Abstract
The coaggregation receptor polysaccharides (RPS) of Streptococcus oralis and related species are recognized by lectin-like adhesins on other members of the oral biofilm community and by RPS-specific antibodies. The former interactions involve β-GalNAc or β-Gal containing host-like motifs in the oligosaccharide repeating units of these polysaccharides, whereas the latter involves features of these molecules that are immunogenic. In the present investigation, the molecular and corresponding structural basis for the serotype specificity of S. oralis ATCC 10557 RPS was determined by engineering the production of this polysaccharide in transformable Streptococcus gordonii 38. This involved the systematic replacement of genes in the rps cluster of strain 38 with different but related genes from S. oralis 10557 and structural characterization of the resulting polysaccharides. The results identify four unique genes in the rps cluster of strain 10557. These include wefI for an α-Gal transferase, wefJ for a GalNAc-1-phosphotransferase that has a unique acceptor specificity, wefK for an acetyl transferase that acts at two positions in the hexasaccharide repeating unit, and a novel wzy associated with the β1-3 linkage between these units. The serotype specificity of engineered polysaccharides correlated with the wefI-dependent presence of α-Gal in these molecules rather than with partial O-acetylation or with the linkage between repeating units. The findings illustrate a direct approach for defining the molecular basis of polysaccharide structure and antigenicity.
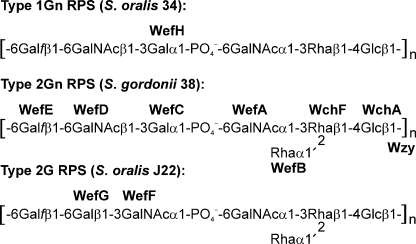
The characteristic presence of different bacteria in naturally occurring biofilm communities (1), such as those that form on host mucosal surfaces, raises the possibility that bacterial surface polysaccharides, in addition to their role as antigens, function as recognition molecules for biofilm development. This possibility is evident from studies of the cell wall polysaccharides on Streptococcus oralis and related viridans group streptococci that function as receptors for lectin-like adhesins on other members of the dental plaque biofilm community (2, 3). Structural characterization of these polysaccharides (4-9) from over 20 different streptococcal strains that coaggregate with Actinomyces naeslundii revealed six coaggregation receptor polysaccharides (RPS),2 three of which are shown in Fig. 1. The presence of a host-like motif, either GalNAcβ1-3Gal (Gn) or Galβ1-3GalNAc (G), in the oligosaccharide repeating units of these molecules accounts for recognition of RPS-bearing streptococci by GalNAc- and/or Gal-reactive surface adhesins of A. naeslundii and various other members of the dental plaque biofilm community (10, 11). In contrast, the reactions of RPS-specific antibodies involve the common l-rhamnose (l-Rha)-branched region in serotype 2 polysaccharides (9) or α-GalNAc in serotype 1 polysaccharides (12). Thus, the determinants of RPS serotype (i.e. 1, 2, 3, etc.) and receptor type (i.e. Gn or G) appear to be distinct.
FIGURE 1.
Types of RPS produced by S. oralis 34, S. gordonii 38, and S. oralis J22 indicating the molecular basis of RPS structure. Synthesis of types 1Gn and 2G RPS depends on genes that are complementary to those in strain 38 except as indicated for the transferases associated with the unique structural features of these polysaccharides.
Studies of RPS structure and function were extended to the molecular level by identification of the chromosomal locus (rps) of Streptococcus gordonii 38 for type 2Gn RPS biosynthesis (13). The rps cluster of this strain was found to contain 14 genes, including seven for putative glycosyltransferases. The first two, wchA and wchF, were associated with the presence Rhaβ1-4Glc in these polysaccharides (Fig. 1) based on studies (14-16) of similar genes for capsular polysaccharide biosynthesis in closely related Streptococcus pneumoniae. Genes for other glycosyl and glycosyl-1-phosphotransferases were identified by their effects on RPS structure in gene replacement experiments. Thus, replacement of the genes designated wefC and wefD in strain 38 with those designated wefF and wefG in S. oralis J22 switched RPS production from type 2Gn to 2G (17). Moreover, the replacement of wefC with wefF or wefD with wefG resulted in the synthesis of modified recognition motifs, either GalNAcβ1-3GalNAc or Galβ1-3Gal, respectively, thereby firmly establishing the donor specificity of each encoded transferase (Fig. 1). In other studies, deletion of wefB eliminated l-Rha branches, converting types 2Gn and 2G RPS to linear types 1Gn and 1G, respectively (18). Further results gained from studies of genetic complementation distinguished the GalNAc-1-phosphotransferases encoded by downstream wefC of S. gordonii 38 and wefH of S. oralis 34 by a subtle difference in acceptor specificity. Both transferases acted on the linear acceptor formed in the absence of wefB, but only WefC acted on the branched acceptor formed in the presence of wefB. Thus, genetic engineering of RPS gene clusters and structural characterization of the resulting polysaccharides provides an approach for distinguishing closely related genes such as wefH, wefC, and wefF by differences in either the donor or the acceptor specificities of their encoded transferases.
Molecular studies of RPS structure and function have now been extended to type 3G RPS of S. oralis ATCC 10557 (5), a polysaccharide that is structurally related but antigenically distinct from those described above. In the present communication, we describe the rps cluster of S. oralis 10557 and the utilization of selected genes from this strain to engineer the production of type 3G RPS in transformable S. gordonii 38. The results associate the unique structural features of this polysaccharide with four presently described genes and identify one of these as a principal determinant of RPS serotype specificity. These findings, which complete the comparative molecular characterization of a major RPS group (8), provide the necessary basis for tracing the evolution of these polysaccharides as recognition molecules for biofilm formation in the host oral environment.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture Conditions—The wild type and mutant streptococci used in this study (see Table 1) were grown at 37 °C in Todd-Hewitt broth (THB) or brain-heart infusion (Difco Laboratories). These media were supplemented with erythromycin at 10 μg ml-1 or spectinomycin at 250 μg ml-1 as needed for the maintenance of antibiotic-resistant streptococcal strains.
TABLE 1.
Streptococcal strains used in this study
| Strain | Description | Source |
|---|---|---|
| S. oralis 10557 | ATCC 10557, human subacute bacterial endocarditis isolate (36), type 3G RPS (5) | Ref. 37 |
| S. oralis TC1 | S. oralis 10557 containing ermAM in place of wefK, RPS+b | This study |
| S. oralis TC2 | S. oralis 10557 containing ermAM in place of the truncated wzy and wzy, RPS−b | This study |
| S. oralis TC3 | S. oralis TC2 containing wzy of S. oralis 10557 in place of ermAM, RPS+ | This study |
| S. gordonii 38 | type 2Gn RPS (9,13) | Ref. 37 |
| S. gordonii GC16 | S. gordonii 38 containing wefF and wefG of S. oralis J22 in place of wefC and wefD, type 2G RPS | Ref. 17 |
| S. gordonii GC25 | S. gordonii GC16 containing ermAM in place of wefA and wefB, RPS− | This study |
| S. gordonii GC27 | S. gordonii GC25 containing wefA of S. gordonii 38 in place of ermAM, type 1G RPS | This study |
| S. gordonii GC30 | S. gordonii GC27 containing ermAM in place of wefA, RPS− | This study |
| S. gordonii GC32 | S. gordonii GC30 containing wefI of S. oralis 10557 in place of ermAM, RPS− | This study |
| S. gordonii GC21 | S. gordonii 38 containing ermAM in place of wefA, RPS− | Ref. 18 |
| S. gordonii GC51 | S. gordonii GC21 containing wefI of S. oralis 10557 in place of ermAM RPS+ | This study |
| S. gordonii GC31 | S. gordonii GC27 containing ermAM in place of wzy, RPS− | This study |
| S. gordonii GC39 | S. gordonii GC31 containing wzy of S. oralis 10557 in place of ermAM, RPS+ | This study |
| S. gordonii GC29 | S. gordonii GC27 containing ermAM in place of wzx, RPS− | This study |
| S. gordonii GC38 | S. gordonii GC29 containing wzx of S. gordonii 38 and wefK of S. oralis 10557 in place of ermAM, RPS+ | This study |
| S. gordonii GC34 | S. gordonii GC32 containing ermAM in place of wzy, RPS− | This study |
| S. gordonii GC35 | S. gordonii GC34 containing wzy of S. oralis 10557 in place of ermAM, RPS+ | This study |
| S. gordonii GC36 | S. gordonii GC35 containing ermAM in place of wzx, RPS− | This study |
| S. gordonii GC37 | S. gordonii GC36 containing wzx of S. gordonii 38 and wefK of S. oralis 10557 in place of ermAM, type 3G RPS | This study |
| S. oralis J22 | Wild type strain, type 2G RPS (4,17) | Ref. 37 |
| S. oralis MC10 | S. oralis J22 containing spc in place of wefF, RPS− | This study |
| S. oralis MC11 | S. oralis MC10 containing ermAM linked to wefJ of S. oralis 10557 in place of spc, type 1G RPS | This study |
| S. oralis MC12 | S. oralis J22 containing ermAM linked to wefJ of S. oralis 10557 between wefF and wefG, RPS+ | This study |
| S. gordonii SK120 | PB179, human oral isolate (38), type 3G RPS (8) | Ref. 39 |
| S. oralis SK23 | PB182, human oral isolate (38), type 3G RPS (8) | Ref. 39 |
| S. oralis H127a | Human periodontal pocket (32), type 3G RPS (8) | Ref. 37 |
| S. oralis 4477 | Human (infant) oral isolate, type 3G RPS (C. A. Bush, unpublished) | M. Cole |
S. mitis H127 (8) has been reclassified as S. oralis (M. Kilian, personal communication).
RPS+ or RPS− indicates that cell surface RPS was either detectable or not detectable, respectively, by dot immunoblotting with RPS-specific IgG.
Antibodies and Immunochemical Methods—RPS serotype-specific antibodies were affinity-purified from selected sera by 4 m MgCl2 elution from small columns of immunoadsorbent prepared by coupling partially oxidized RPS to Affi-Gel Hz (Bio-Rad) as previously described (13). RPS serotype 1-specifc IgG was prepared from rabbit antiserum R26 against S. oralis 34 by elution from coupled type 1Gn RPS (3). RPS serotype 2-specific IgG was prepared from antiserum R102 against S. gordonii 38 by elution from coupled type 2G RPS (18). RPS serotype 3-specific IgG was prepared from antiserum R98 against S. oralis 10557 (8) by elution from coupled type 3G RPS. RPS-specific IgG from antiserum R49 against S. oralis J22 (13) was also used in colony immunoblotting to detect certain RPS-producing mutants including S. gordonii GC51.
Dot immunoblotting was performed as previously described (17) to detect binding of RPS-specific antibodies to streptococci. Briefly, streptococci were harvested, washed with buffer, adjusted to uniform cell densities, and applied to nitrocellulose membranes in decreasing numbers using a Bio-Dot Microfiltration Apparatus (Bio-Rad). The membranes were blocked in Tris-buffered saline containing 0.1% Tween 20 and 2% skim milk and incubated with 12 ng ml-1 RPS-specific IgG for 1 h followed by alkaline phosphatase-conjugated goat anti-rabbit IgG (Bio-Rad) for 1 h prior to development with nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate to detect bound antibody.
Immunodiffusion was performed as previously described with undiluted rabbit antiserum against S. oralis MC2 (18) or S. oralis 10557 (8) and 0.5 mg/ml solutions of different purified polysaccharides.
PCR Amplification and Sequencing of the Type 3G RPS Gene Cluster—The 24,688-bp DNA sequence of the S. oralis 10557 rps cluster and flanking regions (GenBank™ accession number AB289547) was assembled from the sequences of overlapping PCR products. These products were amplified from genomic DNA of strain 10557 using primers designed from sequences in rps clusters of S. gordonii 38 (13) and S. oralis J22 (17). Inverse PCR (19) was performed to extend certain sequences. The sequences were assembled and annotated using Vector NTI software (Invitrogen) and the National Center for Biotechnology Information BLAST program.
Competence-stimulating Peptides (CSP)—The gene comC and flanking regions were PCR-amplified from genomic DNA of S. oralis strains 10557 and J22 using the forward primer tArg2 (20) designed from the DNA sequence of S. pneumonia Arg-tRNA and a reverse primer (i.e. CACCATTATTTCGAGCATAGAC) designed from a common sequence in comD of different streptococci. PCR products were purified, sequenced, and annotated as described above to identify ComC of S. oralis 10557 (i.e. MKNTEKLEQFKEVTEAELQEIRGGDKRLPYFFKHLFSNRTK) and S. oralis J22 (i.e. MKNTEKLEQFKKVTEAELQEIRGGEIRKENNFLFYFFKRK). The position of the putative Gly-Gly cleavage site in each sequence was used to identify the corresponding mature CSP, which is underlined. Each mature CSP was synthesized using automated 9-fluorenylmethoxy carbonyl chemistry and purified by high performance liquid chromatography (CBER Research Central, National Institutes of Health).
Construction of Streptococcal Mutant Strains—Table 1 lists the parent and mutant strains used in this study. All of the mutant strains were prepared following previously described molecular methods (17). This involved the transformation of S. oralis or S. gordonii parental strains with PCR products that contained the ermAM or spc cassette or various intact genes from S. gordonii 38 or S. oralis 10557 flanked by 0.5-1-kb gene targeting sequences for homologous recombination. Transforming DNA was prepared by overlap extension PCR (21) performed as previously described (17). Briefly, PCRs contained KOD Hot Start DNA polymerase (Novagen), a template consisting of three overlapping PCR fragments (i.e. the upstream gene targeting sequence, the gene or genes of interest, and the downstream gene targeting sequence) and appropriate primers complementary to the 5′-end of the upstream gene targeting sequence and 3′-end of downstream gene targeting sequence. Individual PCR fragments were prepared by amplification of ermAM from pKSerm2 (22), spc from pDL278 (23), and various genes of interest and targeting sequences from streptococcal genomic DNA using appropriately designed primers (24). Previous results (18) have shown that insertion of the ermAM cassette, which contains its own promoter but lacks a transcriptional terminator, does not have a polar effect on the expression of downstream genes.
Transformation of S. gordonii 38 and derivatives of this strain was performed as previously described (17) with minor modifications. Briefly, an overnight THB culture was diluted 1/20 in THB containing 5% heat-inactivated horse serum (Sigma) (THB-HS). Following incubation for 2 h at 37 °C, the resulting culture was again diluted 1/20 in fresh THB-HS and incubated for 2 h at 37 °C to obtain competent cells in early log-phase. Transformation reactions containing 50 μl of competent cells, 450 μl of THB-HS, and 2 μg of transforming DNA were incubated for 2 h at 37 °C prior to plating on brain-heart infusion agar containing 5% heat-inactivated horse serum and appropriate antibiotics, which were added as needed. Transformation of S. oralis J22, S. oralis 10557, and derivatives of these strains was performed by the same procedure except that the homologous CSP was added to transformation reactions at a final concentration of 100 μg ml-1. RPS-producing transformants, obtained by replacement of the ermAM or spc cassettes with genes that complemented RPS production, were identified by colony-immunoblotting with RPS-specific IgG (17). Integration of overlap extension PCR products at the expected location in the streptococcal chromosome was confirmed by PCR amplification of specific products across the upstream and downstream boundaries of the insertion using primers designed from flanking sequences that were extraneous to those used for gene targeting or by DNA sequencing of the affected region.
Purification of Polysaccharides—RPS was solubilized by mutanolysin digestion of protease-treated streptococcal cell walls and purified by gradient elution from an anion exchange column (DEAE-Sepharose Fast Flow; GE Healthcare) as previously described (8).
Structural Characterization of Polysaccharides—NMR spectra of purified polysaccharides were recorded as in previous studies (5, 17, 25) with a Bruker DRX 500 MHz spectrometer with a cryoprobe using XWINNMR as the standard acquisition software. The NMR measurements were done at 25 °C. Generally, a 10-mg sample of RPS was exchanged twice with 3 ml of 99.96% D2O, lyophilized, and dissolved in 0.6 ml of 99.99% D2O. The chemical shifts were recorded relative to internal acetone (1H, 2.225ppm; 13C, 31.05ppm). All of the data were processed using NMRPipe, NMRDraw, and NMRView software. Double quantum filtered homonuclear coherence spectroscopy and total correlation spectroscopy (TOCSY) were carried out to assign the scalar coupled proton of each monosaccharide residue. 13C chemical shifts were assigned by heteronuclear single quantum coherence spectroscopy (HSQC). Inter-residual linkages were determined by nuclear Overhauser spectroscopy (NOESY) with mixing times of 100 and 300 ms and in one case by long range C-H heteronuclear multiple bond coherence.
Glycosyl composition and linkage analyses were done by gas chromatography-mass spectrometry of the sugar alditol acetates and of the partially methylated alditol acetates. This work, which was performed at the Complex Carbohydrate Research Center of the University of Georgia, is described in the supplemental data.
RESULTS
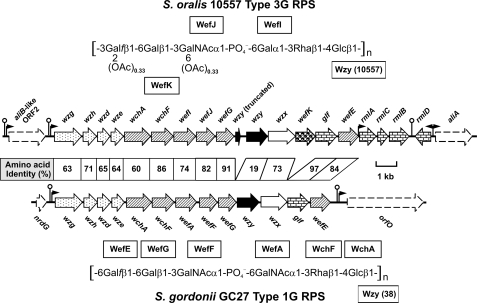
Identification and Molecular Comparison of RPS Gene Clusters—The rps cluster of S. oralis 10557 (Fig. 2), which resembles those of S. oralis J22 (17) and S. oralis 34 (18), was identified downstream of dexB and two aliB-like ORFs and upstream of aliA. The 5′-end of this cluster contained the four expected regulatory genes (i.e. wzg, wzh, wzd, and wze) and the 3′-end contained the first three genes (i.e. rmlA, rmlC, and rmlB) for dTDP-l-Rha biosynthesis. The last gene for this pathway, rmlD, was found immediately downstream of rmlB but appeared to be transcribed in the opposite direction from a bidirectional promoter present between rmlD and aliA.
FIGURE 2.
Molecular comparison of S. oralis 10557 type 3G RPS with S. gordonii GC27 type 1G RPS showing genes for common regulatory proteins (arrow with dots), glycosyl or glycosyl-1-phosphotransferases (hatched arrow), polymerases (black arrow), flippases (white arrow), enzymes for nucleotide sugar biosynthesis (arrow with bricks), and in the case of strain 10557, an acetyl transferase (arrow with circles). Synthesis of type 3G RPS depends on genes that are complementary to those in strain GC27 except as indicated for the transferases and polymerase associated with the unique structural features of type 3G RPS.
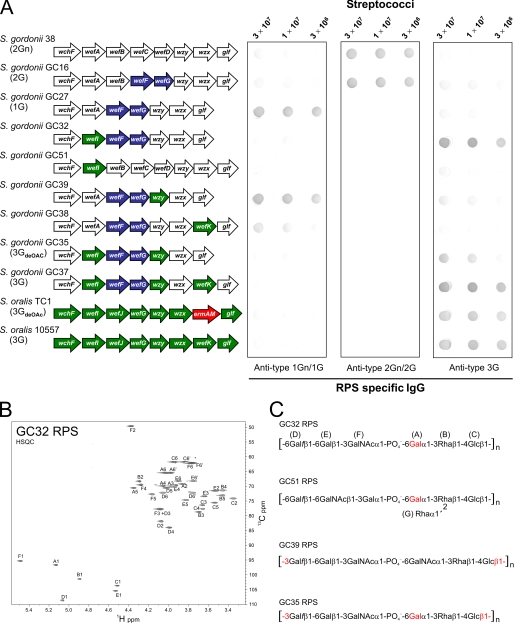
We anticipated that the unique structural features of type 3G RPS would depend on genes in the central region of the S. oralis 10557 rps cluster. To identify these genes, we sought to engineer the production of type 3G RPS in transformable S. gordonii 38. We previously (17) converted type 2Gn RPS of S. gordonii 38 to type 2G RPS of S. gordonii GC16 by replacing wefC and wefD in strain 38 with wefF and wefG from S. oralis J22 (Fig. 3A). In the present study, we deleted wefB from S. gordonii GC16, thereby converting type 2G RPS to type 1G RPS of S. gordonii GC27 (Fig. 3A). This deletion required two steps. The first was replacement of wefA and wefB in strain GC16 with the ermAM cassette to obtain the RPS- transformant, S. gordonii GC25 (Table 1). The second was replacement of the ermAM cassette in strain GC25 with wefA from wild type S. gordonii 38 to obtain the RPS+ transformant, S. gordonii GC27. The 1H and 13C chemical shifts in an HSQC NMR spectrum of strain GC27 RPS (Table 2) were indistinguishable from those of previously characterized type 1G RPS of S. oralis MC2 (18); results from glycosyl composition and linkage analyses of strain GC27 RPS also support the structure shown in Fig. 2. Using the same two-step method, we then replaced selected genes in the rps cluster of strain GC27 with related genes from S. oralis 10557 (Fig. 2) and isolated the resulting polysaccharides for structural characterization.
FIGURE 3.
Carbohydrate engineering of type 3G RPS in S. gordonii. A, the partial ORF diagram of each strain indicates the presence of genes from S. gordonii 38 (white arrows), S. oralis J22 (blue arrows), S. oralis 10557 (green arrows), or ermAM (red arrow). For dot immunoblotting, nitrocellulose membranes were spotted with decreasing numbers of each wild type or mutant streptococcal strain, incubated with RPS-specific rabbit IgG, washed, and developed with alkaline phosphatase-conjugated goat anti-rabbit IgG and substrate. B, C-H correlation spectrum (HSQC) of RPS from strain GC32 showing the anomeric and central regions but not methyl region of the spectrum. C, novel RPS structures indicating the residue letters used for the assignment of HSQC 1H and 13C chemical shifts in B and Table 2.
TABLE 2.
Residue by residue comparison of HSQC 1H and 13C chemical shifts for the RPS of S. gordonii 38 and mutant strains
| Strain | Residue | H-1, C-1 | H-2, C-2 | H-3, C-3 | H-4, C-4 | H-5, C-5 | H-6, H-6′, C-6 | NAc, CH3 |
|---|---|---|---|---|---|---|---|---|
| Sg 38a | GalNAcα (A) | 5.180, 95.81 | 4.233, 50.91 | 3.973, 68.00 | 4.078, 68.80 | 4.330, 70.44 | 4.01, 4.07, 65.18 | 2.087, 23.12 |
| GC27 | GalNAcα (A) | 5.071, 95.13 | 4.224, 50.41 | 4.012, 68.08 | 4.063, 68.67 | 4.353, 70.33 | 3.997, 4.039, 65.05 | 2.054, 22.72 |
| GC32 | Galα (A) | 5.127, 96.67 | 3.851, 69.13 | 3.963, 70.11 | 4.051, 69.72 | 4.344, 70.50 | 3.980, 4.024, 65.42 | |
| GC51 | Galα (A) | 5.148, 97.18 | 3.903, 68.77 | 3.930, 69.89 | 4.068, 69.52 | 4.339, 70.37 | 3.88, 3.96, 64.78 | |
| GC39 | GalNAcα (A) | 5.070, 95.50 | 4.225, 50.58 | 4.015, 68.35 | 4.066, 68.94 | 4.354, 70.70 | 3.996, 4.042, 65.42 | 2.057, 23.04 |
| GC35 | Galα (A) | 5.127, 96.67 | 3.855, 69.13 | 3.963, 70.11 | 4.051, 69.72 | 4.348, 70.50 | 3.980, 4.026, 65.42 | |
| Sg 38 | Rhaβ (B) | 4.925, 100.51 | 4.332, 73.05 | 3.727, 79.63 | 3.546, 71.75 | 3.476, 73.42 | 1.351, 17.70 | |
| GC27 | Rhaβ (B) | 4.844, 101.13 | 4.207, 68.08 | 3.658, 78.33 | 3.428, 71.06 | 3.431, 72.72 | 1.339, 17.47 | |
| GC32 | Rhaβ (B) | 4.888, 101.36 | 4.290, 68.35 | 3.695, 78.70 | 3.453, 71.28 | 3.458, 73.04 | 1.344, 17.77 | |
| GC51 | Rhaβ (B) | 4.932, 101.76 | 4.371, 73.64 | 3.781, 79.11 | 3.525, 71.67 | 3.482, 73.43 | 1.355, 17.45 | |
| GC39 | Rhaβ (B) | 4.845, 101.55 | 4.210, 68.35 | 3.659, 78.70 | 3.424, 71.28 | 3.434, 73.04 | 1.342, 17.77 | |
| GC35 | Rhaβ (B) | 4.888, 101.36 | 4.295, 68.35 | 3.692, 78.70 | 3.455, 71.28 | 3.460, 73.04 | 1.344, 17.77 | |
| Sg 38 | Glcβ (C) | 4.489, 103.53 | 3.341, 74.02 | 3.596, 76.83 | 3.604, 78.07 | 3.490, 75.60 | 3.783, 3.947, 61.80 | |
| GC27 | Glcβ (C) | 4.512, 103.38 | 3.336, 73.79 | 3.650, 76.04 | 3.648, 77.16 | 3.533, 75.21 | 3.826, 3.943, 61.34 | |
| GC32 | Glcβ (C) | 4.510, 103.70 | 3.346, 74.21 | 3.660, 76.36 | 3.656, 77.53 | 3.536, 75.58 | 3.839, 3.941, 61.71 | |
| GC51 | Glcβ (C) | 4.497, 103.74 | 3.353, 74.04 | 3.612, 76.90 | 3.618, 78.15 | 3.489, 75.67 | 3.783, 3.958, 61.76 | |
| GC39 | Glcβ (C) | 4.613, 102.92 | 3.322, 73.82 | 3.656, 76.17 | 3.649, 77.53 | 3.551, 75.58 | 3.847, 3.942, 61.71 | |
| GC35 | Glcβ (C) | 4.615, 102.92 | 3.323, 73.82 | 3.665, 76.16 | 3.658, 77.53 | 3.538, 75.58 | 3.841, 3.941, 61.52 | |
| Sg 38 | Galfβ (D) | 5.068, 108.66 | 4.070, 81.78 | 4.080, 77.53 | 3.989, 83.93 | 4.010, 70.51 | 3.741, 4.053, 72.08 | |
| GC27 | Galfβ (D) | 5.061, 108.36 | 4.070, 81.51 | 4.070, 77.41 | 3.987, 83.61 | 4.014, 70.23 | 3.748, 4.053, 71.79 | |
| GC32 | Galfβ (D) | 5.059, 108.78 | 4.071, 81.83 | 4.085, 77.73 | 3.990, 83.98 | 4.017, 70.50 | 3.751, 4.051, 72.26 | |
| GC51 | Galfβ (D) | 5.067, 108.85 | 4.072, 81.87 | 4.086, 77.55 | 3.995, 84.01 | 4.010, 70.48 | 3.749, 4.061, 72.20 | |
| GC39 | Galfβ (D) | 5.089, 108.58 | 4.247, 80.46 | 4.281, 85.34 | 4.132, 83.00 | 3.954, 71.28 | 3.681, 3.722, 63.67 | |
| GC35 | Galfβ (D) | 5.088, 108.58 | 4.246, 80.46 | 4.280, 85.34 | 4.131, 83.00 | 3.953, 71.28 | 3.680, 3.716, 63.67 | |
| Sg 38 | GalNAcβ (E) | 4.657, 104.01 | 3.956, 53.37 | 3.745, 71.63 | 3.944, 68.78 | 3.815, 74.58 | 3.746, 3.913, 68.03 | 2.045, 23.16 |
| GC27 | Galβ (E) | 4.527, 105.09 | 3.526, 71.16 | 3.638, 73.06 | 3.929, 69.35 | 3.831, 74.28 | 3.760, 3.894, 67.88 | |
| GC32 | Galβ (E) | 4.527, 105.46 | 3.524, 71.48 | 3.634, 73.43 | 3.924, 69.72 | 3.822, 74.60 | 3.753, 3.895, 68.16 | |
| GC51 | GalNAcβ (E) | 4.645, 104.18 | 3.947, 53.34 | 3.751, 71.62 | 3.944, 68.80 | 3.829, 74.62 | 3.790, 3.906, 68.20 | 2.045, 23.16 |
| GC39 | Galβ (E) | 4.533, 105.46 | 3.529, 71.48 | 3.639, 73.43 | 3.932, 69.52 | 3.834, 74.21 | 3.761, 3.893, 67.77 | |
| GC35 | Galβ (E) | 4.529, 105.46 | 3.526, 71.48 | 3.636, 73.43 | 3.926, 69.52 | 3.826, 74.21 | 3.755, 3.897, 67.77 | |
| Sg 38 | Galα (F) | 5.495, 96.58 | 3.895, 68.02 | 3.964, 79.68 | 4.242, 70.05 | 4.128, 72.38 | 3.73, 61.89 | |
| GC27 | GalNAcα (F) | 5.486, 94.93 | 4.375, 49.23 | 4.078, 77.26 | 4.273, 69.15 | 4.163, 72.43 | 3.780, 3.758, 61.78 | 2.054, 22.72 |
| GC32 | GalNAcα (F) | 5.489, 95.30 | 4.371, 49.60 | 4.085, 77.73 | 4.273, 69.52 | 4.158, 72.65 | 3.775, 3.761, 62.10 | 2.047, 23.04 |
| GC51 | Galα (F) | 5.496, 96.73 | 3.895, 68.80 | 3.969, 79.68 | 4.247, 70.12 | 4.128, 72.40 | 3.73, 61.92 | |
| GC39 | GalNAcα (F) | 5.487, 95.30 | 4.374, 49.41 | 4.074, 77.92 | 4.274, 69.33 | 4.159, 72.65 | 3.778, 3.756, 62.10 | 2.057, 23.04 |
| GC35 | GalNAcα (F) | 5.486, 95.30 | 4.370, 49.41 | 4.070, 77.92 | 4.273, 69.33 | 4.156, 72.65 | 3.777, 3.760, 62.10 | 2.047, 23.04 |
| Sg 38 | Rhaα (G) | 5.040, 100.92 | 4.041, 71.29 | 3.850, 71.10 | 3.449, 72.75 | 4.046, 69.47 | 1.265, 17.49 | |
| GC51 | Rhaα (G) | 5.274, 101.58 | 4.084, 71.06 | 3.857, 71.00 | 3.441, 72.88 | 4.070, 69.46 | 1.278, 17.31 |
Sg 38 denotes type 2Gn RPS from S. gordonii 38 (9).
The Allelic Glycosyltransferases Encoded by wefA and wefI Differ in Donor Specificity—The presence of α1-3-linked GalNAc in type 1G RPS of strain GC27 was predicted to depend on wefA (Fig. 2). Consequently, we suspected that the homologue of this gene in S. oralis 10557 accounted for the presence of α1-3-linked Gal in type 3G RPS. To test this hypothesis, we constructed S. gordonii GC32 (Fig. 3A) by precise replacement of wefA in S. gordonii GC27 with the gene that is now designated wefI from S. oralis 10557, via the RPS- transformant, S. gordonii GC30 (Table 1). Because the RPS from strain GC32 was predicted to have a novel structure, we did not expect that its NMR chemical shifts would match those of a previously described polysaccharide. Instead, tentative assignments were made by comparison of chemical shifts for domains of the structure predicted to be identical based on the genes for polysaccharide biosynthesis. Thus, chemical shifts of strain GC32 RPS (Fig. 3B and Table 2) were predicted to be identical to those of strain GC27 RPS for residues C, D, E, and F and similar for residue B with major differences only for residue A, which was predicted to be α-Gal in strain GC32 rather than α-GalNAc in strain GC27. Chemical shifts for residue A of strain GC32 RPS were predicted to be similar to those of residue A in de-O-acetylated strain 10557 RPS (5), which, as described below, is identical to strain GC35 RPS. Consideration of the data in Table 2 indicates that this scheme is reliable to approximately ± 0.02 ppm in 1H and ± 0.2 ppm in 13C. However, because chemical shift analogies do not constitute rigorous proof of a novel polysaccharide structure, the 1H chemical shift assignments for each sugar residue of strain GC32 were verified by 1H coupling coherence using coherence spectroscopy and TOCSY spectra (supplemental Fig. S1). Proton proximity and inter-residue connections were verified by NOESY spectra (supplemental Fig. S2), proving that strain GC32 RPS has a structure identical to that of type 1G RPS except for the presence of α-Gal at position A (Fig. 3C). Glycosyl composition and linkage analyses of the RPS from strain GC32 (supplemental data) are consistent with the proposed structure. The replacement of α-GalNAc with α-Gal had a dramatic effect on immunoreactivity, changing serotype 1 strain GC27 to serotype 3 strain GC32 (Fig. 3A).
In addition to distinguishing the donor specificities of WefA and WefI, the
structures of S. gordonii GC27 RPS and GC32 RPS implied that
α-GalNAc and α-Gal are both potential acceptors for the subsequent
WefF-dependent transfer
GalNAcα-1-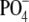 . In view of this,
we wondered whether related WefC, which transfers
Galα-1-
. In view of this,
we wondered whether related WefC, which transfers
Galα-1-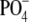 to α-GalNAc in
the synthesis of type 2Gn RPS (Fig.
1) could also utilize α-Gal as an acceptor. To assess this
possibility, we constructed S. gordonii GC51
(Fig. 3A) by precise
replacement of wefA in S. gordonii 38 with wefI of
S. oralis 10557 via the RPS- transformant, S.
gordonii GC21 (Table 1).
The structure of strain GC51 RPS, which was expected to be like that of S.
gordonii 38 RPS (i.e. type 2Gn) except for the presence of
α-Gal at residue A, was determined using a procedure similar to that
described above for strain GC32 RPS. Therefore, chemical shift assignments for
S. gordonii 38 RPS were used to predict those of residues B, C, D, E,
F, and G in strain GC51 RPS, whereas shifts of de-O-acetylated strain
10557 RPS, which is identical to strain GC35 RPS, were used for residue A.
Examination of these data in Table
2 shows that the match of predicted and experimental chemical
shifts (supplemental Fig. S3) is reasonable but not as close as that seen with
strain GC32 RPS. Most notable was a 0.23-ppm discrepancy in the 1H
chemical shift for the anomeric signal of residue G (α-Rha). Therefore,
to verify the chemical shift and linkage assignment, the chemical shift
assignments made by TOCSY (supplemental Fig. S4) and by NOESY (supplemental
Fig. S5) were augmented with a long range C-H correlation by heteronuclear
multiple bond coherence (supplemental Fig. S6). Glycosyl composition and
linkage analyses of strain GC51 RPS (supplemental data) support the proposed
structure shown in Fig.
3C. Surprisingly, strain GC51 was not stained in dot
immunoblotting (Fig.
3A) following incubation with anti-2Gn/2G RPS specific
IgG (i.e. R103). Thus, the reaction of this serotype-specific
antibody with strain 38 must involve not only the l-Rha branch in
type 2Gn RPS (9) but also
adjacent α-GalNAc in this polysaccharide.
to α-GalNAc in
the synthesis of type 2Gn RPS (Fig.
1) could also utilize α-Gal as an acceptor. To assess this
possibility, we constructed S. gordonii GC51
(Fig. 3A) by precise
replacement of wefA in S. gordonii 38 with wefI of
S. oralis 10557 via the RPS- transformant, S.
gordonii GC21 (Table 1).
The structure of strain GC51 RPS, which was expected to be like that of S.
gordonii 38 RPS (i.e. type 2Gn) except for the presence of
α-Gal at residue A, was determined using a procedure similar to that
described above for strain GC32 RPS. Therefore, chemical shift assignments for
S. gordonii 38 RPS were used to predict those of residues B, C, D, E,
F, and G in strain GC51 RPS, whereas shifts of de-O-acetylated strain
10557 RPS, which is identical to strain GC35 RPS, were used for residue A.
Examination of these data in Table
2 shows that the match of predicted and experimental chemical
shifts (supplemental Fig. S3) is reasonable but not as close as that seen with
strain GC32 RPS. Most notable was a 0.23-ppm discrepancy in the 1H
chemical shift for the anomeric signal of residue G (α-Rha). Therefore,
to verify the chemical shift and linkage assignment, the chemical shift
assignments made by TOCSY (supplemental Fig. S4) and by NOESY (supplemental
Fig. S5) were augmented with a long range C-H correlation by heteronuclear
multiple bond coherence (supplemental Fig. S6). Glycosyl composition and
linkage analyses of strain GC51 RPS (supplemental data) support the proposed
structure shown in Fig.
3C. Surprisingly, strain GC51 was not stained in dot
immunoblotting (Fig.
3A) following incubation with anti-2Gn/2G RPS specific
IgG (i.e. R103). Thus, the reaction of this serotype-specific
antibody with strain 38 must involve not only the l-Rha branch in
type 2Gn RPS (9) but also
adjacent α-GalNAc in this polysaccharide.
The Contribution of wzy to RPS Structure—Our model of RPS biosynthesis attributed the presence of Glcβ1-6Galf in type 1G RPS of S. gordonii GC27 to Wzy-dependent polymerization of hexasaccharide repeating units (Fig. 2). Consequently, we suspected that the presence of Glcβ1-3Galf in type 3G RPS resulted from the action of Wzy in S. oralis 10557. The predicted sequences of Wzy in strains 10557 and GC27 are only 19% identical (Fig. 2). To establish the role of these proteins, we constructed S. gordonii GC39 (Fig. 3A) by precisely replacing wzy in S. gordonii GC27 with wzy of S. oralis 10557 via the RPS- transformant, S. gordonii GC31 (Table 1). Strain GC39 RPS was predicted to differ from strain GC27 RPS only in the linkage between β-Glc (residue C) and β-Galf (residue D) being β1-3 in the former and β1-6 in the latter structure. Accordingly, the chemical shifts of strain GC39 RPS (Table 2 and supplemental Fig. S7) matched those of strain GC27 RPS at residues A, B, E, and F and those of de-O-acetylated strain 10557 RPS (5), represented by strain GC35 in Table 2, at residues C and D. The accuracy of this assignment was verified by homonuclear coupling correlation as determined by TOCSY (supplemental Fig. S8) and proton proximity as determined by NOESY (supplemental Fig. S9), proving a structure identical to that of type 1G RPS, except for the β1-3 linkage between Glc and Galf (Fig. 3C). Glycosyl composition and linkage analyses of strain GC39 RPS (supplemental data) confirm the proposed structure. Strains GC27 and GC39 both reacted strongly with anti-type 1Gn/1G RPS-specific IgG and weakly with anti-type 3G RPS-specific IgG in dot immunoblotting (Fig. 3A). Thus, the presence of β1-6 or β1-3 linkages between RPS repeating units did not have a noticeable effect on immunoreactivity.
The Gene wefK Encodes an Acetyl Transferase That Acts at Two Positions
in Type 3G RPS—The novel gene wefK in the rps
cluster of S. oralis 10557 (Fig.
2) encodes a protein with 10 transmembrane helices as defined by
the SOSUI system. The WefK sequence retrieved a group of uncharacterized
proteins from the data base, including closely related WciG, a putative acetyl
transferase involved in the synthesis of several S. pneumoniae
capsular polysaccharide serotypes
(14). Like these
polysaccharides, type 3G RPS of S. oralis 10557 is partially
O-acetylated (Fig. 2).
Previous estimates of this modification (i.e. 33%
O-acetylation at both the 2-OH of Galf and 6-OH of
GalNAcα-1-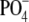 ) were based on
1H and 13C chemical shifts of acetate methyl groups and
chemical shifts at both the site of O-acetylation and at adjacent
sites of polysaccharides isolated from bacteria grown in complex medium
supplemented with Tween 80 (5).
Comparable estimates made in the present study for the same polysaccharide
isolated from bacteria grown in THB were 10% at the 2-OH of Galf and
30% at the 6-OH of GalNAcα-1-
) were based on
1H and 13C chemical shifts of acetate methyl groups and
chemical shifts at both the site of O-acetylation and at adjacent
sites of polysaccharides isolated from bacteria grown in complex medium
supplemented with Tween 80 (5).
Comparable estimates made in the present study for the same polysaccharide
isolated from bacteria grown in THB were 10% at the 2-OH of Galf and
30% at the 6-OH of GalNAcα-1-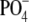 .
Thus, the extent of O-acetylation appeared to vary somewhat with
growth conditions.
.
Thus, the extent of O-acetylation appeared to vary somewhat with
growth conditions.
To assess the role of wefK, we inserted this gene between wzx and glf of S. gordonii GC27 to obtain S. gordonii GC38, via the RPS- transformant, S. gordonii GC29 (Table 1). In dot immunoblotting (Fig. 3A), strain GC38 exhibited reduced anti-type 1 immunoreactivity, compared with that of parental strain GC27. This finding, which suggested that the insertion of wefK reduced cell surface RPS production, was consistent with the relatively low yield of polysaccharide isolated from strain GC38.
The construction of S. gordonii GC35
(Fig. 3A) by
replacement of wzy of S. gordonii GC32 with wzy of
S. oralis 10557 via the RPS- transformant S.
gordonii GC34 (Table 1)
provided a convenient approach for further assessing the role of
wefK. The HSQC 1H and 13C chemical shifts in
NMR spectra of S. gordonii GC35 RPS, which expressed wefI
and wzy from S. oralis 10557, were indistinguishable from
those of de-O-acetylated type 3G RPS prepared by chemical
de-O-acetylation (5).
Glycosyl composition and linkage analyses of strain GC35 RPS (supplemental
data) also agree with the expected structure. We then constructed S.
gordonii GC37 (Fig.
3A) by inserting wefK between wzx and
glf of S. gordonii GC35, via the RPS-
transformant, S. gordonii GC36
(Table 1). NMR spectra recorded
for strain GC37 RPS identified this polysaccharide as type 3G RPS and revealed
partial O-acetylation at both expected positions (i.e. 5% to
10% at the 2-OH of Galf and ∼30% at the 6-OH of
GalNAcα-1-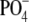 )). The extent of
this modification was comparable with that seen in type 3G RPS isolated from
strain 10557 cultured under the same conditions. As expected, the linkage and
composition analyses of strain GC37 RPS (supplemental data) are in agreement
with the proposed structure.
)). The extent of
this modification was comparable with that seen in type 3G RPS isolated from
strain 10557 cultured under the same conditions. As expected, the linkage and
composition analyses of strain GC37 RPS (supplemental data) are in agreement
with the proposed structure.
We also isolated S. oralis TC1 (Fig. 3A) by replacing wefK in S. oralis 10557 with the ermAM cassette by transformation of this strain in the presence of CSP. In contrast with NMR spectra of strain 10557 RPS, the NMR spectra of strain TC1 RPS showed no evidence of O-acetylation, thereby further associating wefK with this modification. Immunostaining of strains TC1 and 10557 in dot immunoblotting, like that seen with strains GC35 and GC37, was only noted following incubation of these strains with anti-type 3G RPS-specific IgG (Fig. 3A). Thus, O-acetylation, which is well known for its effect on the antigenicity of bacterial polysaccharides (14, 26, 27), did not appear to be a critical determinant of RPS immunoreactivity.
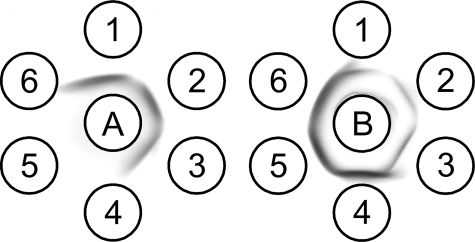
Identification of wefA and wefI as Molecular Determinants of RPS Serotype Specificity—Serotype-specific reactions of different strains in dot immunoblotting (Fig. 3A) were only observed on blots incubated with relatively low concentrations of primary antibody (i.e. 12 ng of IgG/ml). To verify the essential results of these determinations, we performed conventional immunodiffusion experiments (Fig. 4) with the purified polysaccharides of selected strains and high titer rabbit anti-streptococcal serum prepared against type 1G RPS-producing S. oralis MC2 (18) or type 3G RPS-producing S. oralis 10557. Antiserum against type 1G RPS (Fig. 4, left panel) gave a reaction of identity with type 1G RPS of S. gordonii GC27 (well 1), RPS from wefK-containing S. gordonii GC38 (well 2) and RPS from S. gordonii GC39, which expressed wzy of S. oralis 10557 (well 3). In contrast, RPS from wefI-containing S. gordonii GC32 (well 4) was unreactive, thereby indicating that the presence of α-Gal in place of α-GalNAc abolished the reaction of serotype 1-specific antibodies. In a parallel experiment performed with antiserum against type 3G RPS (Fig. 4, right panel), serotype-specific immunoprecipitation was noted with RPS from wefI-containing S. gordonii GC32 (well 4) and with type 3G RPS from S. gordonii GC37 (well 5) or S. oralis 10557 (well 6). The weak reactions seen with other polysaccharides (i.e. those in wells 1, 2, and 3) were not serotype-specific. Thus, RPS serotype 3 immunoreactivity reflected the wefI-dependent presence of α-Gal in different polysaccharides rather than partial O-acetylation or the β1-3 linkage between RPS repeating units.
FIGURE 4.
Molecular basis of RPS serotype specificity revealed by immunodiffusion analysis of wild type and mutant polysaccharides. Immunoprecipitation of rabbit antibody A against type 1G RPS-producing S. oralis MC2 with the RPS of S. gordonii GC27 (circle 1, type 1G), S. gordonii GC38 containing wefK (circle 2), or S. gordonii GC39 containing wzy of strain 10557 (circle 3) is correlated with the wefA-dependent presence of α-GalNAc in these polysaccharides. In contrast, strong immunoprecipitation of rabbit antibody B against type 3G RPS-producing S. oralis 10557 with the RPS of S. gordonii GC32 containing wefI (circle 4), S. gordonii GC37 (type 3G) containing wefI, wefK, and wzy of strain 10557 (circle 5), or S. oralis 10557 (circle 6, type 3G) is correlated with wefI-dependent presence of α-Gal in these polysaccharides.
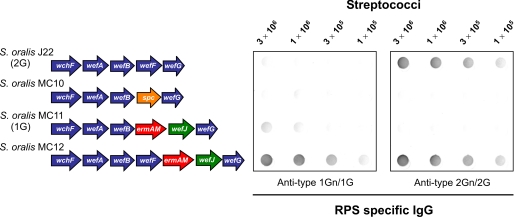
The Allelic Glycosyl-1-Phosphotransferases Encoded by wefF and wefJ Differ in Acceptor Specificity—In view of the subtle difference in the acceptor specificities of WefC and WefH (Fig. 1) for linear versus branched structures, we wondered whether a similar difference might exist between WefF of S. oralis J22 and the homologue of this transferase in S. oralis 10557 (i.e. WefJ in Fig. 2). To examine this possibility, we tested wefJ for its ability to complement type 2G RPS production in S. oralis J22 (Fig. 5). Initially, we replaced wefF of strain J22 with the spc cassette to obtain the RPS- transformant, S. oralis MC10. We then replaced the spc cassette in strain MC10 with wefJ genetically linked to the ermAM cassette. The presence of ermAM did not have a polar effect on the expression of downstream genes. The resulting RPS+ transformant, S. oralis MC11 reacted weakly with anti-type 1Gn/1G RPS-specific IgG (Fig. 5). Consistent with this reaction, the HSQC spectra of S. oralis MC11 RPS was identical to that of type 1G RPS (18), with no evidence for the presence of l-Rha branches. This finding suggested that the action of WefJ was limited to the relatively small amount of linear acceptor that remained available in the presence of WefB. Finally, we constructed S. oralis MC12 (Fig. 5) by inserting wefJ (linked to the nonpolar ermAM cassette) between wefF and wefG of parental S. oralis J22. Strong immunostaining of strain MC12 was noted with either type 1Gn/1G RPS or 2Gn/2G RPS-specific IgG, thereby suggesting the production of a hybrid polysaccharide from the action of both WefF and WefJ in this strain. Thus, the acceptor specificities of these GalNAc-1-phosphotransferases are distinct.
FIGURE 5.
Dot immunoblots of wild type and mutant streptococci with anti-type 1Gn/1G and anti-type 2Gn/2G RPS-specific antibodies showing a difference in the acceptor specificities of the GalNAc-1-phosphotransferases encoded by wefF of S. oralis J22 and allelic wefJ of S. oralis 10557. ORF diagrams of different strains identify genes from S. oralis J22 (blue arrows), S. oralis 10557 (green arrows), spc (orange arrow), or ermAM (red arrow). Strain MC11, obtained by replacing wefF in S. oralis J22 with wefJ (linked to ermAM), produced a relatively small amount of type 1G RPS, which was identified by HSQC spectra of the isolated polysaccharide.
The Predicted Properties of Truncated wzy—The 116-amino acid coding sequence of the small ORF located between wefG and wzy of S. oralis 10557 (Fig. 2) is ∼53% identical to the N-terminal regions of Wzy from S. oralis 34, S. gordonii 38, or S. oralis J22. To determine whether the truncated ORF in strain 10557 represents non essential DNA, we precisely deleted this region to obtain strain TC3, via the RPS- transformant, strain TC2 (Table 1). As anticipated, strain TC3 and wild type S. oralis 10557 were indistinguishable in dot immunoblotting performed with anti-type 3G RPS-specific IgG (results not shown).
We then wondered whether truncated wzy-like ORFs similar to the one in S. oralis 10557 occur in other type 3G RPS-producing isolates. To address this question we PCR-amplified and sequenced the wefG-wzy region in each of four seemingly unrelated type 3G RPS-producing isolates (Table 1), S. oralis SK23, S. gordonii SK120, S. oralis 4477, and S. oralis H127. Interestingly, these strains all contained truncated wzy-like coding sequences, (GenBank™ accession number AB301711, AB301712, AB301713, and AB301714, respectively). The 2922-bp sequence of the wefG-wzy region in strain 10557, including the 780-bp sequence between these genes, was identical to the corresponding region in strain SK23. Greater than 96% nucleotide sequence identity also was noted between the sequences of wefG or intact wzy in these strains and the same genes in strains SK120, 4477, or H127. However, the regions between these genes in the latter three strains differed both in length (i.e. 797, 767, and 1222 bp, respectively) and in the size of their truncated wzy-like ORFs (i.e. 116, 82, and 247 encoded amino acid residues, respectively). The values for the percentages of amino acid sequence identity were 57% for the first 60 amino acid residues of the truncated wzy-like ORFs in the five strains that produced type 3G RPS, 69% for the corresponding regions of Wzy in S. oralis 34, S. gordonii 38, and S. oralis J22, and 72% for the consensus sequences of these two groups. Importantly, strains 34, 38, and J22 Wzy were the only bacterial sequences retrieved from the existing data base by the consensus sequence of truncated Wzy. Thus, the truncated ORFs identified in each of five type 3G RPS-producing strains appeared to be remnants of the same recombinational event, namely the replacement of one wzy with another, resulting in a change in the linkage between repeating units from β1-6 in an ancestral polysaccharide to β1-3 in type 3G RPS.
DISCUSSION
The results of the present study associate the distinct structural and
biological properties of S. oralis 10557 type 3G RPS and types 1Gn,
2Gn, and 2G RPS of other strains with a relatively small number of genes for
different glycosyl or glycosyl-1-phosphotransferases
(Fig. 6). Three of these
(i.e. wchA, wchF, and wefE) account for the common presence
of Rhaβ1-4Glc at one end and Galf at the other end of each RPS
repeating unit (13), whereas
nine others determine the variable features of these polysaccharides. Four
distinct RPS serotypes are associated with the presence of allelic
wefI or wefA and the presence or absence of downstream
wefB in different strains. In the absence of wefB, the
wefA- or wefI-dependent transfer of α-GalNAc or
α-Gal to Rhaβ1-4Glc leads to the formation of linear serotype 1 or
3 polysaccharides, respectively. In contrast, the apparent action of WefA or
WefI followed by that of WefB (i.e. synthesis of the l-Rha
branch) yields branched polysaccharides that react as different serotypes.
Synthesis of the recognition motifs in these polysaccharides depends on genes
from two allelic groups. One group encodes the glycosyl-1-phosphotransferases
(i.e. WefF, WefJ, WefC, and WefH) that add
Gal-1-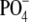 or
GalNAc-1-
or
GalNAc-1-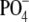 to the branched or linear
acceptors associated with different serotypes. The other group encodes WefG
and WefD, which catalyze the subsequent β1-3 transfer of Gal or GalNAc,
respectively, completing the recognition motifs in these polysaccharides. The
WefE-dependent transfer of Galf to terminal β-Gal or
β-GalNAc sets the stage for Wzx-dependent transport and Wzy-dependent
polymerization by allelic polymerases that join the conserved Glc and
Galf ends of adjacent repeats through β1-6 or β1-3
linkages. Thus, the distinct structural and biological properties of these
polysaccharides depend to a considerable extent on the complementary
activities of different allelic transferases and polymerases.
to the branched or linear
acceptors associated with different serotypes. The other group encodes WefG
and WefD, which catalyze the subsequent β1-3 transfer of Gal or GalNAc,
respectively, completing the recognition motifs in these polysaccharides. The
WefE-dependent transfer of Galf to terminal β-Gal or
β-GalNAc sets the stage for Wzx-dependent transport and Wzy-dependent
polymerization by allelic polymerases that join the conserved Glc and
Galf ends of adjacent repeats through β1-6 or β1-3
linkages. Thus, the distinct structural and biological properties of these
polysaccharides depend to a considerable extent on the complementary
activities of different allelic transferases and polymerases.
FIGURE 6.
Proposed molecular basis of RPS structure and function based on the structures of polysaccharides synthesized from RPS gene clusters of wild type or genetically engineered strains of streptococci. The red arrows indicate the sites for linkage of donor to acceptor.
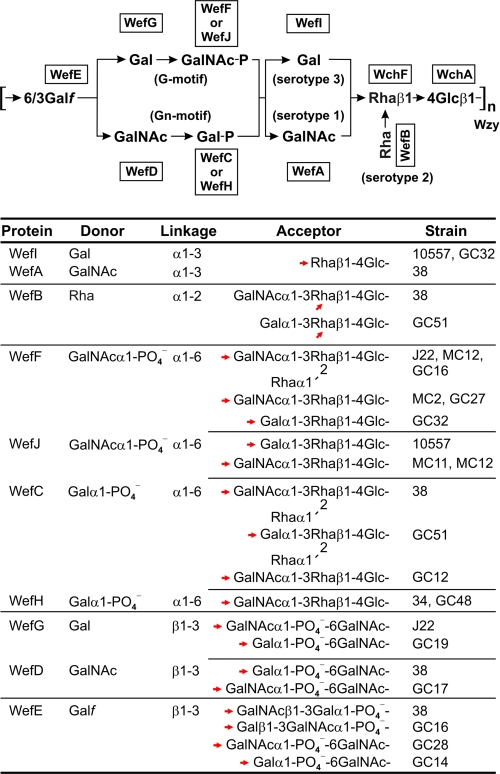
We previously identified several transferases involved in RPS biosynthesis (Fig. 1) by the effects of the corresponding genes on the structures of polysaccharides produced by transformable S. gordonii 38 (17, 18). We have now extended this approach to type 3G RPS of S. oralis 10557. Initially, we converted the type 2Gn rps cluster of S. gordonii 38 to a type 1G rps cluster in S. gordonii GC27 and then transformed this strain and various intermediates with genes from S. oralis 10557 to engineer the production of type 3G RPS. The results obtained firmly establish the donor specificities of allelic WefA and WefI, the different linkage specificities of Wzy in strain 38 and strain 10557 and identify WefK as an acetyl transferase that acts at two positions of the strain 10557 repeat. Additional findings demonstrate the relaxed acceptor specificities of allelic WefF, WefJ, and WefC for Gal- and GalNAc-containing structures by the apparent ability of these encoded proteins to act on the product of either WefI or WefA. They also distinguish the allelic GalNAc-1-phosphotransferases, WefF and WefJ, by their ability to act on the branched acceptor formed by the action of WefB (Fig. 5).
The ability to alter streptococcal polysaccharide production by genetic transformation, which dates back to the discovery of the biological role of DNA (28), was utilized in previous studies of S. pneumoniae capsular polysaccharide serotype 19 biosynthesis (29). Refinements of this approach in our studies of RPS biosynthesis include the precise replacement of individual genes in the rps cluster of one strain with complementary genes from other strains and structural characterization of the resulting polysaccharides. The NMR spectra of such polysaccharides isolated from mutanolysin cell wall digests by anion exchange column chromatography indicate that a homogeneous oligosaccharide repeating subunit comprises at least 80% of each RPS sample. However, unassigned resonances with variable intensities in the 5-20% range are also typically present in HSQC spectra, such as those of strain GC32 RPS (Fig. 3B) and strain GC51 RPS (supplemental Fig. S3). Glycosyl composition and linkage analyses of these and other RPS samples in the present study (supplemental data) revealed the expected sugars as well as small amounts of GlcNAc, terminal Glc, and Rha in linkages that are not expected from the RPS structure determined by NMR. Thus, the unassigned resonances noted in NMR spectra of RPS preparations most likely reflect the presence of extraneous cell wall polysaccharides, such as the putative rhamnose-glucose polysaccharide of S. gordonii 38 (13). Although it remains to be determined whether such polysaccharides are linked to RPS through small fragments of peptidoglycan, a promising approach for their elimination involves deletion of the corresponding genes, thereby creating strains that only produce RPS. Studies to examine this possibility are underway.
In previous studies, we showed that α-GalNAc is the immunodominant sugar of RPS serotype 1 polysaccharides (12) and also that the antigenic difference between serotype 1 and serotype 2 polysaccharides depends on the presence or absence of l-Rha branches (9). We have now established the molecular and corresponding structural basis for the distinct serotype specificity of type 3G RPS by engineering the production of this polysaccharide from type 1G RPS of S. gordonii GC27 (Figs. 2 and 3). Surprisingly, we converted type 1G RPS to a polysaccharide that was antigenically identical to type 3G RPS of S. oralis 10557 simply by swapping wefA and wefI, thereby changing α-GalNAc in type 1G RPS to α-Gal in the RPS of strain GC32. In contrast, polysaccharides obtained by changing the Wzy-dependent linkage from β1-6 to β1-3 or by the WefK-dependent introduction of O-acetyl groups at two positions reacted like type 1G RPS. In related experiments, swapping wefA for wefI changed α-GalNAc in type 2Gn RPS of S. gordonii 38 to α-Gal in the branched RPS of S. gordonii GC51. We expected that this structural change would not have a dramatic effect on antigenicity because of the common presence of immunodominant l-Rha branches (9) in the polysaccharides of these strains. Instead, RPS serotype 2-specific IgG, which reacted with strain 38 as expected, failed to react with strain GC51. Thus, the WefA-dependent presence of α-GalNAc and WefI-dependent presence of α-Gal appear to be critical for the synthesis of different linear as well as branched RPS serotypes (i.e. linear serotypes 1 and 3 and branched serotypes 2 and GC51), each of which can occur in association with either of two receptor types (Fig. 6).
RPS-bearing streptococci, which include strains of S. sanguinis, S. gordonii, and S. oralis, are thought to fill specific niches within the host oral environment. Importantly, this environment includes the characteristic biofilm communities that are passed from one generation to the next in each host species. Within this context, the structural diversity seen in different types of RPS, like that seen in cell surface glycans of higher multicellular organisms (30, 31), can be attributed to a wide range of selection pressures associated with both the negative and positive consequences of specific recognition events. Thus, in theory, the present day repertoire of RPS serotypes can be traced back to the survivors of past lethal encounters between bacteria and the host immune system or perhaps bacteriophage. In contrast, the presence of host-like features in these polysaccharides may depend at least in part on the selective advantage gained from adhesin-mediated recognition of RPS-bearing streptococci by other commensal species. Such interactions, which result in intimate associations between different bacteria, may be a prerequisite for the establishment of mutualism in biofilm communities (3, 33). If so, the evolution of these communities may be associated with the evolution of different RPS types. In addition to the presently considered types, such polysaccharides include type 4Gn RPS, which contains ribitol-5-phosphate but not Glc or l-Rha (6, 8) and the RPS of S. oralis ATCC 55229 (34), which has putative Rhaα1-2Rha recognition motifs (35). The evolutionary history of these polysaccharides, although not defined, may parallel that of the human oral environment. In this regard, the identification of what appears to be a remnant of the same ancestral wzy in the rps clusters of five independent type 3G RPS-producing strains (Fig. 6) suggests that the replacement of this gene with present day wzy is not a recent event. The association of other specific genes with the unique features of type 3G RPS and related structural types provides additional molecular markers for tracing the evolution of these polysaccharides in the biofilm communities of man and related species. The information gained may well provide insight into the role these polysaccharides play in biofilm development and their coevolution with the host oral environment.
Supplementary Material
Acknowledgments
We thank Shuntaro Ito and Kazushi Kunimatsu for contributions and Dwayne Lunsford for helpful review of the manuscript. We acknowledge Biswa Choudhury of the Complex Carbohydrate Research Center at the University of Georgia for performing the composition and linkage analyses and Russell Carlson for advice on interpretation.
This work was supported in part by the Intramural Research Program of the NIDCR, National Institutes of Health, by Grant 96 from the Keiryokai Research Foundation, by Grants-in-Aid for Promoting Technological Seeds and a High-Tech Research Project (2005-2009) from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to Y. Y.), and by Grant 02-12702 from the National Science Foundation (to C. A. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental data and supplemental Figs. S1-S9.
Footnotes
The abbreviations used are: RPS, receptor polysaccharide; CSP, competence stimulating peptide; G motif, Galβ1-3GalNAc; Gn motif, GalNAcβ1-3Gal; HSQC, heteronuclear single quantum coherence spectroscopy; NOESY, nuclear Overhauser spectroscopy; RPS-, no cell surface RPS detected by dot immunoblotting; rps, chromosomal locus for RPS biosynthesis; TOCSY, total correlation spectroscopy; GalNAc, N-acetylgalactosamine; THB, Todd-Hewitt broth; ORF, open reading frame; l-Rha, l-rhamnose.
References
- 1.Davey, M. E., and O'Toole, G. A. (2000) Microbiol. Mol. Biol. Rev. 64 847-867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu, S. D., Cisar, J. O., Sandberg, A. L., and Kilian, M. (1994) Microb. Ecol. Health Dis. 7 125-137 [Google Scholar]
- 3.Palmer, R. J., Jr., Gordon, S. M., Cisar, J. O., and Kolenbrander, P. E. (2003) J. Bacteriol. 185 3400-3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abeygunawardana, C., Bush, C. A., and Cisar, J. O. (1990) Biochemistry 29 234-248 [DOI] [PubMed] [Google Scholar]
- 5.Abeygunawardana, C., Bush, C. A., and Cisar, J. O. (1991) Biochemistry 30 6528-6540 [DOI] [PubMed] [Google Scholar]
- 6.Abeygunawardana, C., Bush, C. A., and Cisar, J. O. (1991) Biochemistry 30 8568-8577 [DOI] [PubMed] [Google Scholar]
- 7.Abeygunawardana, C., Bush, C. A., Tjoa, S. S., Fennessey, P. V., and McNeil, M. R. (1989) Carbohydr. Res. 191 279-293 [DOI] [PubMed] [Google Scholar]
- 8.Cisar, J. O., Sandberg, A. L., Reddy, G. P., Abeygunawardana, C., and Bush, C. A. (1997) Infect. Immun. 65 5035-5041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy, G. P., Abeygunawardana, C., Bush, C. A., and Cisar, J. O. (1994) Glycobiology 4 183-192 [DOI] [PubMed] [Google Scholar]
- 10.Cisar, J. O., Sandberg, A. L., Abeygunawardana, C., Reddy, G. P., and Bush, C. A. (1995) Glycobiology 5 655-662 [DOI] [PubMed] [Google Scholar]
- 11.Takahashi, Y., Ruhl, S., Yoon, J. W., Sandberg, A. L., and Cisar, J. O. (2002) Oral Microbiol. Immunol. 17 257-262 [DOI] [PubMed] [Google Scholar]
- 12.McIntire, F. C., Crosby, L. K., Vatter, A. E., Cisar, J. O., McNeil, M. R., Bush, C. A., Tjoa, S. S., and Fennessey, P. V. (1988) J. Bacteriol. 170 2229-2235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu, D. Q., Thompson, J., and Cisar, J. O. (2003) J. Bacteriol. 185 5419-5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bentley, S. D., Aanensen, D. M., Mavroidi, A., Saunders, D., Rabbinowitsch, E., Collins, M., Donohoe, K., Harris, D., Murphy, L., Quail, M. A., Samuel, G., Skovsted, I. C., Kaltoft, M. S., Barrell, B., Reeves, P. R., Parkhill, J., and Spratt, B. G. (2006) PLoS Genet. 2 262-269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, S. M., Wang, L., and Reeves, P. R. (2001) Infect. Immun. 69 1244-1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kolkman, M. A., van der Zeijst, B. A., and Nuijten, P. J. (1997) J. Biol. Chem. 272 19502-19508 [DOI] [PubMed] [Google Scholar]
- 17.Yoshida, Y., Ganguly, S., Bush, C. A., and Cisar, J. O. (2005) Mol. Microbiol. 58 244-256 [DOI] [PubMed] [Google Scholar]
- 18.Yoshida, Y., Ganguly, S., Bush, C. A., and Cisar, J. O. (2006) J. Bacteriol. 188 4125-4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochman, H., Gerber, A. S., and Hartl, D. L. (1988) Genetics 120 621-623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havarstein, L. S., Gaustad, P., Nes, I. F., and Morrison, D. A. (1996) Mol. Microbiol. 21 863-869 [DOI] [PubMed] [Google Scholar]
- 21.Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K., and Pease, L. R. (1989) Gene (Amst.) 77 61-68 [DOI] [PubMed] [Google Scholar]
- 22.Lunsford, R. D., and London, J. (1996) J. Bacteriol. 178 5831-5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeBlanc, D. J., Lee, L. N., and Abu-Al-Jaibat, A. (1992) Plasmid 28 130-145 [DOI] [PubMed] [Google Scholar]
- 24.Lee, M. S., and Morrison, D. A. (1999) J. Bacteriol. 181 5004-5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abeygunawardana, C., and Bush, C. A. (1993) Adv. Biophysical Chem. 3 199-249 [Google Scholar]
- 26.Orskov, F., Orskov, I., Sutton, A., Schneerson, R., Lin, W., Egan, W., Hoff, G. E., and Robbins, J. B. (1979) J. Exp. Med. 149 669-685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slauch, J. M., Mahan, M. J., Michetti, P., Neutra, M. R., and Mekalanos, J. J. (1995) Infect. Immun. 63 437-441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avery, O. T., MacLeod, C. M., and McCarty, M. (1944) J. Exp. Med. 79 137-158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morona, J. K., Morona, R., and Paton, J. C. (1999) J. Bacteriol. 181 5355-5364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gagneux, P., and Varki, A. (1999) Glycobiology 9 747-755 [DOI] [PubMed] [Google Scholar]
- 31.Varki, A. (2006) Cell 126 841-845 [DOI] [PubMed] [Google Scholar]
- 32.Hawley, R. J., Lee, L. N., and LeBlanc, D. J. (1980) Antimicrob. Agents Chemother. 17 372-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer, R. J., Jr., Kazmerzak, K., Hansen, M. C., and Kolenbrander, P. E. (2001) Infect. Immun. 69 5794-5804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glushka, J., Cassels, F. J., Carlson, R. W., and van Halbeek, H. (1992) Biochemistry 31 10741-10746 [DOI] [PubMed] [Google Scholar]
- 35.Cassels, F. J., Hughes, C. V., and Nauss, J. L. (1995) J. Indust. Microbiol. 15 176-185 [DOI] [PubMed] [Google Scholar]
- 36.White, J. C., and Niven, C. F., Jr. (1946) J. Bacteriol. 51 717-722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cisar, J. O., Kolenbrander, P. E., and McIntire, F. C. (1979) Infect. Immun. 24 742-752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bridge, P. D., and Sneath, P. H. (1983) J. Gen. Microbiol. 129 565-597 [DOI] [PubMed] [Google Scholar]
- 39.Kilian, M., Mikkelsen, L., and Henrichsen, J. (1989) Int. J. Syst. Bacteriol. 39 471-484 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.