Abstract
Post-translational modifications of histone tails direct nuclear processes including transcription, DNA repair, and chromatin packaging. Lysine 20 of histone H4 is mono-, di-, or trimethylated in vivo, but the regulation and significance of these methylations is poorly understood. The SET domain proteins PR-Set7 and Suv4-20 have been implicated in mono- and trimethylation, respectively; however, enzymes that dimethylate lysine 20 have not been identified. Here we report that Drosophila Suv4-20 is a mixed product specificity methyltransferase that dimethylates ∼90% and trimethylates less than 5% of total H4 at lysine 20 in S2 cells. Trimethylation, but not dimethylation, is reduced in Drosophila larvae lacking HP1, suggesting that an interaction with HP1 regulates the product specificity of Suv4-20 and enrichment of trimethyllysine 20 within heterochromatin. Similar to the Drosophila enzyme, human Suv4-20h1/h2 enzymes generate di- and trimethyllysine 20. PR-Set7 and Suv4-20 are both required for normal levels of methylation, suggesting they have non-redundant functions. Alterations in the level of lysine 20 methylation following knock-down or overexpression of Suv4-20 did not affect lysine 16 acetylation, revealing that these two modifications are not competitive in vivo. Depletion of Suv4-20h1/h2 in HeLa cells impaired the formation of 53BP1 foci, suggesting dimethyllysine 20 is required for a proper DNA damage response. Collectively, the data indicate that Suv4-20 generates nearly ubiquitous dimethylation that facilitates the DNA damage response and selective trimethylation that is involved in heterochromatin formation.
Histone post-translational modifications (PTMs)3 play key roles in regulating genomic processes including gene transcription, chromatin assembly, DNA replication, recombination, and DNA repair (1–3). Diverse, context-specific regulation of these processes has been proposed to be facilitated by interacting regulatory factors that recognize specific histone PTMs or combinations of PTMs, individually or collectively, and direct distinct regulatory outcomes (4). Lysine residues targeted by methyltransferases (and demethylases) afford the greatest potential combinatorial and functional complexity among histone PTMs because the identity of the site, whether it is unmodified (0m) or mono- (1m), di- (2m), or trimethylated (3m), and the presence of additional PTMs at nearby sites can potentiate the binding of site-specific regulatory factors (5, 6).
Only one lysine residue, Lys-20 (K20), is known to be methylated in histone H4 (7, 8). A single SET domain-containing protein, Set9, forms all 1m, 2m, and 3mK20-H4 in Schizosaccharomyces pombe, and the function of Lys-20 methylation seems to be limited to DNA damage responses in this organism (9). This role appears to be conserved in higher eukaryotes because Crb2 and 53BP1, the homologous DNA damage checkpoint signaling proteins of S. pombe and humans, both recognize Lys-20 dimethylation (10). However, additional functions have been ascribed to Lys-20 methylation in higher eukaryotes, and the SET domain-containing proteins PR-Set7, Suv4-20, Ash1, and NSD1 have all been implicated in methylating Lys-20 in such organisms (11–15). Work by several laboratories has established that PR-Set7 forms 1mK20-H4 exclusively in vitro (13, 14, 16–18). Analyses with antisera specific for 1m, 2m, or 3mK20-H4 suggest that the product specificity of the murine Suv4-20h1 and Suv4-20h2 isoforms is predominantly Lys-20 trimethylation (11). Systematic characterization of the product specificities of Ash1 and NSD1 has not been reported.
Recent evidence suggests that 1m, 2m, and 3mK20-H4 are functionally distinct in higher eukaryotes although several contradictory findings have yet to be resolved. In mammals, immunocytological analyses suggest that although 1mK20-H4 is enriched on the inactive X chromosome, it is also distributed throughout euchromatin and heterochromatin, 2mK20-H4 is preferentially localized to inactive euchromatin and 3mK20-H4 is enriched in constitutive heterochromatin (19, 20). The phenotype of Drosophila embryos lacking PR-Set7 suggests a role for this enzyme in mitosis (21, 22). Similarly, depletion of PR-Set7 in U2OS cells and normal human fibroblasts is associated with defects in DNA replication and repair that lead to aberrant mitosis and growth inhibition, but surprisingly these phenotypes are not apparent in HeLa cells (23, 24). It is not clear whether these are due to a requirement for Lys-20 monomethylation or for other aspects of PR-Set7 function. In contrast, chromatin immunoprecipitation analyses showing preferential association of 1mK20-H4 with transcriptionally active genes imply a role in transcriptional regulation (25–27). Similarly, preferential localization of 3mK20-H4 to constitutive heterochromatin in mammals and dominant suppression of position effect variegation by Suv4-20 in Drosophila suggest that Lys-20 trimethylation is involved in gene silencing (11, 28), even though ChIP analyses suggest that 3mK20-H4 is not enriched at the promoters of inactive genes (26, 29).
To better understand the regulation of H4-K20 methylation, we have used Top Down mass spectrometry (TDMS) for direct identification and relative quantification of singly and multiply modified forms of intact H4. We show here that nearly all H4 in asynchronous Drosophila S2 cells is methylated at Lys-20 with dimethylation present on ∼90% of the molecules. To identify the enzyme responsible for this abundant dimethylation, we performed an RNAi screen targeting SET domain protein-encoding genes in Drosophila. Depletion of Suv4-20 led to a marked reduction in the level of 2mK20-H4, revealing that Drosophila Suv4-20 is a dual product specificity methyltransferase which forms most, if not all, of the abundant 2mK20-H4 and forms only small amounts of 3mK20-H4 by comparison in vivo. Heterochromatin Protein 1 (HP1) is required for normal levels of 3mK20-H4 in Drosophila larvae, suggesting that the product specificity of Suv4-20 may be modulated by interactions with HP1. Depletion of PR-Set7 led to increased levels of 0mK20-H4, suggesting that PR-Set7 methylates 0mK20-H4 to form 1mK20-H4 in vivo. Comparisons of the results obtained for depletion of PR-Set7 and Suv4-20 individually and in combination suggest that PR-Set7 mediates most Lys-20 monomethylation in vivo and that the majority of 1mK20-H4 formed by PR-Set7 subsequently serves as the physiological substrate for Suv4-20. However, 0mK20-H4 can also serve as a substrate for Suv4-20 in the absence of PR-Set7. We show that Lys-20 methylation is regulated similarly in human cells and that Lys-20 dimethylation mediated by Suv4-20h1/h2 is required for efficient 53BP1 foci formation following DNA damage.
EXPERIMENTAL PROCEDURES
Drosophila S2 Cell Culture and RNAi—S2 cells were grown in Schneider's medium containing 10% fetal bovine serum (FBS) and penicillin/streptomycin at 26 °C. RNAi was performed as described previously (30) with some modifications. 6-well plates were seeded with 1 ml of S2 cells per well (2 × 106 cells per ml in medium lacking serum and antibiotics), and then 30 μg of double-stranded RNA (dsRNA) were added to each well. After 1 h, 2 ml of complete medium containing 15% FBS was added to each well. On day 4, cells were harvested and portions processed for molecular analysis or resuspended as above in medium lacking serum and antibiotics and a second dsRNA addition performed for analysis on day 8. Aliquots of cells (∼106) treated in parallel were harvested at regular intervals to monitor cell cycle progression by flow cytometry as described previously (31).
Templates representing 400–700-bp portions of the 31 known or potential Drosophila SET domain-containing genes listed in supplemental Table S1 were amplified using gene-specific primers fused to T7 promoters. Double-stranded RNAs were synthesized from these templates using the T7 MEGA-script Kit (Ambion). Double-stranded RNA targeting bases 773–1368 of the coding region of the firefly luciferase gene was used as a negative control. The concentration and quality of dsRNA was assessed using absorbance at 260 nm and electrophoresis on 1% agarose gels.
HeLa S3 Cell Culture and siRNA—HeLa or HeLa S3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS at 37 °C. siRNA was performed using 21-bp siRNA as described previously (32). Briefly, 6-well plates were seeded with 1 × 105 Hela or 3 × 105 Hela S3 cells/2 ml of DMEM containing 10% FBS per well. After 24 h, cells were transfected with 10 μl of 20 μm Suv4-20 siGENOME siRNA (Dharmacon) mixed with 5 μl of oligofectamine (Invitrogen). This treatment was repeated after a further 24 h and subsequently at 4-day intervals as required. siRNA targeting the sequence 5′-CUUACGCUGAGUACUUCGA-3′ in the firefly luciferase gene was used as a negative control. Cell cycle progression of cells treated in parallel was monitored by flow cytometry.
Generation of Larvae Lacking HP1—Drosophila HP1 is encoded by the Su(var)2–5 gene. To examine the levels of K20-H4 methylation in flies lacking HP1, a cross was performed between y,w67c23; Su(var)2–504/Cy0, GFP and y,w67c23; Su(var)2–505/Cy, GFP flies. The resulting non-GFP progeny are trans-heterozygous for two null mutations in Su(var)2–5. Non-GFP third instar larvae were collected, homogenized in SDS sample buffer, heated at 95 °C for 5 min, and the clarified supernatant was used for immunoblot analysis. Similar stage y,w67c23 larvae with wild-type levels of HP1 were used as a control.
FLAG-Suv4-20h2-expressing HeLa Stable Cell Lines—HeLa S3 cells were transfected with a pcDNA3 plasmid containing FLAG-Suv4-20h2 using Lipofectamine 2000 (Invitrogen) and the manufacturer's protocol. Transfected cells were selected with G418 (1 mg/ml). Clones with high expression were selected and maintained in DMEM containing 10% FBS and G418 (500 μg/ml).
Immunofluorescence Microscopy—FLAG-Suv4-20h2-expressing cells grown on coverslips were fixed with 4% paraformaldehyde in phosphate-buffered saline, pH 7.5 containing 4% sucrose for 15 min and then permeabilized with 0.2% Triton X-100 in Tris-buffered saline (TBS) for 15 min. Coverslips were blocked with 2% bovine serum albumin, 2% newborn calf serum, 0.1% Tween-20 in TBS, and then incubated with mouse anti-FLAG M2 antibody (Sigma, 1:5000) or rabbit anti-3mK20-H4 (Abcam, 1:2000) at room temperature for 2 h. Coverslips were then incubated with fluorescein-conjugated or Cy5-conjugated secondary antibodies (Jackson Immuno-Research, 1:800). Extensive washes with TBS-0.1% Tween-20 were performed between incubations. DAPI (0.1 μg/ml final) was included in the last wash solution to stain DNA. Images were captured with Leica DM RXA2 microscope and analyzed using Openlab software. Cells treated with bleocin (1 μg/ml for 1 h in normal medium) were fixed and permeabilized simultaneously with 4% paraformaldehyde, 0.25% Triton X-100, and 2.5 mm MgCl2 in phosphate-buffered saline and then stained with mouse anti-53BP1 (Upstate, 1:250) and modified histone antibodies as above. Images were acquired with a Zeiss LSM 510 laser scanning confocal microscope. The average number of 53P1 foci per cell was determined from 6 randomly selected fields each containing 30–100 cells from two separate experiments.
RT-PCR—Total RNA was prepared using TRIzol reagent (Invitrogen) following the manufacturer's protocol. Reverse transcription was performed using a First Strand Synthesis Kit (Invitrogen) and oligo-dT primers. Specific cDNAs were then PCR-amplified with gene-specific primers (supplemental Table S2).
Immunoblotting—Cultured cells were washed twice with TBS, lysed in SDS sample buffer, and heated at 95 °C for 10 min. Proteins were separated on SDS-PAGE gels and transferred to polyvinylidene difluoride or nitrocellulose membranes. The antisera to 1mK20-H4 (Abcam), 2mK20-H4 (Upstate), and 3mK20-H4 (Abcam) were diluted 1:3000. Antisera to individual acetylation sites in H4 (Upstate) were diluted 1:2000. Antisera to α-tubulin (Sigma) were used at 1:5000. Antisera to Drosophila PR-Set7 were raised against residues 1–100 expressed in bacteria as a C-terminal His tag fusion, affinity-purified using antigen coupled to N-hydroxysuccinamide-activated agarose beads, and the purified preparation diluted 1:1500. Monoclonal antibody to Drosophila HP1 was obtained from the Developmental Studies Hybridoma Bank (University of Iowa) and used diluted 1:5000.
Histone Purification and Mass Spectrometry—Drosophila S2 cell nuclei were isolated using DNIB buffer (10 mm Tris-HCl, 3 mm MgCl2, 0.25 m sucrose, 0.3% Nonidet P-40, pH 8.0) freshly supplemented with 1 mm dithiothreitol, 10 mm sodium butyrate, 5 nm microcystin LR, 0.5 mm aminoethyl benzenesulfonyl fluoride (AEBSF). Cells were lysed in cold DNIB buffer for 5 min on ice and nuclei collected by low speed centrifugation. Nuclei were washed with DNIB buffer lacking Nonidet P-40, and then histones were extracted out with 0.4 n H2SO4. Nuclei were isolated from HeLa S3 cells and histones extracted by a similar procedure using NIB buffer (15 mm Tris-HCl, 60 mm KCl, 15 mm NaCl, 5 mm MgCl2, 1 mm CaCl2, 0.25 m sucrose, 0.3% Nonidet P-40, pH 7.5) supplemented as above. Histones were recovered by 20% trichloroacetic acid precipitation followed by washes with acetone/0.1% HCl and acetone. Crude histone was resuspended in H2O and oxidized in 3% H2O2 and 3% formic acid for 4 h at room temperature prior to reverse phase (RP) HPLC to facilitate analysis of Lys-20 methylation by mass spectrometry (33). Drosophila and human histones were separated on a Vydac C18 column (2.1 mm inner diameter × 250 mm, Grace Discovery Sciences) with a multistep gradient from buffer A (5% CH3CN, 0.1% trifluoroacetic acid) to buffer B (90% CH3CN, 0.094% trifluoroacetic acid) and pooled fractions representing the entire single peak obtained for H4 recovered by drying in a SpeedVac. Purified H4 was resuspended in electrospray solvent and mass spectrometry performed as described previously (33).
RESULTS
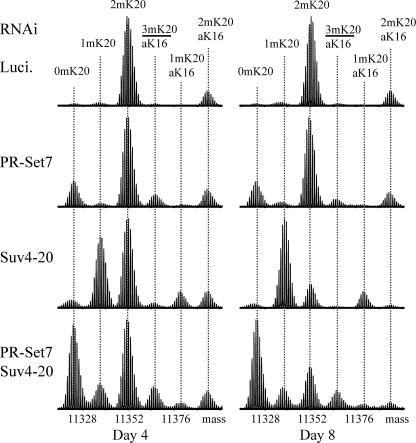
RNAi Screen for H4-K20 Methyltransferases—Searching the SMART data base (34) and BLAST analyses (35) with relaxed restrictions identified 32 candidate SET domain-containing genes in Drosophila melanogaster, and we performed RNAi for 31 of these candidates in S2 cells (supplemental Table S1). However, only depletion of PR-Set7 or Suv4-20 affected Lys-20 methylation in immunoblots performed as an initial assay for the effects of RNAi (supplemental Fig. S1). To confirm and extend these findings, we used TDMS to characterize the modification of H4 recovered from cultures on days 4 and 8 of dsRNA treatment targeting firefly luciferase (control), PR-Set7 alone, Suv4-20 alone, or PR-Set7 and Suv4-20 in combination (Fig. 1). A major advantage of using TDMS over immunochemical or other approaches for histone modification analysis is that the relative abundance of differently modified forms can be assessed without bias because of the nature or number of modifications present on individual molecules (36).
FIGURE 1.
Fourier transform mass spectrometry of histone H4 from Drosophila S2 cells depleted of Suv4-20 confirms that Suv4-20 mediates the formation of abundant 2mK20-H4. Typical results for days 4 and 8 of RNAi treatment to deplete firefly luciferase (negative control), PR-Set7, Suv4-20, or both PR-Set7 and Suv4-20 are shown. The post-translational modifications determined to be associated with each species by tandem mass spectrometry are indicated at the top of the figure. The fourth largest component labeled in each spectrum is a mixture of the nearly isobaric 3mK20-H4 and aK16-H4 forms. Slightly different amounts of sample were analyzed in each case, so the spectra are normalized according to the height of the tallest peak. The relative abundances of the forms in each sample are listed in Table 1.
Direct TDMS analysis of H4 prepared from luciferase dsRNA-treated S2 cells resolved six components differing in molecular mass by multiples of 14 Da because of either methylation or acetylation. Gas phase isolation of individual components followed by fragmentation and tandem MS enabled modifications to be assigned to each form as shown in Fig. 1. All molecules were α-N-acetylated at Thr-1, a co-translational modification, and molecules bearing only this modification are considered to be unmodified. The most abundant form was dimethylated at Lys-20 without additional PTMs (2mK20-H4). The next most abundant form was dimethylated at Lys-20 and acetylated at Lys-16 (aK16, 2mK20-H4). Together, these forms accounted for ∼90% of all H4 in untreated asynchronous S2 cells, and this was not altered by treatment with luciferase dsRNA (Table 1). The remaining four components: 0mK20-H4, 1mK20-H4, aK16,1mK20-H4, and a mixture containing the nearly isobaric 3mK20-H4 and aK16-H4 forms each accounted for less than 5% percent of the sample in both untreated asynchronous and luciferase dsRNA-treated cells (Table 1). We estimate that 3mK20-H4 represented ∼1% of total H4 in these control samples, slightly lower than we typically find in mammalian cells (Table 2) (8) and just below our threshold for precise direct quantitation from mixtures (36).
TABLE 1.
Global changes in H4 modification following depletion of PR-Set7 or Suv4-20 in Drosophila S2 cells
The relative abundances, in percentage of the total of H4 characterized by Fourier transform mass spectrometry in each sample, were determined from the recorded spectra. The data represent the average of four independent experiments ± S.E.
|
PTMs
|
Asynch
S2a
|
Luciferase
|
PR-Set7
|
Suv4-20
|
PR-Set7 + Suv4-20
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Day 4 | Day 8 | Day 4 | Day 8 | Day 4 | Day 8 | Day 4 | Day 8 | ||
| 0mK20 | 1.3 ± 0.2 | 1.9 ± 0.5 | 1.3 ± 0.4 | 15.4 ± 1.5 | 16.5 ± 1.2 | 2.4 ± 0.4 | 2.2 ± 0.2 | 30.6 ± 3.8 | 51.9 ± 0.3 |
| 1mK20 | 2.2 ± 0.4 | 2.4 ± 0.4 | 2.0 ± 0.2 | 2.0 ± 0.3 | 2.1 ± 0.4 | 33.4 ± 1.9 | 59.4 ± 4.6 | 11.9 ± 1.8 | 10.3 ± 0.6 |
| 2mK20 | 75.8 ± 2.8 | 76 ± 4.2 | 79.1 ± 2.0 | 62.1 ± 1.6 | 63.8 ± 2.0 | 45.5 ± 0.5 | 18.6 ± 1.4 | 36.4 ± 1.6 | 24.3 ± 0.8 |
| 3mK20 or aK16 | 3.6 ± 0.7 | 4.6 ± 3.3 | 1.6 ± 0.5 | 7.7 ± 1.2 | 6.2 ± 1.2 | 1.7 ± 0.5 | 2.0 ± 0.9 | 11.4 ± 0.9 | 9.0 ± 0.8 |
| aK16 & 1mK20 | 1.5 ± 0.3 | 1.4 ± 1.2 | 0.6 ± 0.3 | 0.6 ± 0.3 | 0.6 ± 0.3 | 8.0 ± 0.5 | 14.2 ± 1.8 | 3.1 ± 0.6 | 1.6 ± 0.1 |
| aK16 & 2mK20 | 15.6 ± 2.0 | 13.8 ± 1.4 | 15.5 ± 1.6 | 12.2 ± 1.1 | 10.8 ± 0.4 | 8.9 ± 0.9 | 3.5 ± 1.1 | 6.5 ± 0.8 | 3.0 ± 0.3 |
Asynch S2, asynchronous growing cells.
TABLE 2.
Global changes in H4 modification in HeLa S3 cells constitutively expressing FLAG-Suv4-20h2
The relative abundances, in percentage of the total of H4 characterized by Fourier transform mass spectrometry in each sample, were determined from the recorded spectra. Data representative of two independent analyses are reported.
| PTMs | Asynch. HeLa S3a | Asynch. FLAG-Suv4-20h2 HeLa S3b |
|---|---|---|
| 0mK20 | 4.4 | 4.8 |
| 1mK20 | 3.3 | 2.0 |
| 2mK20 | 57.8 | 50.1 |
| 3mK20 | 3.9 | 18.9 |
| aK16 | 4.0c | 4.4c |
| 1mK20 & aK16 | 1.0 | 1.1 |
| 2mK20 & aK16 | 25.7 | 18.7 |
Asynch. HeLa S3, asynchronous growing non-transfected cells.
Asynch. FLAG-Suv4-20h2 HeLa S3, asynchronous growing clonal cell line stably expressing FLAG-tagged Suv4-20h2.
Monoacetylation at Lys-16 and Lys-12 are both detected at a ratio of approximately 4:1 for aK16/aK12.
Depletion of PR-Set7 led to a readily apparent increase in the level of 0mK20-H4 (Fig. 1 and Table 1), suggesting that PR-Set7 is responsible for forming at least a portion of 1mK20-H4 in vivo even though the global levels of 1mK20-H4 did not change appreciably (see below). In contrast, depletion of Suv4-20 caused the levels of 1mK20-H4 to increase, and those of 2mK20-H4 to decrease markedly, providing dramatic evidence that Suv4-20 forms most, if not all, 2mK20-H4 in vivo. Immunoblotting with antisera to 3mK20-H4 confirmed that depletion of Suv4-20 also led to decreased levels of 3mK20-H4 (supplemental Fig. S1). Combined RNAi against mRNAs encoding PR-Set7 and Suv4-20 appeared to prevent the majority of Lys-20 methylation such that 0mK20-H4 ultimately became the predominant form (Fig. 1 and Table 1).
The persistence of 1mK20-H4 following PR-Set7 depletion suggested the possibility that other enzymes might contribute to Lys-20 monomethylation. However, depletion of Ash1 or NSD1 did not affect global levels of 1m, 2m, or 3mK20-H4 assessed by immunoblotting (data not shown) or by TDMS (supplemental Fig. S2). Moreover, combined RNAi targeting mRNAs encoding Ash1 or NSD1 in conjunction with PR-Set7 resulted in H4 modification profiles that were essentially identical to those obtained following RNAi for PR-Set7 alone (compare Fig. 1 and supplemental Fig. S2), suggesting that neither factor is redundant with PR-Set7 at the global level.
Our finding that 0mK20-H4 predominated when PR-Set7 and Suv4-20 were depleted together, whereas 1mK20-H4 predominated when only Suv4-20 was depleted, clearly suggests that PR-Set7 forms most 1mK20-H4 in vivo. Thus, the apparent perduration of 1mK20-H4 and limited decrease in 2mK20-H4 levels following depletion of PR-Set7 alone indicates that 0mK20-H4 serves as a substrate for Suv4-20 in the absence of PR-Set7, forming detectable amounts of 1mK20-H4 as an intermediate in the formation of 2m/3mK20-H4. This is consistent with the ability of the murine Suv4-20h1/h2 SET domains to methylate bacterially expressed H4 in recombinant nucleosomes in vitro (11).
Taken together, the results in Fig. 1 suggest that PR-Set7 is the major activity responsible for forming 1mK20-H4, most of which is methylated further by Suv4-20 to form predominantly 2mK20-H4 and only a small amount of 3mK20-H4, representing ∼90 and <5% of total H4, respectively, in S2 cells.
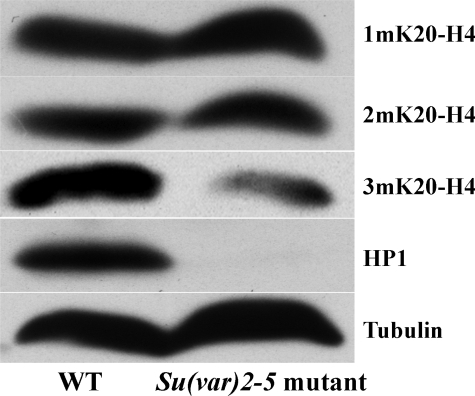
HP1 Is Required for the Normal Levels of 3mK20-H4—Polytene chromosome staining with 3mK20-H4 antisera is diminished in HP1-null larvae, consistent with the notion that interactions with HP1 contribute to the preferential localization of Suv4-20 to pericentric heterochromatin (11). Given our finding that Suv4-20 forms 2mK20 on ∼90% of all H4 in S2 cells, we investigated whether HP1 was required for global levels of either 2m or 3mK20-H4. Remarkably, immunoblot analyses revealed that the level of 3mK20-H4 was significantly reduced in Drosophila larvae lacking HP1, but that the levels of 1m and 2mK20-H4 were unaffected (Fig. 2). These data suggest the intriguing possibility that interactions with HP1 regulate both the product specificity of Suv4-20 and the enrichment of 3mK20-H4 in heterochromatin.
FIGURE 2.
HP1 is required for normal levels of 3mK20-H4 in Drosophila. Whole cell extracts from wild type and Su(var)2–5 mutant larvae lacking HP1 were analyzed on immunoblots using H4-K20 methylation state-specific antisera. Extracts were also analyzed with antisera to HP1 to confirm the absence of HP1 in the mutant larvae and antisera to α-tubulin to ensure equivalent loading.
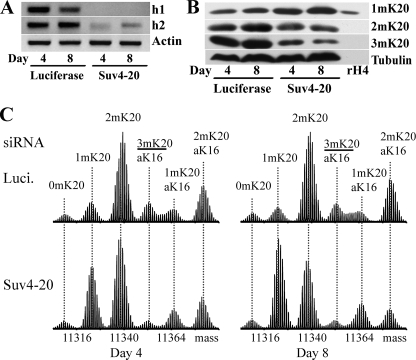
Human Suv4-20 Proteins Mediate Lys-20 Dimethylation—High levels of 2mK20-H4 are also present in normal human diploid cells, cancer cell lines, and normal animal tissues (8). To determine whether the dual di- and trimethylation product specificity of Drosophila Suv4-20 is conserved in humans, we used a mixture of siRNA sequences to deplete HeLa cells of both the h1 and h2 isoforms of Suv4-20 (Fig. 3A). An increase in the level of 1mK20-H4 and decreased levels of both 2m and 3mK20-H4 were apparent in immunoblots from Suv4-20 siRNA-treated cells (Fig. 3B). The changes in the mass spectra of H4 from Suv4-20 siRNA-treated HeLa cells (Fig. 3C) were similar to those observed for S2 cells following Suv4-20 depletion except that the reduction in 2mK20-H4 on day 8 of treatment was not quite as great. Nonetheless, these results demonstrate that the human Suv4-20 proteins also mediate the formation of 2m and 3mK20-H4.
FIGURE 3.
Human Suv4-20 proteins also mediate formation of 2mK20-H4 in vivo. Typical results are shown for days 4 and 8 of treatment of HeLa cells with siRNA targeting firefly luciferase (negative control) or pooled siRNA targeting both Suv4-20h1 and Suv4-20h2. A, relative levels of mRNA for Suv4-20h1 and Suv4-20h2 detected by RT-PCR. The levels of actin mRNA were used to ensure equivalent loading. B, relative levels of 1m, 2m, and 3mK20-H4 detected by immunoblotting with H4 Lys-20 methylation state-specific antisera. The levels of α-tubulin were used to ensure equivalent loading. C, Fourier transform mass spectra of H4 from siRNA-treated cells. Slightly different amounts of sample were analyzed in each case, so the spectra are normalized according to the height of the tallest peak.
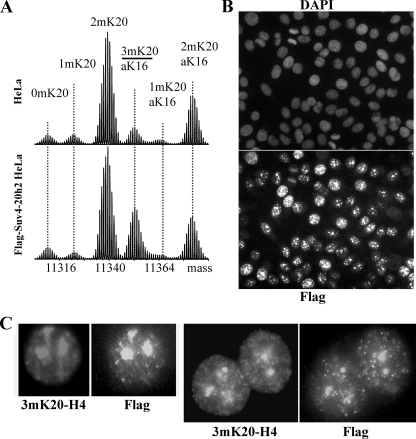
To further investigate the regulation of Lys-20 di- and trimethylation, we created a HeLa S3 cell line stably expressing FLAG-Suv4-20h2. The level of 3mK20-H4 in these cells is ∼4-fold greater compared with parental HeLa S3 cells, and this is accompanied by decreases in the relative levels of 1mK20-H4, 2mK20-H4, and aK16,2mK20-H4, but not other forms (Fig. 4A and Table 2). Because Lys-20 methylation in HeLa cells is progressive with 1mK20-H4 formed by PR-Set7 serving as the precursor to 2m- and 3mK20-H4 (8), the decreased level of 1mK20-H4 presumably reflects methylation by Suv4-20h2 to form 2m/3mK20-H4 whereas the reduction in 2mK20 forms presumably reflects methylation by Suv4-20h2 to form 3mK20-H4. The finding that the levels of unmethylated H4 (0mK20-H4 and aK16-H4) are unaltered in these Suv4-20h2-overexpressing cells also supports the notion that 2m- and 3mK20-H4 are formed primarily from 1mK20-H4 in vivo. In agreement with previous observations (11), FLAG-Suv4-20h2 and 3mK20-H4 localized preferentially to intensely DAPI-stained heterochromatin in these cells (Fig. 4, B and C).
FIGURE 4.
Constitutive expression of Suv4-20h2 preferentially increases 3mK20-H4. A, Fourier transform mass spectra of H4 from asynchronous control HeLa S3 cells (upper panel) and cells stably expressing FLAG-Suv40–20h2 (lower panel). The spectra are normalized according to the height of the tallest peak. B, immunofluorescence microscopy reveals that most cells in this line express FLAG-Suv4-20h2 and that it localizes to nuclei. The upper panel shows staining with DAPI to reveal nuclei; the bottom panel shows anti-FLAG staining. C, double staining with antisera to 3mK20-H4 and anti-FLAG reveals that FLAG-Suv4-20h2 and 3mK20-H4 are largely co-localized in heterochromatin.
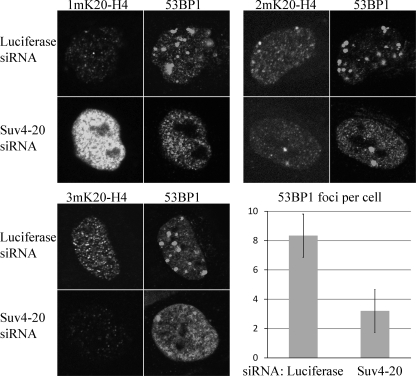
Dimethyllysine 20 Formed by Suv4-20 Is Involved in the DNA Damage Response—An earlier study suggested that depletion of PR-Set7 impairs recruitment of the 53BP1 DNA damage checkpoint signaling protein into damage-associated foci following exposure of HeLa cells to ionizing radiation (10). Immunoblotting indicated that global levels of both 1m and 2mK20-H4 were reduced in PR-Set7-depleted cells, suggesting that the defect in 53BP1 recruitment could be caused by the reduction in either 1mK20-H4 or 2mK20-H4, or possibly both forms, because the estimated affinity of 53BP1 in vitro for 2mK20-H4 is only slightly higher than that for 1mK20-H4 (10). Because our results indicate that depletion of Suv4-20h1/h2 affects the levels of 2mK20-H4 more than depletion of PR-Set7, we determined whether depleting Suv4-20h1/h2 affected 53BP1 foci formation. HeLa cells treated with control or Suv4-20h1/h2 siRNA were exposed briefly to the radiomimetic compound bleocin to induce double strand DNA breaks (37). 53BP1 foci formation and H4-K20 methylation status were analyzed by immunofluorescence microscopy (Fig. 5). Consistent with our immunoblot and TDMS analyses (Fig. 3, B and C), depletion of Suv4-20h1/h2 lead to a marked increase in the level of 1mK20-H4 and significant decreases in the levels of 2m- and 3mK20-H4. Despite this marked increase in the level of 1mK20-H4, significantly fewer 53BP1 foci were formed following bleocin treatment in cells depleted of Suv4-20h1/h2, suggesting that 2mK20-H4 preferentially mediates 53BP1 foci formation in vivo compared with 1mK20-H4.
FIGURE 5.
53BP1 recruitment to DNA damage foci is impaired in Suv4-20 siRNA cells. HeLa cells transfected with luciferase siRNA or Suv4-20 siRNA targeting both Suv4-20h1 and Suv4-20h2 for 8 days were treated with 1 μg/ml bleocin for 1 h to induce DNA damage prior to staining with the antisera to Lys-20-methylated H4 or 53BP1 as shown. The histogram displays the average number of 53BP1 foci observed per cell in six randomly selected fields (each containing 30–100 cells) from each group in two different experiments. Error bars represent the S.E. The Student's t test with unequal variance shows that the change is significant with p < 0.05.
DISCUSSION
Recent data implicate 1m, 2m, and 3mK20-H4 in regulating transcription, DNA damage responses, and heterochromatin function, respectively, suggesting that different states of Lys-20 methylation are functionally distinct (10, 11, 26). The discovery that 2mK20-H4 is far more abundant than 1m and 3mK20-H4 in both Drosophila and human cells (Figs. 1 and 3) compelled us to investigate which enzyme(s) mediate formation of 2mK20-H4. Combining RNAi depletion of SET domain proteins in Drosophila S2 cells with immunoblot and TDMS analyses of H4 modification, we established that Suv4-20 mediates the formation of most, if not all 2mK20-H4 in vivo (Fig. 1, Table 1, and supplemental Fig. S1). PR-Set7 appears to be the major source of 1mK20-H4, which serves as the primary substrate for Suv4-20. Additional proteins may possibly contribute to low levels of Lys-20 methylation, but our results suggest that although Ash1 and NSD1 have been implicated in Lys-20 methylation previously (12, 15), they do not mediate Lys-20 methylation detectable at the global level (supplemental Fig. S2). These proteins may mediate levels of Lys-20 methylation that are too small to be detected using the approach employed here. Alternatively, their physiological substrates may not include H4 (38, 39).
The murine Suv4-20h1 and h2 isoforms both localize preferentially to pericentric heterochromatin (11). However, our finding that Suv4-20 proteins direct Lys-20 dimethylation that affects ∼80 and 90% of total H4 in HeLa and S2 cells, respectively, implies that Drosophila Suv4-20 and human Suv4-20h1/h2 act widely throughout chromatin. Among the possible explanations for this discrepancy, we favor the hypothesis that Suv4-20 proteins associate with chromatin in a dynamic fashion, giving rise to 2mK20-H4 at most loci, but that they interact differently with sites in constitutive heterochromatin and others scattered sparsely throughout the genome to give rise to 3mK20-H4. Our finding that HP1 is required for normal levels of 3mK20-H4 (Fig. 2) suggests that it, and possibly other heterochromatin-associated proteins, may interact with Suv4-20 proteins to favor formation of 3mK20-H4 at heterochromatic loci. This view is supported by the ability of murine Suv4-20h2 to interact directly with HP1 in vitro and the observation that 3mK20-H4 staining of the chromocenter is diminished in polytene chromosomes from HP1-null flies (11). Moreover, members of the retinoblastoma (RB) protein family interact with Suv4-20h1/h2 in vivo. Global levels of 3mK20-H4 are reduced in RB1/RBL1/RBL2 triple knock-out MEFs (40, 41), and mutations in the LXCXE motif of the RB1 “binding pocket” diminish the pericentric heterochromatin localization of 3mK20-H4 without disrupting the interaction ability of RB1 with Suv4-20 proteins (41). Together, these observations suggest that Suv4-20 is regulated by multiple factors.
Based on evidence that H4 acetylation at Lys-16 and methylation at Lys-20 are competitive in enzyme assays in vitro, are localized differently in vivo, and that Lys-20 methylation activity is greatest during mitosis (13, 42), Lys-20 methylation has been proposed to function as an epigenetic mark, which enables the transcriptionally silent state of genes to be transmitted to daughter cells through mitosis (43). Our finding that the most abundant form of H4 that is acetylated at Lys-16 in control cultures of S2 cells and HeLa cells is also dimethylated at Lys-20 clearly argues against this (Figs. 1 and 3, Tables 1 and 2). Moreover, we did not detect any changes in acetylation at lysines 5, 8, 12, and 16 in S2 cells by immunoblotting, despite the significant decreases in the levels of 2m and 3mK20-H4 that followed depletion of Suv4-20 alone or in combination with PR-Set7 (supplemental Fig. S1). TDMS analyses revealed that depletion of Suv4-20 in S2 cells led to decreases in the level of aK16,2mK20-H4 and corresponding increases in the level of aK16,1mK20-H4 relative to the luciferase RNAi controls (Fig. 1 and Table 1). This is most consistent with the notion that the increased abundance of aK16,1mK20-H4 is due to the persistence of Lys-20 monomethylation during Suv4-20 depletion on molecules which would otherwise become aK16,2mK20-H4 rather than a global enhancement in H4 acetylation. We found that global H4-K16 acetylation, approximated by the sum of the abundances of aK16,1mK20-H4, aK16,2mK20-H4, and the mixture containing 3mK20-H4+aK16-H4 (Table 1), differed little between control and Suv4-20-depleted S2 cells. The degree of Lys-20 methylation does not appear to be a significant factor in this regard because global acetylation was also not enhanced following RNAi for PR-Set7 (Fig. 1 and Table 1). Similarly, depletion of Suv4-20h1/h2 (Fig. 3) or PR-Set7 (data not shown), did not enhance global H4 acetylation in HeLa cells. Overexpression of FLAG-Suv4-20h2 in HeLa cells led to a dramatic increase in 3mK20-H4 and concomitant decreases in the level of 2mK20-H4 and aK16,2mK20-H4 (Fig. 4 and Table 2). However, we believe that the decreased abundance of aK16,2mK20-H4 in this case, like that of 2mK20-H4, is a consequence of the high level of 3mK20-H4 attained, rather than antagonism between Suv4-20h2 and Lys-16 acetyltransferases, because all methylation states compete for the same Lys-20 residue. Taken together, our data support the conclusion that H4 acetylation and Lys-20 methylation are not competitive with each other at the global level in vivo.
Our findings that Suv4-20 mediates ubiquitous Lys-20 dimethylation in Drosophila and human cells and that 53BP1 appears to preferentially recognize 2mK20-H4 compared with 1mK20-H4 following DNA damage suggest a role for Suv4-20 in genome maintenance that has only partially been recognized. Suv4-20 deficiency in mice results in abnormal telomere elongation and recombination that may be linked to oncogenesis or cancer progression (33, 44), and decreased expression of Suv4-20h2 has been suggested to occur in human breast cancer cells (45). However, it is not clear if these links reflect the importance of Lys-20 di/trimethylation or other aspects of Suv4-20 function. Evidence suggesting that marked reductions in the levels of 3mK20-H4 and aK16-H4 are common in human cancers has been described (46), but the levels of 3mK20-H4 assessed in both normal and cancer samples by these authors far exceed those we have found in diverse cell types using TDMS (8, 33). Further analyses are required to fully address this issue. However, because nearly all Lys-20 dimethylation occurs preferentially on newly synthesized H4 in the M and G1 phases of the cell cycle in the absence of DNA damage (8), we suggest that other aspects of Suv4-20 function are also likely to be significant to the role of Suv4-20 in genome maintenance. Recent evidence that p53 activity is negatively regulated by PR-Set7-mediated monomethylation suggests that non-histone substrates may also be important in Suv4-20 function (47). The data presented here suggesting that the mixed methylation product specificity of Suv4-20 is regulated by interactions with HP1 in Drosophila should serve as the basis for further investigations of the regulation of Lys-20 methylation in chromatin function and the pathogenesis of cancer or other diseases.
Supplementary Material
Acknowledgments
We thank Dr. A. Belmont for access to the Drosophila RNAi library.
This work was supported in part by Grant 04-76 from the Roy J. Carver Charitable Trust and Basil O'Connor Scholar Award FY05-1232 from the March of Dimes (to C. A. M.), Grant GM 61513 from the National Institutes of Health (to L. L. W.), and the Packard Foundation, the Sloan Foundation, a Cottrell Scholar Award, and National Institutes of Health Grant GM 067193-06 (to N. L. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
Footnotes
The abbreviations used are: PTMs, post-translational modifications; 0m, unmethylated; 1m, monomethylated; 2m, dimethylated; 3m, trimethylated; DAPI, 4′,6-diamidino-2-phenylindole; FTMS, Fourier transform mass spectrometry; TDMS, Top Down mass spectrometry; ds, double-stranded; FBS, fetal bovine serum; K20, lysine 20; siRNA, short interfering RNA; GFP, green fluorescent protein; RNAi, RNA interference.
References
- 1.Berger, S. L. (2007) Nature 447 407–412 [DOI] [PubMed] [Google Scholar]
- 2.Downs, J. A., Nussenzweig, M. C., and Nussenzweig, A. (2007) Nature 447 951–958 [DOI] [PubMed] [Google Scholar]
- 3.Groth, A., Rocha, W., Verreault, A., and Almouzni, G. (2007) Cell 128 721–733 [DOI] [PubMed] [Google Scholar]
- 4.Jenuwein, T., and Allis, C. D. (2001) Science 293 1074–1080 [DOI] [PubMed] [Google Scholar]
- 5.Martin, C., and Zhang, Y. (2005) Nat. Rev. Mol. Cell. Biol. 6 838–849 [DOI] [PubMed] [Google Scholar]
- 6.Ruthenburg, A. J., Allis, C. D., and Wysocka, J. (2007) Mol. Cell 25 15–30 [DOI] [PubMed] [Google Scholar]
- 7.DeLange, R. J., Fambrough, D. M., Smith, E. L., and Bonner, J. (1969) J. Biol. Chem. 244 319–334 [PubMed] [Google Scholar]
- 8.Pesavento, J. J., Yang, H., Kelleher, N. L., and Mizzen, C. A. (2008) Mol. Cell. Biol. 28 468–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders, S. L., Portoso, M., Mata, J., Bahler, J., Allshire, R. C., and Kouzarides, T. (2004) Cell 119 603–614 [DOI] [PubMed] [Google Scholar]
- 10.Botuyan, M. V., Lee, J., Ward, I. M., Kim, J. E., Thompson, J. R., Chen, J., and Mer, G. (2006) Cell 127 1361–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schotta, G., Lachner, M., Sarma, K., Ebert, A., Sengupta, R., Reuter, G., Reinberg, D., and Jenuwein, T. (2004) Genes Dev. 18 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rayasam, G. V., Wendling, O., Angrand, P. O., Mark, M., Niederreither, K., Song, L., Lerouge, T., Hager, G. L., Chambon, P., and Losson, R. (2003) EMBO J. 22 3153–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishioka, K., Rice, J. C., Sarma, K., Erdjument-Bromage, H., Werner, J., Wang, Y., Chuikov, S., Valenzuela, P., Tempst, P., Steward, R., Lis, J. T., Allis, C. D., and Reinberg, D. (2002) Mol. Cell 9 1201–1213 [DOI] [PubMed] [Google Scholar]
- 14.Fang, J., Feng, Q., Ketel, C. S., Wang, H., Cao, R., Xia, L., Erdjument-Bromage, H., Tempst, P., Simon, J. A., and Zhang, Y. (2002) Curr. Biol. 12 1086–1099 [DOI] [PubMed] [Google Scholar]
- 15.Beisel, C., Imhof, A., Greene, J., Kremmer, E., and Sauer, F. (2002) Nature 419 857–862 [DOI] [PubMed] [Google Scholar]
- 16.Yin, Y., Liu, C., Tsai, S. N., Zhou, B., Ngai, S. M., and Zhu, G. (2005) J. Biol. Chem. 280 30025–30031 [DOI] [PubMed] [Google Scholar]
- 17.Xiao, B., Jing, C., Kelly, G., Walker, P. A., Muskett, F. W., Frenkiel, T. A., Martin, S. R., Sarma, K., Reinberg, D., Gamblin, S. J., and Wilson, J. R. (2005) Genes Dev. 19 1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couture, J. F., Collazo, E., Brunzelle, J. S., and Trievel, R. C. (2005) Genes Dev. 19 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohlmaier, A., Savarese, F., Lachner, M., Martens, J., Jenuwein, T., and Wutz, A. (2004) PLoS Biol. 2 E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sims, J. K., Houston, S. I., Magazinnik, T., and Rice, J. C. (2006) J. Biol. Chem. 281 12760–12766 [DOI] [PubMed] [Google Scholar]
- 21.Karachentsev, D., Sarma, K., Reinberg, D., and Steward, R. (2005) Genes Dev. 19 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakaguchi, A., and Steward, R. (2007) J. Cell Biol. 176 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tardat, M., Murr, R., Herceg, Z., Sardet, C., and Julien, E. (2007) J. Cell Biol. 179 1413–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorgensen, S., Elvers, I., Trelle, M. B., Menzel, T., Eskildsen, M., Jensen, O. N., Helleday, T., Helin, K., and Sorensen, C. S. (2007) J. Cell Biol. 179 1337–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talasz, H., Lindner, H. H., Sarg, B., and Helliger, W. (2005) J. Biol. Chem. 280 38814–38822 [DOI] [PubMed] [Google Scholar]
- 26.Barski, A., Cuddapah, S., Cui, K., Roh, T. Y., Schones, D. E., Wang, Z., Wei, G., Chepelev, I., and Zhao, K. (2007) Cell 129 823–837 [DOI] [PubMed] [Google Scholar]
- 27.Vakoc, C. R., Sachdeva, M. M., Wang, H., and Blobel, G. A. (2006) Mol. Cell. Biol. 26 9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kourmouli, N., Jeppesen, P., Mahadevhaiah, S., Burgoyne, P., Wu, R., Gilbert, D. M., Bongiorni, S., Prantera, G., Fanti, L., Pimpinelli, S., Shi, W., Fundele, R., and Singh, P. B. (2004) J. Cell Sci. 117 2491–2501 [DOI] [PubMed] [Google Scholar]
- 29.Mikkelsen, T. S., Ku, M., Jaffe, D. B., Issac, B., Lieberman, E., Giannoukos, G., Alvarez, P., Brockman, W., Kim, T. K., Koche, R. P., Lee, W., Mendenhall, E., O'Donovan, A., Presser, A., Russ, C., Xie, X., Meissner, A., Wernig, M., Jaenisch, R., Nusbaum, C., Lander, E. S., and Bernstein, B. E. (2007) Nature 448 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clemens, J. C., Worby, C. A., Simonson-Leff, N., Muda, M., Maehama, T., Hemmings, B. A., and Dixon, J. E. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Juan, G., and Darzynkiewicz, Z. (1998) Methods Mol. Biol. 91 67–75 [DOI] [PubMed] [Google Scholar]
- 32.Julien, E., and Herr, W. (2004) Mol. Cell 14 713–725 [DOI] [PubMed] [Google Scholar]
- 33.Pesavento, J. J., Garcia, B. A., Streeky, J. A., Kelleher, N. L., and Mizzen, C. A. (2007) Mol. Cell. Proteomics 6 1510–1526 [DOI] [PubMed] [Google Scholar]
- 34.Letunic, I., Copley, R. R., Schmidt, S., Ciccarelli, F. D., Doerks, T., Schultz, J., Ponting, C. P., and Bork, P. (2004) Nucleic Acids Res. 32 D142–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pesavento, J. J., Mizzen, C. A., and Kelleher, N. L. (2006) Anal. Chem. 78 4271–4280 [DOI] [PubMed] [Google Scholar]
- 37.Nabatiyan, A., Szuts, D., and Krude, T. (2006) Mol. Cell. Biol. 26 1839–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byrd, K. N., and Shearn, A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 11535–11540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka, Y., Katagiri, Z., Kawahashi, K., Kioussis, D., and Kitajima, S. (2007) Gene 397 161–168 [DOI] [PubMed] [Google Scholar]
- 40.Gonzalo, S., Garcia-Cao, M., Fraga, M. F., Schotta, G., Peters, A. H., Cotter, S. E., Eguia, R., Dean, D. C., Esteller, M., Jenuwein, T., and Blasco, M. A. (2005) Nat. Cell Biol. 7 420–428 [DOI] [PubMed] [Google Scholar]
- 41.Isaac, C. E., Francis, S. M., Martens, A. L., Julian, L. M., Seifried, L. A., Erdmann, N., Binne, U. K., Harrington, L., Sicinski, P., Berube, N. G., Dyson, N. J., and Dick, F. A. (2006) Mol. Cell. Biol. 26 3659–3671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice, J. C., Nishioka, K., Sarma, K., Steward, R., Reinberg, D., and Allis, C. D. (2002) Genes Dev. 16 2225–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Margueron, R., Trojer, P., and Reinberg, D. (2005) Curr. Opin. Genet. Dev. 15 163–176 [DOI] [PubMed] [Google Scholar]
- 44.Benetti, R., Gonzalo, S., Jaco, I., Schotta, G., Klatt, P., Jenuwein, T., and Blasco, M. A. (2007) J. Cell Biol. 178 925–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tryndyak, V. P., Kovalchuk, O., and Pogribny, I. P. (2006) Cancer Biol. Ther. 5 65–70 [DOI] [PubMed] [Google Scholar]
- 46.Fraga, M. F., Ballestar, E., Villar-Garea, A., Boix-Chornet, M., Espada, J., Schotta, G., Bonaldi, T., Haydon, C., Ropero, S., Petrie, K., Iyer, N. G., Perez-Rosado, A., Calvo, E., Lopez, J. A., Cano, A., Calasanz, M. J., Colomer, D., Piris, M. A., Ahn, N., Imhof, A., Caldas, C., Jenuwein, T., and Esteller, M. (2005) Nat. Genet. 37 391–400 [DOI] [PubMed] [Google Scholar]
- 47.Shi, X., Kachirskaia, I., Yamaguchi, H., West, L. E., Wen, H., Wang, E. W., Dutta, S., Appella, E., and Gozani, O. (2007) Mol. Cell 27 636–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.