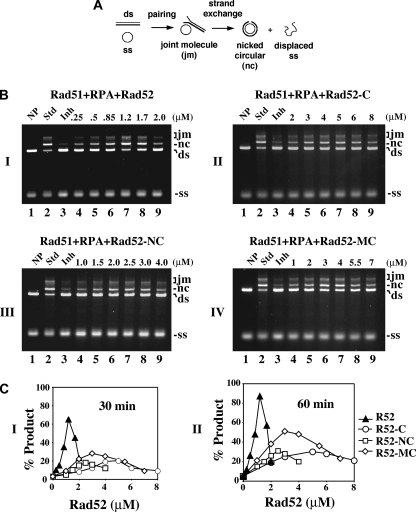
Rad52-C Has Recombination Mediator Activity—We used the His6-tagged form of the Rad52 variants and a well established DNA strand exchange assay to test for recombination mediator activity (Fig. 3A and Refs. 15–17 and 41). In this assay, efficient DNA strand exchange is seen when the φX174 ssDNA is preincubated with Rad51 before the addition of RPA (Fig. 3B, lanes 2), whereas incubation of the ssDNA with Rad51 and RPA simultaneously leads to a pronounced attenuation of the reaction because of interference of presynaptic filament assembly by RPA (Fig. 3B, lanes 3).
FIGURE 3.
The C terminus of Rad52 possesses recombination mediator activity. A, schematic representation of the homologous DNA pairing and strand exchange reaction. Homologous pairing between the circular ssDNA (ss) and linear duplex DNA (ds) substrates yields a DNA joint molecule (jm), which is converted into a nicked circular duplex molecule (nc) by DNA strand exchange. B, DNA strand exchange reactions conducted to examine the recombination mediator activity of the Rad52 species: Rad52 in panel I, Rad52-C in panel II, Rad52-N/C in panel III, and Rad52-M/C in panel IV. The standard reaction (Std, lane 2) involved preincubating the ssDNA with Rad51 to allow for the formation of the presynaptic filament before RPA was added. Co-incubating the ssDNA with Rad51 and RPA resulted in severe inhibition of the DNA strand exchange reaction (Inh, lane 3). The inclusion of the various Rad52 species during the incubation of ssDNA with Rad51 and RPA led to restoration of DNA strand exchange. NP indicates no protein added. C, the results of the 30- and 60-min time points in B were plotted in panels I and II, respectively.
As expected, the inclusion of Rad52 effectively alleviated the suppressive effect of RPA on DNA strand exchange (Fig. 3B, panel I, lanes 4–9). Under the conditions used (10 μm Rad51, 30 μm ssDNA, and 2 μm RPA), the optimal concentration of Rad52 to see full restoration of DNA strand exchange was 1.2 μm. Reproducibly, Rad52-C could also restore a significant level of DNA strand exchange with RPA co-addition, although, when compared with Rad52, a higher amount (2–8 μm) was needed (Fig. 3C). The maximal extent of restoration that could be achieved with Rad52-C was, however, significantly lower, about one-third of that obtained with Rad52 (Fig. 3C). Full time course experiments demonstrating DNA strand exchange restoration by Rad52 and Rad52-C are presented in supplemental Fig. S1. We also found that neither GST-tagged Rad52-N nor GST-tagged Rad52-M is capable of restoring DNA strand exchange over a wide concentration range of these protein species (data not shown). In contrast, GST-Rad52 and GST-Rad52-C possess mediator activity comparable with that of the His6-tagged version of these Rad52 species (data not shown). As anticipated, neither Rad52 nor any of the Rad52 domains (N, M, and C) exhibited any DNA strand exchange activity with or without RPA (data not shown).
Both Rad52 N and M Contribute to Recombination Mediator Function—Because Rad52 is considerably more effective than Rad52-C in DNA strand exchange restoration (Fig. 3C), we asked whether the N and M portions contribute toward the recombination mediator function. To address this, we fused either Rad52 N or M to the C portion. When expressed in E. coli, the Rad52-N/C and Rad52-M/C species are soluble, and we were able to devise procedures for their purification (Fig. 1). Relative to Rad52-C, significant restoration of DNA strand exchange was seen with a lower concentration of Rad52-N/C, but the maximal level of restoration was not higher than what could be achieved with Rad52-C (Fig. 3, B and C). Interestingly, Rad52-M/C was more effective than Rad52-C in restoring DNA strand exchange (Fig. 3B). Although restoration of DNA strand exchange by Rad52-M/C occurred over the same concentration range as Rad52-C, the final level was significantly higher. Full time course experiments involving Rad52-N/C and Rad52-M/C are presented in supplemental Fig. S1. As expected, neither Rad52-N/C nor Rad52-M/C has any DNA strand exchange activity with or without RPA (data not shown).
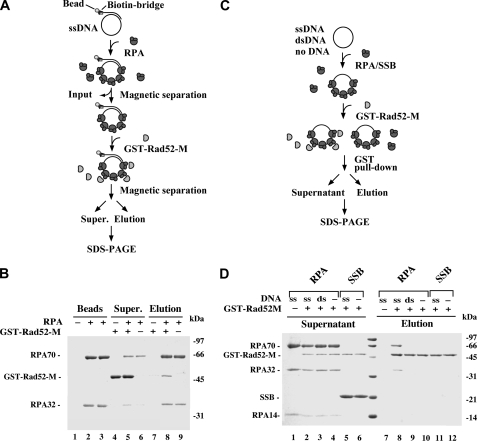
Rad52-M Interacts with DNA-bound RPA—Because the M portion of Rad52 has no DNA binding activity (Fig. 2) and does not interact with Rad51 (32, 35), we wondered whether its enhancement of the Rad52-C recombination mediator activity (Fig. 3B and supplemental Fig. S1) could be due to an ability to associate with RPA. However, only a very weak interaction between GST-Rad52-M and RPA was observed in an affinity pulldown assay (Fig. 4D, compare lanes 4 and 10). The result, although negative, is entirely consistent with the observation that Rad52 does not associate strongly with RPA in solution (data not shown). We next considered the possibility that interaction between Rad52-M and RPA occurs when RPA is DNA-bound. Two separate affinity pulldown assays were employed to address this possibility. In the first assay, RPA was incubated with biotinylated ssDNA immobilized on streptavidin magnetic beads to produce a RPA-ssDNA affinity matrix, which was then used to test for a possible interaction with GST-Rad52-M (Fig. 4A). A significant amount of Rad52-M was retained on the RPA-ssDNA beads (Fig. 4B). As expected, Rad52-M did not associate with DNA magnetic beads without RPA (Fig. 4B, lane 7). In the second pulldown assay, Rad52-M was first incubated with RPA or E. coli SSB without DNA or with ssDNA or dsDNA, and then the reactions were mixed with glutathione-Sepharose beads to capture GST-Rad52-M and associated RPA or SSB (Fig. 4C). In agreement with the results from the first pulldown assay (Fig. 4B), a stoichiometric amount of RPA was found associated with Rad52-M upon inclusion of ssDNA, whereas no retention of RPA occurred without DNA (Fig. 4D, compare lanes 8 and 10). We also verified that GST-Rad52-M does not associate with E. coli SSB (Fig. 4D) and that dsDNA is ineffective in mediating complex formation of RPA with Rad52-M (Fig. 4D). Taken together, the results revealed an ability of Rad52-M to associate avidly with DNA-bound RPA, and it does so in a species-specific manner. As anticipated, Rad52-M does not interact with free or ssDNA-bound Rad51 (data not shown).
FIGURE 4.
Rad52-M interacts with DNA-bound RPA. A, in this affinity pulldown assay, RPA is incubated with streptavidin magnetic beads harboring φX174 ssDNA, and the resulting RPA-containing beads are used to bind Rad52-M. B, three reactions were set up. In the first reaction, ssDNA magnetic beads that did not harbor RPA (lane 1) were mixed with GST-Rad52-M (lanes 4 and 7); in the second reaction, ssDNA magnetic beads harboring RPA (lane 2) were mixed with GST-Rad52-M (lanes 5 and 8); and in the third reaction, ssDNA magnetic beads harboring RPA (lane 3) were mixed in buffer without GST-Rad52-M (lanes 6 and 9). After mixing, the beads were captured with a magnet, and associated proteins were eluted with SDS. The supernatants (Super.) that contained unbound proteins and the SDS eluates (Elution) were analyzed by SDS-PAGE with Coomassie Blue staining. The RPA content of the magnetic beads (Beads) used in the three reactions is also shown. C, in this affinity pulldown assay, GST-Rad52-M is incubated with RPA or E. coli SSB either in the absence of DNA or in the presence of ssDNA or dsDNA before being mixed with glutathione-Sepharose to capture GST-Rad52-M and any associated RPA. D, the various supernatants containing unbound proteins and the SDS eluates harboring the captured proteins were analyzed by SDS-PAGE with Coomassie Blue staining. GST-Rad52-M was omitted from the control reaction in lane 1.
Examination of Recombination Mediator Activity by Electron Microscopy—We incubated φX174 circular ssDNA with Rad51 and RPA in the presence or absence of His6-tagged Rad52 or Rad52-M/C. These reactions were then examined by electron microscopy. We analyzed over 100 randomly picked nucleoprotein complexes in each case and classified these complexes according to the extent of the Rad51 presynaptic filament. In the reaction that had Rad51 and RPA only, ∼40% of the nucleoprotein complexes contained mostly or entirely RPA, and only very few (∼10% of the total) of the DNA molecules had 60% or higher coverage by Rad51 (Fig. 5 and supplemental Fig. S2). As expected, the addition of Rad52 effectively restored presynaptic filament formation, with almost all of the nucleoprotein complexes possessing 60% or higher Rad51 coverage (Fig. 5 and supplemental Fig. S2). The inclusion of Rad52-MC had a strong impact on Rad51 presynaptic filament assembly, with a significant fraction (>35%) of the nucleoprotein complexes having ≥60% Rad51 coverage. Overall, the results from the electron microscopy analysis provided independent verification of Rad52-M/C possessing a recombination mediator activity. However, like for DNA strand exchange, Rad52-M/C is not as effective as Rad52 in the promotion of Rad51 presynaptic filament assembly (Fig. 5B).
FIGURE 5.
Electron microscopy analysis of recombination mediator action. A, micrographs showing an example of full Rad51 filament (panel i), a RPA-ssDNA nucleoprotein complex (panel ii), and ssDNA molecules covered partly by Rad51 and partly by RPA (panels iii and iv). The RPA-ssDNA complexes in panel ii are circled, and the arrows in panels iii and iv mark the junctions of Rad51-coated DNA and RPA-coated DNA. The black scale bars in the panels denote 200 nm. B, quantification of Rad51 presynaptic filament formation in the three reactions: Rad51+RPA, Rad51+RPA+ Rad52-M/C, and Rad51+RPA+Rad52.
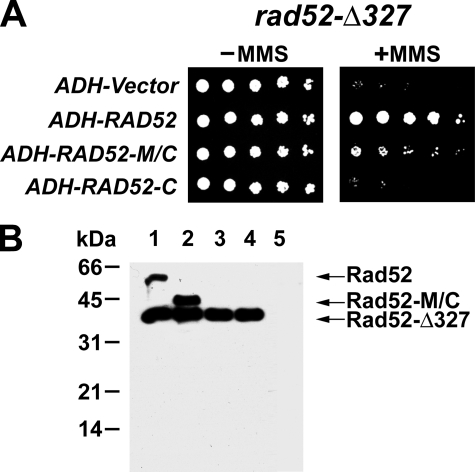
Rad52-M/C Is Biologically Efficacious in the rad52Δ327 Mutant—The above results indicated that the C portion of Rad52 has a recombination mediator activity that is augmented by the RPA-binding domain located within the M portion of the protein. To ask whether Rad52-M/C has biological activity in vivo, we examined the ability of plasmids containing either RAD52 or RAD52-M/C inserted downstream of the ADH promoter for their ability to complement the sensitivity of rad52 mutant strains to the genotoxin MMS. To ensure that Rad52-M/C is transported into the nucleus, its C terminus was joined to the nuclear localization signal sequence derived from the SV40 large T antigen. Protein expression was verified by immunoblot analysis of cell extracts using anti-Rad52 antibodies (Fig. 6B and supplemental Fig. S3). In addition to the rad52Δ mutant, we included the rad52-Δ327 mutant, which does not express the C-terminal third of Rad52, in the analysis, because it is known that the N-terminal portion of the Rad52 contributes to HR in a Rad51-independent fashion (42, 43). RAD52-M/C partially complemented the MMS sensitivity of the rad52-Δ327 mutant (Fig. 6A), but little or no complementation of the MMS sensitivity of the rad52Δ mutant was seen (data not shown). In addition, we did not observe any complementation of the rad52-Δ327 or rad52Δ strain transformed with a similar plasmid expressing RAD52-C (Fig. 6A and data not shown). This result is likely due to a lack of Rad52-C protein expression as revealed by immunoblot analysis (Fig. 6B). Finally, the presence of a His6 tag at either the N or the C terminus of Rad52 has no effect on protein expression in yeast cells or on the complementation of the rad52-Δ327 mutant (supplemental Fig. S3). Likewise, attaching a His6 tag to the N terminus of Rad52-M/C does not affect its expression or biological efficacy (supplemental Fig. S3).
FIGURE 6.
Complementation of the rad52-Δ327 mutant by Rad52-M/C. A, rad52-Δ327 strains harboring the empty ADH vector, ADH-RAD52, ADH-RAD52-M/C, or ADH-RAD52-C were serially diluted and spotted onto SC-Ura medium with or without 0. 005% MMS. B, extracts of rad52-Δ327 cells harboring ADH plasmids expressing Rad52 (lane 1), Rad52-M/C (lane 2), Rad52-C (lane 3), or empty ADH vector (lane 4) or rad52Δ cells harboring empty ADH vector (lane 5) were subjected to immunoblot analysis with anti-Rad52 antibodies.
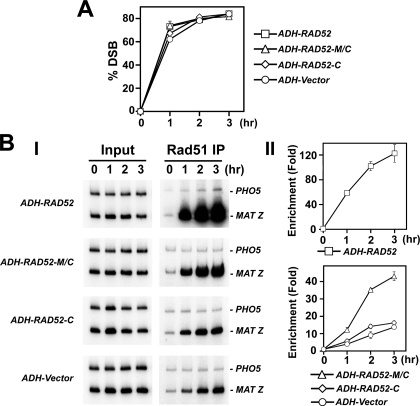
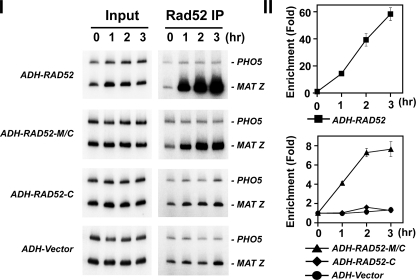
Rad52-M/C Targets Rad51 to DNA Double-strand Breaks in Cells—The recruitment of Rad51 to DSBs in cells is strongly dependent on Rad52 (18–21). We employed ChIP to examine the targeting of Rad51 to a site-specific HO-induced break in rad52Δ and rad52-Δ327 mutant cells that expressed either Rad52, Rad52-C, or Rad52-M/C from the plasmids described in the previous section. We used yeast strains where the DSB is engaged by the HR machinery but cannot be repaired because of the absence of the homologous donor sequence (20).
The expression of the HO-endonuclease was induced with galactose, and DSB formation was monitored by PCR (Fig. 7A). At various times after DSB induction, cell extracts were prepared and subjected to immunoprecipitation with anti-Rad51 or anti-Rad52 antibodies. The target sequence (MAT-Z) and an internal control sequence (PHO5) were amplified from the precipitated DNA by PCR. In agreement with published work (20, 21), we observed rapid recruitment of Rad51 upon DSB formation in wild-type cells, reaching a maximum of 40-fold by 3 h (data not shown). In contrast, the amount of Rad51 at the DSB increased only about 2–3-fold in rad52Δ cells (data not shown). In rad52-Δ327 cells, Rad51 recruitment was diminished, but not ablated, because there was a 12-fold enrichment by 3 h (Fig. 7B). Importantly, expression of Rad52-M/C and Rad52 in rad52-Δ327 cells led to a 40-fold and >100-fold enrichment of Rad51 at the DSB site by 3 h, respectively (Fig. 7B). Moreover, Rad51 recruitment to the DSB was enhanced more then 10-fold in rad52Δ cells expressing Rad52-M/C (data not shown). We did not find any Rad51 or Rad52-C enrichment at the HO-induced break in rad52-Δ327 cells that harbor ADH-RAD52-C (Figs. 7B and 8). This was very likely due to a lack of Rad51-C protein expression (Fig. 6B). Lastly, as expected, both full-length Rad52 and Rad52-M/C are enriched at the DSB site (Fig. 8). Taken together, the ChIP data are also indicative of the presence of a recombination mediator activity in Rad52-M/C. We note that the recruitment of Rad52-M/C to the HO breaks site is less robust than that of Rad52 (Fig. 8), which could be a contributing factor as to why this Rad52 species is not as biologically efficacious as the full-length protein.
FIGURE 7.
Rad52-M/C promotes DSB recruitment of Rad51. A, kinetics of DSB induction. Donorless rad52-Δ327 cells harboring GAL10-HO and also the empty ADH vector or a ADH plasmid expressing Rad52, Rad52-M/C, or Rad52-C were grown in the presence of galactose to induce HO-expression and a DSB at MAT. The induction of the HO break was quantified by PCR (20, 21). B, recruitment of Rad51 to the HO-induced break in rad52-Δ327 cells harboring the ADH vector or ADH-RAD52, ADH-RAD52-M/C, or RAD52-C. These cells were subjected to ChIP with anti-Rad51 antibodies. The MAT Z target sequence and control PHO5 sequence were amplified using the appropriate primer sets (21). Panels I and II show the PCR products and the quantification of the results, respectively. Enrichment was determined by dividing the PCR signal of MAT Z by the PHO5 PCR signal and normalizing to time zero value.
FIGURE 8.
DSB recruitment of Rad52 species in rad52-Δ327 cells. Recruitment of Rad52 to the HO breaks in rad52-Δ327 cells harboring the ADH vector or ADH-RAD52, ADH-RAD52-M/C, or ADH-RAD52-C. These cells were subjected to ChIP with anti-Rad52 antibodies. The MAT Z target sequence and control PHO5 sequence were amplified using the appropriate primer sets (21). Panels I and II show the PCR products and the quantification of the results, respectively. Enrichment was determined by dividing the PCR signal of MAT Z by the PHO5 PCR signal and normalizing to time zero value.
DISCUSSION
New Insights Concerning the Role of Rad52 in HR—Rad52 plays a central role in various HR processes, including Rad51-dependent and Rad51-independent reactions (6, 7). Biochemical and cell-based analyses have unveiled a recombination mediator activity in Rad52 (15–17, 19–21). These studies have served as the model for understanding the function of this class of HR factors, including the human tumor suppressor BRCA2. Several properties of Rad52 are likely germane for its recombination mediator function, including the abilities to bind DNA, assemble into an oligomeric structure, and associate with Rad51 and RPA. Previous work has established that complex formation with Rad51 is critical for the recombination mediator function of Rad52 (16, 35). Here we have provided evidence that in addition to binding Rad51 (32, 35), the C-terminal third of Rad52 possesses a DNA binding activity and can also nucleate Rad51 onto a RPA-coated DNA template. Moreover, we have shown that the middle portion of Rad52 mediates an interaction with ssDNA-bound RPA and enhances the recombination mediator activity of the C-terminal portion. Consistent with these results, expression of Rad52-M/C partially complements the DNA damage sensitivity of rad52-Δ327 mutant cells.
Genetic experiments by the Livingston group (42, 44) have demonstrated intragenic complementation involving expression of Rad52 fragments in the same cell. This is reminiscent of the complementation of the DNA repair and Rad51 recruitment deficiency by Rad52-M/C in the rad52-Δ327 mutant seen here. Our biochemical analyses have further provided molecular information to explain this intragenic complementation phenomenon.
Biological Role of the Rad52 N Terminus—It is intriguing that the N terminus of Rad52 is in fact not absolutely required for the recombination mediator function in vitro. This finding clearly indicates that functional interactions with Rad51 and RPA can occur without the DNA binding activity and protein oligomerization domain that reside within this region. However, cells that lack the Rad51 interaction domain (as in rad52-Δ327) are still partially capable of Rad51 recruitment to a DSB. Accordingly, these cells are not as sensitive to DNA-damaging treatment as rad52Δ cells (this study and Ref. 32). This observation points to a key function of Rad52 in HR that does not entail a direct interaction with Rad51 but yet has an important impact upon the ability of cells to assemble or maintain the Rad51 presynaptic filament. Furthermore, the inability of the Rad52-M/C species to complement the rad52Δ mutation is also indicative of a vital HR function of the Rad52 N-terminal region.
The Rad55-Rad57 heterodimer also possesses a recombination mediator activity (18, 41, 45), and Rad54 is capable of stabilizing the Rad51 presynaptic filament (21, 46). In addition, the Rad59 protein binds DNA (47) and associates with Rad51, Rad52, and RPA in a complex (48). It remains to be determined whether Rad52, through its N-terminal and middle portions, functionally synergizes with these other HR factors to promote the assembly or stabilization of the Rad51 presynaptic filament. Likewise, the Rdh54 protein, which is structurally and functionally related to Rad54, might act in conjunction with Rad52 in reactions that either directly or indirectly (e.g. in chromatin remodeling) influence Rad51 presynaptic filament assembly or maintenance. It will be interesting to examine whether the Rad51 recruitment ability of the rad52-Δ327 mutant is dependent on any of the aforementioned HR factors.
An amount of Rad52-N/C less than that of Rad52-C can achieve the same level of DNA strand exchange restoration (Fig. 3C), indicating that the N terminus makes a significant contribution toward the recombination mediator efficacy of Rad52. Because Rad52-N does not enhance the recombination mediator activity of Rad52-M/C (supplemental Fig. S4), these two Rad52 parts do not functionally cooperate in trans. Rad52 protein oligomerization that is mediated by the N terminus could confer the ability to bind ssDNA in a cooperative fashion. However, future studies will be needed to determine whether the protein oligomerization or the DNA binding activity of the N terminus is critical for the enhancement of recombination mediator activity. The available collection of N-terminal rad52 mutants predicted to be compromised for DNA binding will be valuable in this regard (49).
Role of RPA Binding in Rad52 Function—S. cerevisiae Rad52 was previously shown to interact with RPA in the yeast two-hybrid system and in an affinity pulldown assay (24, 33). Moreover, human Rad52 and RPA readily form a complex in the absence of DNA (50). Taken together, the available evidence points to RPA interaction as an evolutionarily conserved property of Rad52. We have presented data showing the ability of the Rad52 middle portion to interact with RPA. This interaction is strongly dependent on ssDNA and is also species-specific. In addition, we have shown that the middle portion of Rad52 is able to enhance the mediator activity of Rad52-C.
Rad52 appears to make contacts with all three subunits of RPA (24, 33). Because the C terminus of Rad52 alone exhibits recombination mediator activity and can anneal RPA-coated complementary DNA strands, it too may harbor an ability to bind RPA. Future studies involving protein interaction domain mapping and mutant isolation will ascertain whether different surfaces of Rad52 contact the individual subunits of RPA and will delineate the functional significance of these contacts.
Implications for the Recombination Mediator Function of BRCA2—Cells harboring mutations in the tumor suppressor BRCA2 are profoundly deficient in the homology-directed repair of damaged chromosomes (51, 52). Several lines of evidence are consistent with the premise that BRCA2, like Rad52 in S. cerevisiae, regulates Rad51 activity by providing a recombination mediator function: 1) it physically interacts with Rad51 through several copies of the BRC repeat (53, 54) and also through its C terminus (55), 2) it binds ssDNA (56), 3) it interacts with RPA (57), and 4) BRCA2 mutant cells are deficient in assembling DNA damage-induced Rad51 foci (52). Direct demonstration of the BRCA2 recombination mediator activity has been achieved in recent studies using a polypeptide derived from this protein (39) and also the Ustilago maydis BRCA2 orthologue Brh2 (58). The molecular dissection of Rad52 being conducted in our laboratory and by others should continue to provide useful information and complement parallel studies on related HR factors such as BRCA2.
Supplementary Material
This work was supported by National Institutes of Health Grants P01CA92584, RO1ES07061, and RO1GM57814; Wellcome Trust Grant GR076476; Ministry of Education Youth and Sport of the Czech Republic Grants MSM 0021622413, ME 888, and MSMT LC06030; and funds from the Danish Research Council for Technology and Production Sciences and the Hartmann Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: HR, homologous recombination; ssDNA, single-strand DNA; dsDNA, double-strand DNA; MMS, methylmethane sulfonate; DSB, double-strand break; GST, glutathione S-transferase; ChIP, chromatin immunoprecipitation; RPA, replication protein A.
References
- 1.Cox, M. M., Goodman, M. F., Kreuzer, K. N., Sherratt, D. J., Sandler, S. J., and Marians, K. J. (2000) Nature 404 37–41 [DOI] [PubMed] [Google Scholar]
- 2.Symington, L. S. (2002) Microbiol. Mol. Biol. Rev. 66 630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heller, R. C., and Marians, K. J. (2006) Nat. Rev. Mol. Cell Biol. 7 932–943 [DOI] [PubMed] [Google Scholar]
- 4.Jasin, M. (2002) Oncogene 21 8981–8993 [DOI] [PubMed] [Google Scholar]
- 5.D'Andrea, A. D. (2003) Genes Dev. 17 1933–1936 [DOI] [PubMed] [Google Scholar]
- 6.Paques, F., and Haber, J. E. (1999) Microbiol. Mol. Biol. Rev. 63 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krogh, B. O., and Symington, L. S. (2004) Annu. Rev. Genet. 38 233–271 [DOI] [PubMed] [Google Scholar]
- 8.Sung, P., and Klein, H. (2006) Nat. Rev. Mol. Cell Biol. 7 739–750 [DOI] [PubMed] [Google Scholar]
- 9.Sung, P. (1994) Science 265 1241–1243 [DOI] [PubMed] [Google Scholar]
- 10.Bianco, P. R., Tracy, R. B., and Kowalczykowski, S. C. (1998) Front. Biosci. 3 D570–D603 [DOI] [PubMed] [Google Scholar]
- 11.Sung, P., Krejci, L., Van Komen, S., and Sehorn, M. G. (2003) J. Biol. Chem. 278 42729–42732 [DOI] [PubMed] [Google Scholar]
- 12.Sugiyama, T., Zaitseva, E. M., and Kowalczykowski, S. C. (1997) J. Biol. Chem. 272 7940–7945 [DOI] [PubMed] [Google Scholar]
- 13.Van Komen, S., Petukhova, G., Sigurdsson, S., and Sung, P. (2002) J. Biol. Chem. 277 43578–43587 [DOI] [PubMed] [Google Scholar]
- 14.Eggler, A. L., Inman, R. B., and Cox, M. M. (2002) J. Biol. Chem. 277 39280–39288 [DOI] [PubMed] [Google Scholar]
- 15.Sung, P. (1997) J. Biol. Chem. 272 28194–28197 [DOI] [PubMed] [Google Scholar]
- 16.Shinohara, A., and Ogawa, T. (1998) Nature 391 404–407 [DOI] [PubMed] [Google Scholar]
- 17.New, J. H., Sugiyama, T., Zaitseva, E., and Kowalczykowski, S. C. (1998) Nature 391 407–410 [DOI] [PubMed] [Google Scholar]
- 18.Gasior, S. L., Wong, A. K., Kora, Y., Shinohara, A., and Bishop, D. K. (1998) Genes Dev. 12 2208–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lisby, M., Barlow, J. H., Burgess, R. C., and Rothstein, R. (2004) Cell 118 699–713 [DOI] [PubMed] [Google Scholar]
- 20.Sugawara, N., Wang, X., and Haber, J. E. (2003) Mol Cell 12 209–219 [DOI] [PubMed] [Google Scholar]
- 21.Wolner, B., van Komen, S., Sung, P., and Peterson, C. L. (2003) Mol. Cell 12 221–232 [DOI] [PubMed] [Google Scholar]
- 22.Wang, X., and Haber, J. E. (2004) PLoS Biol. 2 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortensen, U. H., Bendixen, C., Sunjevaric, I., and Rothstein, R. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 10729–10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinohara, A., Shinohara, M., Ohta, T., Matsuda, S., and Ogawa, T. (1998) Genes Cells 3 145–156 [DOI] [PubMed] [Google Scholar]
- 25.Sugiyama, T., Kantake, N., Wu, Y., and Kowalczykowski, S. C. (2006) EMBO J. 25 5539–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song, B., and Sung, P. (2000) J. Biol. Chem. 275 15895–15904 [DOI] [PubMed] [Google Scholar]
- 27.Stasiak, A. Z., Larquet, E., Stasiak, A., Muller, S., Engel, A., Van Dyck, E., West, S. C., and Egelman, E. H. (2000) Curr. Biol. 10 337–340 [DOI] [PubMed] [Google Scholar]
- 28.Cortes-Ledesma, F., Malagon, F., and Aguilera, A. (2004) Genetics 168 553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kagawa, W., Kurumizaka, H., Ishitani, R., Fukai, S., Nureki, O., Shibata, T., and Yokoyama, S. (2002) Mol. Cell 10 359–371 [DOI] [PubMed] [Google Scholar]
- 30.Singleton, M. R., Wentzell, L. M., Liu, Y., West, S. C., and Wigley, D. B. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13492–13497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinohara, A., Ogawa, H., and Ogawa, T. (1992) Cell 69 457–470 [DOI] [PubMed] [Google Scholar]
- 32.Milne, G. T., and Weaver, D. T. (1993) Genes Dev. 7 1755–1765 [DOI] [PubMed] [Google Scholar]
- 33.Hays, S. L., Firmenich, A. A., Massey, P., Banerjee, R., and Berg, P. (1998) Mol. Cell. Biol. 18 4400–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sugiyama, T., New, J. H., and Kowalczykowski, S. C. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krejci, L., Song, B., Bussen, W., Rothstein, R., Mortensen, U. H., and Sung, P. (2002) J. Biol. Chem. 277 40132–40141 [DOI] [PubMed] [Google Scholar]
- 36.Lee, S. E., Moore, J. K., Holmes, A., Umezu, K., Kolodner, R. D., and Haber, J. E. (1998) Cell 94 399–409 [DOI] [PubMed] [Google Scholar]
- 37.Mumberg, D., Muller, R., and Funk, M. (1995) Gene (Amst.) 156 119–122 [DOI] [PubMed] [Google Scholar]
- 38.Krejci, L., Van Komen, S., Li, Y., Villemain, J., Reddy, M. S., Klein, H., Ellenberger, T., and Sung, P. (2003) Nature 423 305–309 [DOI] [PubMed] [Google Scholar]
- 39.San Filippo, J., Chi, P., Sehorn, M. G., Etchin, J., Krejci, L., and Sung, P. (2006) J. Biol. Chem. 281 11649–11657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Antunez de Mayolo, A., Lisby, M., Erdeniz, N., Thybo, T., Mortensen, U. H., and Rothstein, R. (2006) Nucleic Acids Res. 34 2587–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sung, P. (1997) Genes Dev. 11 1111–1121 [DOI] [PubMed] [Google Scholar]
- 42.Asleson, E. N., Okagaki, R. J., and Livingston, D. M. (1999) Genetics 153 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsukamoto, M., Yamashita, K., Miyazaki, T., Shinohara, M., and Shinohara, A. (2003) Genetics 165 1703–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boundy-Mills, K. L., and Livingston, D. M. (1993) Genetics 133 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fortin, G. S., and Symington, L. S. (2002) EMBO J. 21 3160–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mazin, A. V., Alexeev, A. A., and Kowalczykowski, S. C. (2003) J. Biol. Chem. 278 14029–14036 [DOI] [PubMed] [Google Scholar]
- 47.Petukhova, G., Stratton, S. A., and Sung, P. (1999) J. Biol. Chem. 274 33839–33842 [DOI] [PubMed] [Google Scholar]
- 48.Davis, A. P., and Symington, L. S. (2003) DNA Repair (Amst.) 2 1127–1134 [DOI] [PubMed] [Google Scholar]
- 49.Lettier, G., Feng, Q., de Mayolo, A. A., Erdeniz, N., Reid, R. J., Lisby, M., Mortensen, U. H., and Rothstein, R. (2006) PLoS Genet. 2 1773–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park, M. S., Ludwig, D. L., Stigger, E., and Lee, S. H. (1996) J. Biol. Chem. 271 18996–19000 [DOI] [PubMed] [Google Scholar]
- 51.Xia, F., Taghian, D. G., DeFrank, J. S., Zeng, Z. C., Willers, H., Iliakis, G., and Powell, S. N. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 8644–8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moynahan, M. E., Cui, T. Y., and Jasin, M. (2001) Cancer Res. 61 4842–4850 [PubMed] [Google Scholar]
- 53.Wong, A. K., Pero, R., Ormonde, P. A., Tavtigian, S. V., and Bartel, P. L. (1997) J. Biol. Chem. 272 31941–31944 [DOI] [PubMed] [Google Scholar]
- 54.Chen, P. L., Chen, C. F., Chen, Y., Xiao, J., Sharp, Z. D., and Lee, W. H. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5287–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharan, S. K., Morimatsu, M., Albrecht, U., Lim, D. S., Regel, E., Dinh, C., Sands, A., Eichele, G., Hasty, P., and Bradley, A. (1997) Nature 386 804–810 [DOI] [PubMed] [Google Scholar]
- 56.Yang, H., Jeffrey, P. D., Miller, J., Kinnucan, E., Sun, Y., Thoma, N. H., Zheng, N., Chen, P. L., Lee, W. H., and Pavletich, N. P. (2002) Science 297 1837–1848 [DOI] [PubMed] [Google Scholar]
- 57.Wong, J. M., Ionescu, D., and Ingles, C. J. (2003) Oncogene 22 28–33 [DOI] [PubMed] [Google Scholar]
- 58.Yang, H., Li, Q., Fan, J., Holloman, W. K., and Pavletich, N. P. (2005) Nature 433 653–657 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.