Abstract
Nuclear respiratory factors NRF1 and NRF2 regulate the expression of nuclear genes encoding heme biosynthetic enzymes, proteins required for mitochondrial genome transcription and protein import, and numerous respiratory chain subunits. NRFs thereby coordinate the expression of nuclear and mitochondrial genes relevant to mitochondrial biogenesis and respiration. Only two of the nuclear-encoded respiratory chain subunits have evolutionarily conserved tissue-specific forms: the cytochrome c oxidase (COX) subunits VIa and VIIa heart/muscle (H) and ubiquitous (L) isoforms. We used genome comparisons to conclude that the promoter regions of COX6AH and COX7AH lack NRF sites but have conserved myocyte enhancer factor 2 (MEF2) elements. We show that MEF2A mRNA is induced with forced expression of NRF1 and that the MEF2A 5′-regulatory region contains an evolutionarily conserved canonical element that binds endogenous NRF1 in chromatin immunoprecipitation (ChIP) assays. NRF1 regulates MEF2A promoter-reporters according to overexpression, RNA interference underexpression, and promoter element mutation studies. As there are four mammalian MEF2 isotypes, we used an isoform-specific antibody in ChIP to confirm MEF2A binding to the COX6AH promoter. These findings support a role for MEF2A as an intermediary in coordinating respiratory chain subunit expression in heart and muscle through a NRF1 → MEF2A → COXH transcriptional cascade. MEF2A also bound the MEF2A and PPARGC1A promoters in ChIP, placing it within a feedback loop with PGC1α in controlling NRF1 activity. Interruption of this cascade and loop may account for striated muscle mitochondrial defects in mef2a null mice. Our findings also account for the previously described indirect regulation by NRF1 of other MEF2 targets in muscle such as GLUT4.
The electron transport chain (ETC)4 consists of four multisubunit enzyme complexes within the inner mitochondrial (mito) membrane. These act in concert to transfer electrons from succinate or NADH to molecular oxygen while pumping protons from the matrix to the intermembranous space, establishing the electrochemical gradient required for oxidative phosphorylation (OXPHOS) (1). Nuclear genes encode all of the components of complex II, but the other complexes have subunits encoded by both mito (ETCmito) and nuclear (ETCnucl) genes (1, 2). Appropriate ETC subunit stoichiometry requires the coordinate expression of genes on the two genomes and an accounting for a variable number of mito genomes per cell (2, 3). This is orchestrated by the nuclear respiratory (transcription) factors, NRF1 and NRF2 (2–5). These structurally unrelated factors, encoded by nuclear genes, regulate the transcription of TFAM, TFB1M, and TFB2M, nuclear genes of the mito transcription factor Tfam (mtTFA) (6) and Tfbm specificity factors (7). Tfam and Tfbm proteins are imported into mito where they direct transcription from both heavy and light strands of mito DNA (mtDNA). These transcripts are processed to yield the various ETCmito mRNAs, as well as rRNAs, tRNAs, and a primer for the RNA-dependent activity of DNA polymerase γ and mtDNA replication. This NRF → [TFAM, TFBM] → ETCmito transcriptional cascade functions in parallel with the direct control of promoters of overlapping sets of ETCnucl genes by NRF1 and NRF2 (2, 3, 8). As NRFs also regulate nuclear genes encoding complex V (F1F0-ATPase) subunits, heme biosynthetic enzymes, and mito protein import machinery (2, 3, 9, 10), they are the central regulators of mito biogenesis and cellular respiration. This is underscored by the mito deficiency and peri-implant lethality of nrf1 null mouse embryos (11).
Although the entire ETC can be regarded as functioning as a single unit, the reaction catalyzed by cytochrome c oxidase (COX, complex IV) involves the largest free energy change among ETC reactions (1). Subunits I, II, and III that together form the catalytic core of COX are encoded in the mito genome, whereas the remaining 10 subunits serve structural or regulatory functions and are products of nuclear genes. Among ETC enzymes, COX alone has tissue-specific subunit isoforms that are the products of separate nuclear genes. Thus, there are both ubiquitous (L, for liver) and heart and muscle-specific (H) isoforms of subunits VIa and VIIa5 in all mammals, and some species also have L and H variants of COX VIII (2, 8, 12). Multiple COX VIa and VIIa isoforms and cognate genes are also present in lower metazoan species including Drosophila (13). Biochemical studies have suggested that COXH may confer sensitivity of muscle COX activity and respiration to cellular energy demands (14).
In this study, we began with the question as to how the COXH subunit isoforms are coordinately expressed with one another in a tissue-specific manner but also in harmony with other nuclear-encoded proteins that are critical to mito function in muscle. We show that MEF2A, the gene encoding the transcription factor myocyte enhancer factor 2A (MEF2A), is a target of NRF1 regulation. COXH subunit genes are in turn targets of MEF2 (15, 16), and we show binding of endogenous myocyte MEF2A protein to the cox6aH gene promoter. We therefore propose that a transcriptional cascade exists (NRF1 → MEF2A → COXH) that functions with the NRF → [TFAM, TFBM] → ETCmito cascade and direct NRF → ETCnucl regulation to provide coordinate control of respiratory chain component expression in muscle. Our observations also place MEF2A with NRF1 and PGC1α in a mutually reinforcing transcriptional network.
EXPERIMENTAL PROCEDURES
RNA Analyses—Human tissue RNA was obtained from Ambion. Murine tissue and C2C12 and 10T1/2 cell RNA was isolated as described (17). Ribonuclease protection assays (RPA) and radiolabeled cRNA probe syntheses were carried out as described (17, 18). Human MEF2A and murine mef2a, mef2c, and mef2d cRNA probes have been described (17–19). Templates for other cRNA probes were PCR amplicon fragments subcloned into pBluescript, oriented to permit cRNA production from the vector T7 promoter. The 149-bp template for murine nrf1 cRNA used mouse heart RNA and reverse transcription-PCR with primers 5′-cccggatCCCAGGCTCAGCTTCGGGCA-3′ and 5′-cccgaattcGCTCTTCTGTGCGGACATCAC-3′. The underlined letters are restriction sites used in subcloning. The uppercase letters are cDNA sequences lowercase are extraneous to the cDNA but used to generate the PCR amplicon and restriction site. Templates for murine cox subunits were made using PCR on expressed sequence-tagged cDNAs or reverse transcription-PCR using liver and heart RNA. Primers and expressed sequence tag clones were: cox6aL (143 bp) IMAGE clone 3487598, 5′-gggcgcggatcCTCGGATGTGGAAGGCCCTC-3′ and 5′-gggcgcgaattCTTGGTCCTGATGCGCAGG-3′; cox6aH (120 bp) IMAGE clone 695934, 5′-gggcgcggatCCAACACCTGGCGCCTC-3′ and 5′-gggcgcgaattcGGTGGTGATACGGGATGAAC-3′; cox7aL (206 bp) 129 strain heart cDNA and IMAGE clone 1248366, 5′-gggcgcggatccGAGGATAATGGGATGCCAG-3′ and 5′-gggcgcgaatTCAGATTCCTGGTCCATCG-3′; cox7aH (171 bp) IMAGE clone 463628, 5′-gggcgcggaTCCCAGGCTCTGGTCCGG-3′ and 5′-gggcgcgaattcGCCCCCCAGAGTCAGCGTC-3′; cox7aR (102 bp) IMAGE clone 678445, 5′-gggcgcggaTCCAGAAGGCTGATGGTTTCC-3′ and 5′-gggcgcgaatTCAGGCAGTAGATGGTCCCTC-3′.
Reverse transcriptase reactions (Promega) used 1 μg of total RNA and an oligo(dT) primer. Real-time PCR was performed using a MX-3000 multiplex thermal cycler and SYBR Green with reaction conditions according to the master mix reagent supplier (Stratagene). Quantitative polymerase chain reaction (QPCR) primers for human cDNAs were: (GAPDH, 238 bp) 5′-GAGTCAACGGATTTGGTCGT-3′ and 5′-TTGATTTTGGAGGGATCTCG-3′; (NRF1, 281 bp) 5′-GTACAAGAGCATGATCCTGGA-3′ and 5′-GCTCTTCTGTGCGGACATC-3′; (MEF2A, 187 bp) 5′-GTGTACTCAGCAATGCCGAC-3′ and 5′-AACCCTGAGATAACTGCCCTC-3′; (CYCS, 106 bp) 5′-GTTGAAAAGGGAGGCAAGCA-3′ and 5′-TGTTCTTATTGGCGGCTGTG-3′; and (ACTB, 233 bp) 5′-GGACTTCGAGCAAGAGATGG-3′ and 5′-AGCACTGTGTTGGCGTACAG-3′. QPCR primers for murine cDNAs were: (gapdh, 223 bp) 5′-CTGGAGAAACCTGCCAAGTA-3′ and 5′-TGTTGCTGTAGCCGTATTCA-3′; (cycs, 177 bp) 5′-TTCAGAAGTGTGCCCAGTGC-3′ and 5′-CTCCAAATACTCCATCAGGGTATC-3′; (cox5b, 217 bp) 5′-CAAGGTTACTTCGCGGAGTG-3′ and 5′-TCCTTGGTGCCTGAAGCTG-3′; and (actb, 160 bp) 5′-AAGAGCTATGAGCTGCCTGA-3′ and 5′-TACGGATGTCAACGTCACAC-3′. Mouse mef2a and nrf1 primer pairs were identical to the respective human MEF2A and NRF1 primers. Real-time PCR results were analyzed using MX-3000 software, and the various mRNA quantities were normalized to that of β-actin (ACTB or actb). In brief, the fractional difference in the expression of a gene of interest (goi) mRNA in experimental (e) versus control (c) samples was determined using the formula ΔmRNA (arbitrary units) = 2{Δgoi –Δactb}, where Δgoi is the difference ([e] – [c]) in cycle number at critical threshold for the goi and Δactb is the difference ([e] – [c]) in cycle number at critical threshold for actb. Fluorescence determinations were also used to establish conditions (cycle number) to retrieve aliquots of parallel reactions for direct visualization of amplicon levels by agarose gel electrophoresis. All reported results were repeated in three independent small interfering RNA (siRNA) transfections.
Reporter Plasmids—ptkLuc and the MEF2Ap1-Luc and MEF2Ap2-Luc deletion series and p1[m1MEF2]-Luc and p2[m1MEF2]-Luc have been described (19). tata-Luc (Gift of Grace Gill) has been referred to as E1BLuc (20). p1[m1NRF1]-Luc and p1[m2NRF1]-Luc were created using PCR mutagenesis, exploiting the MEF2A promoter FspI site within the NRF1 element (TGC ↓ GCACGCGCA). Mutations corresponded to those used in mobility shift assay probes (Fig. 3C). A reverse luciferase primer was used in combination with 5′-gggcgctgg ↓ cCACGgGCAGCACA-3′ (m1NRF1) or 5′-gggcgcttt ↓ aaACttGCAGCACA-3′ (m2NRF1) on a MEF2A-Luc template. Amplicons were cut with NcoI and either MscI (m1NRF1) or DraI (m2NRF1), and the NcoI-blunt fragments were substituted into FspI-and NcoI-cut S-MEF2Ap1-Luc. A BamHI-XbaI fragment was substituted into S-MEF2Ap2-Luc to give p2[m1NRF1]-Luc. This strategy was also used with p1[m1MEF2]-Luc and p2[m1MEF2]-Luc (19) to construct reporters containing dual NRF1 and MEF2 element mutations, p1[m1MEF2/m1NRF1]-Luc and p2[m1MEF2/m1NRF1]-Luc. II-E-Luc was made by isolating and subcloning a RPCI-98-01L01 BAC clone EcoRI fragment, using this as a template with forward vector and reverse (5′-gggcgcgtcgacTCTCCTCTCATTGCGTTTTCC-3′) primers in PCR, and subcloning the EcoRI- and SalI-restricted amplicon into tata-Luc. An analogous construct containing an EWG element mutation, II-E[m1EWG]-Luc, was made by combining PCR with vector primers and 5′-GGTTTTTTGCcCATGgGCTCCTTCCCCACAG-3′ and 5′-GAAGGAGCcCATGgGCAAAAAACCGTAAACTC-3′. Bold letters indicate the binding site for the relevant transcription factor.
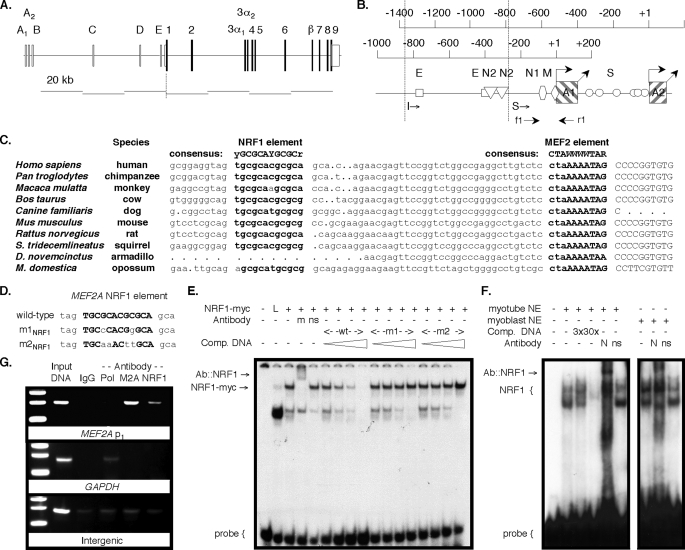
FIGURE 3.
The MEF2A promoter has a conserved canonical NRF1 element. Mammalian MEF2A gene (A) and promoter (B) diagrams. E, E box; M, MEF2; N2, NRF2; N1, NRF1; S, Sp1 elements. Locations of ChIP primers f1 and f2 are shown. Dotted vertical lines indicate locations of the 5′ extent of the short (S)- and intermediate (I)-length promoter fragments (19). C, alignment of mammalian MEF2A proximal promoter 1 (lowercase) and transcript template (uppercase) sequences. A consensus MEF2 element overlies the TSS in each gene. Sequences are from GenBank™ entries AC013526 (human), AADA01045866 (chimpanzee), AANU01176656 (rhesus monkey), AC164694 (cow), CE302601 (dog), AC120123 (mouse), AC134737 (rat), AAQQ01634896 (squirrel), AAGV01302117 (armadillo), and AAFR03022775 (opossum). D, sense strand of the ds-MEF2A NRF1 element probe and competitor oligonucleotides used in mobility shift assays. Residues that conform to the consensus NRF1 element are in bold uppercase. Mobility shift assays using the MEF2A probe and in vitro translated NRF1myc (E) or C2C12 myoblast or myotube nuclear extracts (F). L, unprogrammed lysate; wt, wild type. Antibodies: m, anti-myc; ns, unrelated antigen; N, anti-NRF1. Some reactions included competitor oligonucleotides in 3-, 10-, 30-, or 100-fold molar excess as indicated by triangles. G, ChIP assays using anti-RNA polymerase II (Pol), anti-MEF2A (M2A), and anti-NRF1 (NRF1) antibody chromatin co-precipitates and primers specific for the amplification of mef2a p1, the gapdh proximal promoter or an intergenic region.
Eukaryotic Expression Vectors—pCDNA-MEF2A α2/β, -MEF2C α2/β, and -MEF2D α2/β have been described (17, 18). pAc, pAc-EWG, and pAc-EWGΔN144 were gifts of Grace Gill (Tufts University) and have been described (20). The human NRF1 coding region was PCR-amplified from IMAGE cDNA clone 591311 template using primers (forward) 5′-gggcgcaagcttgccaccATGGAGGAACACGGAGTGAC-3′ and (reverse) 5′-gaggtggcggccgcttcaCTGTTCCAATGTCACCAC-3′, and the HindIII- and NotI-restricted amplicon was subcloned to make pCDNA-NRF1. pCDNA-NRF1ΔN85 was made using PCR on this template with forward primer 5′-gggcgcaagcttgccaccATGGCAACAGGAAAGAAACG-3′ and a reverse vector primer followed by substitution of a HdIII- and EcoRI-restricted amplicon. pCDNA-NRF1VP16 was constructed using PCR with reverse primer 5′-gaggcgccctgcaggCTGTTCCAATGTCACCACC-3′ to delete the NRF1 stop codon and then reintroducing the coding region into a modified pCDNA3 containing sequences encoding the herpes simplex virus VP16 transactivation domain downstream of an SbfI site. pCDNA3-EWG and -EWGΔN144 and pAc-NRF1 and -NRF1ΔN85 were constructed by swapping coding regions between pAc and pCDNA vectors. A derivative of pCDNA3 containing a viral internal ribosome entry site and enhanced green fluorescent protein was used in pCDNA-NRF1VP16/GFP. pCDNA-PGC1α included the IMAGE clone 30094033 SalI to NotI insert.
Mobility Shift Assays—The NRF1 coding region from pCDNA-NRF1 was subcloned into pET28 (Novagen) to give pET-NRF1. pET-NRF1myc was created by substituting a NRF1 insert lacking a stop codon from pCDNA-NRF1VP16 with a SbfI to NotI fragment containing sequence encoding the c-myc epitope (EQKLISEEDLN) and a stop codon. In vitro transcription/translation reactions used these plasmid templates with the TnT system (Promega). Muscle cell nuclear extracts were prepared, and electrophoretic mobility shift assays were conducted as described previously (19). Probe and competitor oligo sequences were: MEF2A NRF1, 5′-gatccGGTAGTGCGCACGCGCAGCACAa-3′ and 5′-gatctTGTGCTGCGCGTGCGCACTACCg-3′; MEF2A NRF1 m1NRF1, 5′-gatccGGTAGTGCcCACGgGCAGCACAa-3′ and 5′-gatctTGTGCTGCcCGTGgGCACTACCg-3′; MEF2A NRF1 m2NRF1, 5′-gatccGGTAGTGCaaACttGCAGCACAa-3′ and 5′-gatctTGTGCTGCaaGTttGCACTACCg-3′; DMef2 EWG, 5′-gatccTTTTTTGCGCATGCGCTCCTTCa-3′ and 5′-gatctGAAGGAGCGCATGCGCAAAAAAg-3′; and MEF2C NRF1, 5′-gatccTTCGGTGCGCGCGCGAATGCGCAAGCCCa-3′ and 5′-gatctGGGCTTGCGCATTCGCGCGCGCACCGAAg-3′.
Chromatin Immunoprecipitation—Three 15-cm dishes of C2C12 or HEK 293 cells were grown to confluence prior to cross-linking in situ, nuclear isolation, DNA shearing, preclearing, and immunoprecipitation. Procedures used were modified from the ChIP-IT kit (Active Motif) using enzymatic DNA shearing as described previously in detail (19). Results were taken only when validated with both positive and negative precipitation and PCR controls. Processing of three independent cross-linked samples gave similar results. Primers for human sequences were: chromosome (chr.) 12 intergenic (174 bp), 5′-atggttgccactggggatct-3′ and 5′-tgccaaagcctaggggaaga-3′; GAPDH, chr. 12 (166 bp), 5′-tactagcggttttacgggcg-3′ and 5′-tcgaacaggaggagcagagagcga-3′; MEF2A, chr. 15 (199 bp) 5′-accgagaggataattcagtcctg-3′ and 5′-acatccgcgcacggatc-3′; PPARGC1A, chr. 4 (204 bp), 5′-gagatggacaatgaagaacagtg-3′ and 5′-agttcccaggagatgtacacg-3′; and GLUT4, chr. 17 (242 bp), 5′-aaggcgtcatctccctgtc-3′ and 5′-aactctgcgggtctggac-3′. Primers for murine sequences were: intergenic, chr. 6 (248 bp), 5′-aacctcatggttgccacag-3′ and 5′-accacgagatctgtaggcaag-3′; gapdh, chr. 6 (207 bp), 5′-agctactcgcggctttacg-3′ and 5′-tcacctggcactgcacaag-3′; mef2a, chr. 7 (233 bp), 5′-accgagagcagaaatatacccta-3′ and 5′-gagccgcctccttcagc-3′; ppargc1a, chr. 5 (124 bp), 5′-gagcacattaaattaacctcagtgg-3′ and 5′-ccagctcatttcctttacttgac-3′; glut4, chr. 11 (215 bp), 5′-taaggctccatctcctttgc-3′ and 5′-gtatgggctacatgtacttgcc-3′; and (cox6ah, chr. 7, (179 bp) 5′-ggatctcctgccagtcaagac-3′ and 5′-ttagaggcagagccattgtca-3′. Immunoprecipitations used rabbit anti-NRF1 (Abcam, ab34682), mouse anti-RNA polymerase II (Upstate Biologicals, clone 8WG16), rabbit anti-MEF2A (19), or control IgG.
Cultured Cell Transfection—C2C12 cells were maintained and differentiated as described (17–19). HeLa and HEK 293 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum. S2 cells were maintained at 25 °C in DS2 medium with 10% fetal bovine serum. HEK 293 cells in 15-cm dishes were transfected with 10 μgof pCDNA-NRF1VP16/GFP prior to GFP+ versus GFP– fluorescence-activated cell sorting after 48 h. Promoter reporter assays in mammalian cells in 12-well plates were performed as described using Superfect (Qiagen) (19). S2 cells in 6-well plates were transfected at 50% confluence. Triplicate wells received 3 μg of reporter plasmid and 1 μg of expression vector, and cells were harvested for luciferase assays after 48 h. 10T1/2 cells stably transformed with a MyoD-ER fusion were maintained in medium with G418 and 10% charcoal-stripped fetal bovine serum, and myogenesis was induced using 10 nm estradiol as described (17).
RNA Interference—siRNA transfections were performed using HiPerFect (Qiagen) according to the supplier instructions. C2C12 cells were transfected every 24 h for 2 days with subdivisions to maintain cell subconfluence, and RNA was harvested after a total elapsed time of 72 h. HEK 293 cells were transfected once 48 h before RNA harvesting at 80% cell confluence. 75 ng/well (12-well plate) or 150 ng/well (6-well plate) of dsRNA was used in transfections. Double-stranded siRNAs all had 19 bp corresponding to the targeted mRNA and 3′ UU extensions on each strand. Human siRNA sense sequences were: GAPDH, 5′-GUCAACGGAUUUGGUCGUAUU-3′; NRF1, 5′-GAAACGGCCUCAUGUAUUUUU-3′, 5′-UAGUAUAGCUCAUCUUGUAUU-3′, 5′-CACAUUGGCUGAUGCUUCAUU-3′, and 5′-GCUAUUGUCCUCUGUAUCUUU-3′. Mouse siRNA sequences were: gapdh, 5′-GUGUGAACCACGAGAAAUAUUUU-3′; nrf1, 5′-GAAUGAACGCCACCGAUUUUU-3′, 5′-CAGUAUAGCUCAUCUCGUAUU-3′, 5′-UGAAAUAAGCCUCCCGAUAUU-3′, and 5′-CAACAGGGAAGAAACGGAAUU-3′. A control siRNA that fails to recognize any human or mouse targets (Qiagen catalog No. 1027280) was used as a negative control. An Alex Fluor 488-labeled negative control ds-siRNA (Qiagen) was used initially to optimize transfection conditions.
Miscellaneous Reagents and Procedures—All plasmid segments derived from PCR were verified by dideoxy sequencing (21). Mouse anti-myc epitope (clone 9E10) was from United States Biologicals, and anti-NRF1 antiserum was the generous gift of Richard Scarpula (Northwestern).
RESULTS
MEF2 Regulatory Elements Are Common to Heart/Muscle-specific Respiratory Chain Subunit Gene Promoters—We examined the distribution of cox6a and cox7a isoform mRNA in adult mouse tissues using RPA to discriminate between the highly similar mRNA sequences. Consistent with previous reports (22), cox6aH was present exclusively in skeletal muscle and heart, whereas cox6aL was expressed in all other tissues examined (Fig. 1). cox7aH was also expressed exclusively in striated muscle, and although cox7aL was highest in non-muscle tissues, it was present in skeletal muscle and quite abundant in heart. cox7aR, a third isoform of this subunit (23), was expressed ubiquitously at a relatively low level. Using 10T1/2 MyoD-ER cells wherein myogenesis is induced by estrogen (24), isoform switching from cox6aL to cox6aH coincided with the appearance of a myosin heavy chain skeletal muscle differentiation marker (not shown).
FIGURE 1.
Tissue-specific COX subunit 6A and 7A isoform genes have conserved promoter elements. cRNA probes (p) specific for murine cox6a and cox7a isoforms were used in RPA with 2 μg of mouse brain (B), heart (H), liver (L), skeletal muscle (M), and testis (T) total RNA. Assays presented in the two cox7aL panels used probes from C57BL/6J (left) and 129 strain templates (right), which differ at 2 nucleotides. RNA was isolated from 129 (heart) and C57BL/6J (other tissues) mice. The locations of MEF2 (diamonds) and NRF1 (hexagons) elements in the COX subunit gene promoters are shown below each RPA.
The COX6A, COX7A, and COX8 (H and L forms of each) are compact genes, each having a single promoter and splicing pattern and one initiation codon within the first exon (8). We compared 5′-flanking region sequences of each of these genes among mammals to identify conserved elements (Fig. 1). Transgenic promoter-reporter studies in mice have shown that MEF2 and E box elements provide heart- and muscle-specific expression of mouse cox6aH (16). The corresponding region of other mammalian COX6AH genes is highly conserved (25). Specifically, the region has either one canonical MEF2 element or two sites separated by 7–10 bp (supplemental Table S1). COX7AH and COX8H gene regulatory regions also have functional MEF2 elements (15, 16). No sequences corresponding to NRF1 or NRF2 elements were detected in COXH promoters. By contrast, both the COX6AL and COX7AL promoters have putative or documented NRF1 or NRF2 elements (2, 3, 8), but none has a MEF2 site. Although the COX7AR gene promoter region has not been functionally characterized, we found NRF1 and MEF2 elements within 1.2 kb of the transcription start site (TSS). No other transcription factor binding sites that are conserved and selectively present in the COXH versus COXL promoters were found.
Forced Expression of NRF1 Induces MEF2A Expression—Four mammalian MEF2 genes encode MEF2 factors6 (18, 26). These genes have overlapping but distinct patterns of expression, and the respective protein products also have both unique and redundant functions (26, 27). Our in silico analyses and the prior mouse cox6ah promoter-reporter transgene studies (16) led us to suspect that COXH transcription is regulated by one or more MEF2 factors. Coordinated expression with other ETC components could then be given by a NRF if it controls expression of the cognate MEF2 gene(s). nrf1 expression is known to precede expression of the mef2 genes during mouse embryonic development and to be present in differentiating muscle (11), establishing one prerequisite for such a cascade.
Overexpression of native NRF1 does not typically give robust activation of target genes either in vivo (28) or in cultured mammalian cells (2). We therefore forced expression of a NRF1 fusion to the strong transactivating domain from viral VP16 (NRF1VP16) to determine whether MEF2 gene expression might be controlled by NRF1. Enhanced green fluorescent protein was co-expressed with NRF1VP16 from a viral promoter using an internal ribosome entry site between the fusion and marker coding sequences (Fig. 2A). Human HEK 293 cells were used because they express all MEF2 isotype mRNAs at modest levels under control conditions (29). Transfected cells (GFP+) were separated from untransfected cells (GFP–) by flow cytometry, and gene expression was compared in the populations. MEF2A mRNA was strongly induced in the NRF1VP16/GFP+ cells as confirmed by RPA (Fig. 2B), implicating MEF2A as a target of NRF1 regulation.
FIGURE 2.
NRF1 activity induces MEF2A expression. A, diagram of the construct used to co-express NRF1VP16 and enhanced green fluorescent protein (EGFP) and the scheme for RNA isolation from control and experimental cell populations. B, RPA using RNA from NRF1VP16/GFP-expressing and control cells using a human MEF2A cRNA probe (19).
MEF2A Promoter 1 Contains a Conserved Canonical NRF1 Element—We mapped the 5′-regulatory region of human MEF2A. The gene has two closely approximated alternative first exons, A1 and A2, and cognate promoters ∼65 kb upstream of the first coding exon 1 (Fig. 3, A and B). A MEF2 element overlies the major promoter 1 TSS (TSS p1), and we have shown that this site provides for MEF2A transcriptional autoregulation (19). Of particular interest to our hypothesis, a canonical NRF1 (2, 4) element is centered at –47 relative to TSS p1 (Fig. 3C and supplemental Fig. S1). Two E boxes and two potential NRF2 sites (2, 5) are located further upstream (Fig. 3B and supplemental Fig. S1). The MEF2A 5′-regulatory region within ∼0.8 kb upstream of TSS p1 is highly conserved among mammals, and the MEF2, NRF, and E box elements are each conserved in sequence and location (supplemental Fig. S1). The proximal promoter 1 location of the putative NRF1 element and its evolutionary conservation strongly suggested functional relevance.
We performed both mobility shift and chromatin immunoprecipitation (ChIP) assays to verify NRF1 binding to the putative MEF2A promoter element. In the former, one specific retarded complex was formed in resolved binding reactions of recombinant epitope-tagged NRF1 (NRF1myc) and a dsDNA probe containing the element (Fig. 3E). This complex was not competed by otherwise analogous probes containing either two (m1NRF1) or four (m2NRF1) substitutions within the GC core repeats of the element (Fig. 3D). The complex was supershifted in reactions containing anti-myc antibody, confirming a recombinant NRF1 component. Specific complexes were also formed with the probe and C2C12 myoblast and myotube nuclear protein extracts, and these were supershifted in resolved reactions containing anti-NRF1 antibody (Fig. 3F).
Endogenous nuclear NRF1 interaction with the MEF2A promoter element was verified using ChIP. PCR primers were designed to amplify the region surrounding TSS p1 that includes the NRF1 and MEF2 elements (Fig. 3B), or a downstream region. Anti-NRF1 antibody specifically co-precipitated the MEF2A promoter fragment from HEK 293 (human) cells (Fig. 3G). Similar findings were obtained in ChIP assays using C2C12 (murine) cells and primers designed to detect the analogous region of mef2a (not shown). We had previously shown that MEF2A factors also bind this promoter region (19). NRF1 is therefore positioned to directly regulate MEF2A transcription and to influence its autoregulation.
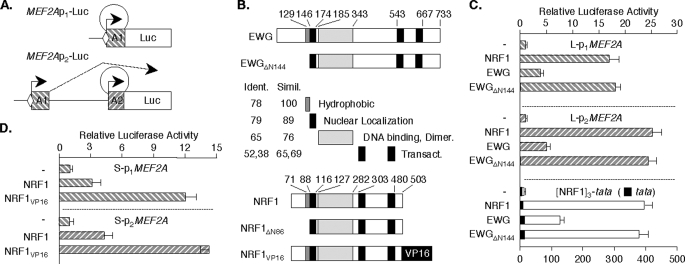
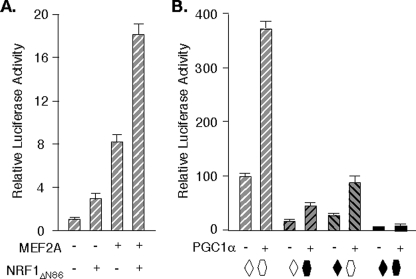
Forced NRF1 Expression Induces MEF2A Promoter Activity—To evaluate the influence of the NRF1 element and factor on MEF2A promoter 1 and 2 activities, we used the MEF2Ap1-Luc and MEF2Ap2-Luc reporters, respectively (19) (Fig. 4A). In general, target gene promoters are only modestly sensitive to NRF1 overexpression in mammalian cells (30). By contrast, forced expression of NRF1 (or its ortholog, EWG) has been reported to have robust activity in cultured Drosophila cells (20, 30). We therefore began by examining the effects of NRF1 expression on the MEF2A reporters in Drosophila S2 cells. Our focus on the NRF1 element led us to use the S-MEF2Ap1-Luc and S-MEF2Ap2-Luc constructs that contain only proximal promoter fragments in order to exclude superfluous upstream enhancers (19). We expressed either human NRF1 or its Drosophila ortholog, EWG, the product of erect wing (20, 31), or a fusion or mutant of either (Fig. 4B) from the Drosophila β-actin promoter. In these studies, MEF2Ap1-Luc activity was induced 17-fold and MEF2Ap2-Luc 25-fold in cells expressing full-length NRF1 (Fig. 4C). Forced expression of EWG gave less potent effects, but an EWG mutant deleted for the N terminus (EWGΔN144) (20) activated both MEF2A promoters to a similar extent as NRF1. These transcription factors gave up to 400-fold stimulation of [NRF1MEF2A]3-tata-Luc, a reporter containing 3 copies of the NRF1 site, but had minimal effect on the parent tata-Luc control. The MEF2A promoters are highly responsive to forced expression of NRF1 in the nuclear milieu provided by this cell type.
FIGURE 4.
Forced expression of NRF1 stimulates MEF2A promoter activity. A, schematic of MEF2A promoter-reporters (19). B, diagram of the domain structures of NRF1 and EWG, and mutants and fusions used. Drosophila S2 cells in 6-well plates (C) and HeLa cells in 12-well plates (D) were transfected in triplicate for each condition with indicated reporters (S2, 3 μg/well; HeLa, 1 μg/well) and SV40-βgal (0.3 μg/well). After 48 h, cell extract luciferase activities were determined, and values were normalized for transfection efficiency using β galactosidase. Data represent the average ± S.E. of 3 independent transfections.
To evaluate MEF2A promoter responses to NRF1 overexpression in mammalian cells, reporter activity was studied in cells expressing intact NRF1, a deletion mutant analogous to EWGΔN144 (NRF1ΔN86), or the NRF1VP16 fusion. In HeLa cells, NRF1 expression produced ∼3-fold activation of MEF2Ap1-Luc and ∼4-fold activation of MEF2Ap2-Luc (Fig. 4D). Although modest, these responses are equal to or greater than those reported for other promoter reporters in mammalian cells (7, 30). As expected, more robust effects were produced by NRF1VP16, which stimulated these reporters 12- and 15-fold, respectively. These studies suggested that NRF1 co-regulates the MEF2A gene promoters through its proximal promoter 1 element.
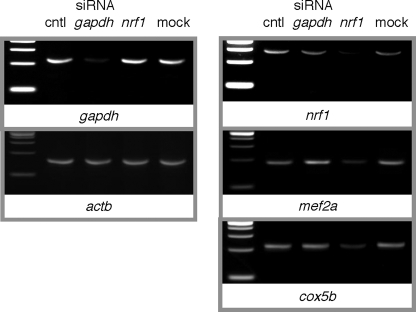
Myocyte mef2a mRNA Is Reduced with nrf1 RNA Interference—We used RNAi to study the effects of nrf1 knockdown on expression of endogenous mef2a mRNA in muscle cells. C2C12 cells were transfected with dsRNA targeted to nrf1. Control cells received an siRNA that targets gapdh, one that fails to hybridize with any known message, or no dsRNA in a mock transfection. Transfection conditions were optimized using Alexa Fluor 488-tagged control siRNA. Because myoblasts proliferate rapidly, we performed serial transfections with a low concentration (5 nm) of the siRNAs. Total cell RNA, harvested at cell confluence after three transfections at doubling time intervals, was used in reverse transcriptase reactions. Validated primer pairs specific for gapdh and nrf1 were used in quantitative PCR, with values normalized to sample β-actin (actb) mRNA levels. Messenger RNAs of these targets were each selectively down-regulated to 8 and to 15% of control levels, respectively, with transfection of the cognate siRNA, as can be seen in stained electrophoretic gels of the reaction products (Fig. 5). Levels of the non-cognate siRNA-transfected cell and control cell mRNAs were not affected and were indistinguishable from those in mock-transfected cells.
FIGURE 5.
nrf1 RNA interference leads to down-regulation of mef2a mRNA in cultured myocytes. Subconfluent C2C12 cells were transfected with 5 nm ds-siRNA directed at no gene (cntl, control), gapdh, or nrf1 at daily intervals for 3 days followed by RNA harvesting at cell confluence. QPCR was used on cDNA produced from these samples to determine nrf1, gapdh, mef2a, cycs, and cox5b mRNA levels in samples, each normalized to the respective actb level. One set of parallel reactions in a confirmatory QPCR was stopped at the midpoint of exponential amplification of the control condition sample, and reaction products were electrophoresed on 4% agarose gels containing ethidium bromide. Similar results were obtained in three independent siRNA transfections.
Having established the conditions under which nrf1 expression in myocytes could be down-regulated, we tested the consequences for mef2a expression. As controls, we examined effects on the expression of established targets of NRF1, including the cycs and cox5b genes that encode the somatic form of cytochrome c and the cytochrome c oxidase Vb subunit, respectively (4). Validated primer pairs for mef2a, cycs, and cox5b were used, with the former pair designed to detect all splicing variants of mef2a (18). In an experimental series, the respective mef2a, cycs, and cox5b mRNA expression levels were reduced in nrf1 siRNA-transfected cells to 33, 30, and 25% of control levels (Fig. 5). Together with the NRF1 overexpression study findings, this clearly demonstrates a strong correlation between cellular NRF1 levels and MEF2A gene expression.
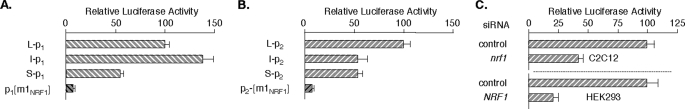
MEF2A Promoter Activity Is Sensitive to NRF1 Element Mutation and to RNAi-mediated Down-regulation of nrf1 Expression—C2C12 transfections with a deletion series of both MEF2A promoter-reporters, including short (S), intermediate (I), and long (L) promoter constructs, showed that preferential activity in myocytes was maintained in the (S) reporters (19). These constructs each retain nearly 50% of full-length promoter function (Fig. 6, A and B). MEF2A reporters with an NRF1 element mutation corresponding to one that failed in binding competition assays (p1- and p2-[m1NRF1]-Luc) had markedly compromised activity compared with the cognate wild-type promoter reporters, consistent with positive control of MEF2A transcription from this element. Similar results were obtained in HEK 293 cells (not shown). This element therefore appears to be crucial to full promoter activity in a variety of cell types in which MEF2A is expressed.
FIGURE 6.
MEF2A promoter NRF1 element mutation and nrf1 RNAi reduce promoter activity in myocytes. Indicated MEF2A promoter 1 (A) or promoter 2 (B) reporters were co-transfected with SV40-βgal into C2C12 cells. Data were analyzed as described for Fig. 4D. C, subclonfluent C2C12 cells (upper panel) in a 12-well plate were transfected with 5 nm ds-siRNA directed at no gene (control) or nrf1. After 72 h, cells were co-transfected with S-MEF2ap2-Luc and SV40-βgal followed by analyses as described for Fig. 4D. Human HEK 293 cells (lower panel) were transfected once with 5 nm ds-siRNA directed at no gene (control) or NRF1. After 2 days, cells were cotransfected with S-MEF2Ap2-Luc and SV40-βgal and analyzed as described for Fig. 4D except that cell extracts were harvested after 24 h.
To confirm that RNAi-mediated NRF1 underexpression reduced mef2a mRNA by a transcriptional mechanism, we evaluated MEF2A promoter-reporter activity in C2C12 and HEK 293 cells. In each case, cells were transfected with control or NRF1 siRNAs as for the mRNA studies followed after 48 h by co-transfection of the S-MEF2Ap2-Luc reporter. NRF1 knockdown led to significant reductions in MEF2A promoter activity to 42 and 25% of controls in C2C12 and 293 cells, respectively (Fig. 6C). The rapid proliferation rate of these cells probably means that this understates the impact of NRF1 down-regulation on MEF2A promoter activity. Taken together with the NRF1 element mutation studies, these results confirm that NRF1 plays a critical role in MEF2A transcription in two diverse cell types, including differentiating muscle cells.
PGC1α Co-activates the MEF2A Promoters from the NRF1 and MEF2 Elements—The close proximity of the MEF2A promoter NRF1 and MEF2 elements suggested the possibility of an interaction. Indeed, the activities of reporters with or without mutations in the MEF2 and NRF1 sites were consistent with element cooperation (Fig. 7B). Under conditions where activity of MEF2Ap2-Luc was ∼5-fold higher than that of p2[m1NRF1]-Luc and 3-fold higher than a reporter with a MEF2 site mutation (p2[m1MEF2]-Luc) (19), it was nearly 20-fold higher than a reporter with mutations in both elements. Forced expression of MEF2A α2/β and NRF1ΔN86 synergistically activated the MEF2A promoter in HeLa cells, also suggesting cooperation of the elements (Fig. 7A). No direct protein-protein interaction between MEF2 and NRF1 was noted in a mammalian two-hybrid system (not shown), eliminating one possible mechanism for element synergy.
FIGURE 7.
PGC1α regulates MEF2A transcription from the adjacent MEF2 and NRF1 promoter elements. A, HeLa cells were transfected as described for Fig. 4G with p2MEF2A-Luc and expression vectors for MEF2A α2/β and NRF1ΔN86 as indicated. Values are normalized to the activity of the reporter in the absence of co-transfection (= 1.0). Averages from three independent experiments are shown. B, C2C12 cells were transfected with S-MEF2Ap2-Luc reporters containing wild-type (open symbols) or mutated (solid symbols) MEF2A (19) and NRF1 elements, with or without pCDNA-PGC1α, and analyzed as in A, except that activities were normalized to wild-type promoter-reporter (= 100%).
NRF1 and MEF2 are each known to interact with PPARγ coactivator 1α (PGC1α) (32–36). This suggested that MEF2A transcription might be sensitive to the abundance of this cofactor. We therefore examined the sensitivity of MEF2A promoter activity to forced expression of this coactivator. As shown in Fig. 7B, PGC1α stimulated MEF2Ap2-Luc activity ∼4-fold in C2C12 cells, which is similar to PGC1α activities on other promoters such as those of the TFBM genes (7). This effect is mediated from the NRF1 and MEF2 elements, as mutation of either site resulted in attenuation in the PGC1α effect, and the response was nearly abolished on p2[m1NRF1/m1MEF2]-Luc. Co-recruitment of PGC1α is one possible mechanism for cooperative control of MEF2A transcription by MEF2 and NRF1.
Endogenous Muscle Cell MEF2A Binds the Promoters of Genes Encoding COX VIaH, PGC1α, and MEF2A—There are four MEF2 isotype genes in all mammals. These genes have distinct but overlapping expression patterns and functions (18, 26, 27). These genes encode either one protein (MEF2B)6 or multiple splicing variant proteins (MEF2A, MEF2C, and MEF2D) that have a common N-terminal DNA-binding and dimerization domain (17, 18), such that all MEF2 proteins can heterodimerize promiscuously. Expression of the MEF2 isotypes is induced at different stages of cultured myoblast differentiation, but each is highly expressed in differentiated myotubes (18). We therefore used C2C12 myotubes as substrate for ChIP assays to confirm occupation of gene promoters relevant to our hypothesis by MEF2A.
We developed a highly specific anti-MEF2A antibody that recognizes all splicing variants of MEF2A but no other MEF2 form. This antibody had been used by us previously to demonstrate MEF2A binding to the MEF2 element in the MEF2A promoter (19). We performed ChIP assays with C2C12 myotubes to verify that MEF2A proteins also bind the cox6aH and ppargc1a gene promoters at the respective MEF2 element regions. The gapdh gene promoter, which is not regulated or bound by MEF2 proteins, served as a negative control. As shown in Fig. 8, MEF2A co-precipitated with chromatin containing the cox6aH and ppargc1a gene fragments but failed to precipitate gapdh. Thus, in this setting of co-expressed MEF2 isotype and splicing variant proteins, MEF2A forms occupy the promoters of MEF2A, COXH, and PPARGC1A. This work does not exclude co-occupation by other MEF2 forms.
FIGURE 8.
Myocyte MEF2A protein isoforms recognize promoter elements of the cox6ah and ppargc1a genes. ChIP assays were performed using C2C12 myotube nuclear extracts and an antibody that recognizes all four splicing variants of MEF2A but no other MEF2 proteins (19). This antibody was used previously to show MEF2A binding to the MEF2 element in the MEF2A gene (19). Primers specific for promoter regions of murine cox6aH, ppargc1a, and gapdh were used to detect co-precipitated gene fragments.
DISCUSSION
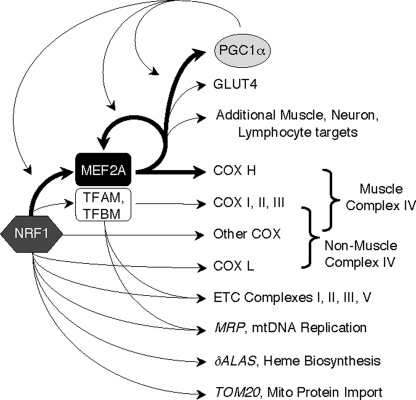
Mito density is particularly rich in cardiac and skeletal muscle, where OXPHOS is brisk. The ETC is unique in these tissues by virtue of the expression of tissue-specific complex IV subunits, COX VIaH and COX VIIaH. Although the functions of these and other non-catalytic COX subunits are incompletely understood, there is evidence to suggest that COXH confer sensitivity of COX to the cellular energy state. In specific, activity of COX isolated from striated muscle is activated by ADP, whereas enzyme purified from other tissues is not (14). In addition, an antibody that specifically recognizes COX VIaH neutralizes this ADP sensitivity. It is therefore speculated that COX VIaH provides for rapid adaptation of ETC activity to energy stores in muscle, where energy demands can vary acutely and dramatically (1, 14). We explored a mechanism that could orchestrate tissue-specific expression of COXH subunits at levels commensurate with other ETC components and the mito and mtDNA density in muscle. We found that COXH gene promoters have evolutionarily conserved MEF2 elements, that myocyte MEF2A occupies the cox6ah promoter, and that MEF2A expression is in turn regulated by NRF1. We therefore propose that a NRF1 → MEF2A → COXH cascade functions in parallel with the NRF → TFAM → ETCmito and NRF → ETCnucl pathways to control respiration in striated muscle (Fig. 9).
FIGURE 9.
MEF2A, with NRF1 and PGC1α, is part of a triad of transcriptional regulators of striated muscle metabolism. Key transcriptional pathways and representative targets of processes controlled by NRF1, MEF2A, and PGC1α are shown, including ramifications of MEF2A regulation by these three factors. Bold arrows indicate transcriptional control demonstrated in this or a recent publication (19). δALAS, δ-aminolevulinic acid synthase; MRP, mito RNA processing RNA; TOM20, translocase of outer membrane component 20.
We have provided multiple lines of evidence that the closely approximated alternative MEF2A promoters are co-regulated by NRF1, the first limb of the proposed transcriptional cascade. We confirmed the evolutionary conservation of a canonical NRF1 binding site in MEF2A promoter 1. Both mobility shift assays with muscle cell nuclear extracts and myotube ChIP assays verified endogenous nuclear NRF1 protein association with this promoter. NRF1 homodimers recognize a 12-bp site, YGCGCAYGCGCR, and there is little tolerance for sequence degeneracy (4). By consequence of this and the presence of multiple CpG doublets, canonical NRF1 sites are predicted to appear in a mammalian genome at a frequency of only 10–8 bp.
This location at –47 relative to the promoter 1 TSS (and ∼600 bp upstream of the promoter 2 TSS) and our DNA binding study results strongly support a role for this site in regulating MEF2A transcription. We saw no change in NRF1 binding to the site during myoblast differentiation using either assay. This was not surprising because this factor is constitutively bound to its DNA targets (37, 38).
A second line of evidence in support of MEF2A as a target of NRF1 was provided by experiments using forced expression of NRF1, N-terminal deletion mutants of NRF1 and its Drosophila ortholog EWG, or a NRF1VP16 fusion protein. We initially showed that the MEF2A mRNA level in cultured HEK cells was strongly induced with expression of NRF1VP16. We used the VP16 transactivation domain fusion in these exploratory studies because the forced expression of native NRF1 is known to induce rather small changes in target gene expression. For example, NRF1 overexpression led to only an ∼2-fold increases in TFAM promoter activity (32), and a synthetic minimal promoter containing four NRF1 elements is typically used to demonstrate transactivity (30, 32). Likewise, transgenic expression of NRF1 in muscle to a level that exceeded the endogenous level by a factor of 10 produced only 1.5- and 2-fold increases in cytochrome c (cycs) and δ-aminolevulinic acid synthase (δ-alas) mRNAs, respectively (28). NRF1VP16 was therefore a valuable tool in the detection of MEF2A as a potential NRF1 target. Unlike the case in cultured mammalian cells, exogenous expression of NRF1 or EWG has been shown by other investigators to produce potent activation of target gene promoters in Drosophila cells (20, 30). We were able to exploit this model system to develop further evidence for the authenticity of MEF2A as a target of NRF1. We also saw modest but significant effects of NRF1 overexpression on MEF2A promoter-reporters in mammalian cells that were similar to those reported for other promoters.
Evidence that specifically addressed the role of NRF1 in controlling MEF2A transcription in muscle came from studies of the function of MEF2A promoter-reporters in C2C12 myocytes. Mutation of the NRF1 element produced a drastic decrease in reporter activity. Furthermore, nrf1 RNAi in this and other cell types led to coincident reductions in nrf1 and mef2a mRNA levels, as well as diminished MEF2A promoter activity. We believe that the composite of evidence reported here confirms that MEF2A transcription is controlled by NRF1. MEF2A is expressed in neurons, adipocytes, in various immune cell types, and in smooth muscle, in addition to skeletal and cardiac myocytes. We have previously provided evidence that MEF2A has only two promoters, p1 and p2, which do not have tissue-specific activities (19). Because NRF1 is ubiquitously expressed (39) and we have shown here that p1 and p2 are coregulated by this factor, NRF1 can probably control MEF2A expression in non-muscle cells as well as in skeletal muscle and heart. The effect of NRF1 RNAi on MEF2A expression and promoter activity in HEK 293 cells supports this contention.
In differentiating mouse myoblasts in vitro, there is a quantum burst in mef2a expression at the time of cell cycle withdrawal (18). There is recent evidence that the transactivity of NRF1 is sensitive to cell cycle-regulated phosphorylation (40). In specific, Cdk4 or Cdk6, in association with the regulatory subunit cyclin D1, can phosphorylate NRF1 Ser-47, which leads to diminished transactivity. Disinhibition then occurs with cell cycle withdrawal because of the associated reduction in cyclin D1 levels and NRF1poS47. Negative regulation of NRF1 activity by this mechanism is consistent with previous studies of the EWGΔN144 N-terminal deletion mutant (20), as well as with our present findings with this and the analogous NRF1ΔN86 mutant. In each case, the mutants were more potent transactivators of NRF1-responsive promoters than the full-length native factors in proliferating cells. We speculate that cyclin D1/Cdk4 activity on NRF1 may account for or be involved in the upsurge in mef2a expression noted at cell cycle withdrawal. This stated, NRF1 transactivity and DNA binding are also positively regulated by incompletely characterized serum-responsive phosphorylation events (30, 38) and possibly in response to cellular redox state (41). In addition, this factor may repress transcription of some or all target genes under some circumstances (37). This suggests additional regulatory complexity with relevance to MEF2A transcription that will require further study.
nrf1 mRNA is detectable very early in embryonic development and is ubiquitously expressed in developing and mature tissues (11, 39). Disruption of murine nrf1 leads to embryonic failure at a very early stage (3.5–6.5 days postcoitus) (11), prior to the appearance of mef2a during normal development. Tests of the role of NRF1 in controlling temporal and spatial expression of MEF2A in vivo will therefore require regulated interference with nrf1 expression and/or conditional nrf1 knockout, complemented by paired transgenic MEF2A and MEF2A[mNRF1] promoter studies. In addition to the NRF1 element, the MEF2A 5′-regulatory region has other conserved elements (supplemental Fig. S1). A canonical MEF2 element overlies the major TSS of one of the alternative TATA-less promoters, and MEF2 activity at this site confers transcriptional autoregulation and sensitivity of MEF2A expression to stress signaling (19). Additional sites of potential relevance to both muscle-specific expression and mito function are also present, including E boxes and putative NRF2 sites. Explorations of the functions of each of these sites are progressing in our laboratory.
Despite evidence presented here that NRF1 controls MEF2A expression as part of a transcriptional cascade, we cannot exclude a role for other MEF2 genes in this network. Indeed, one of the several MEF2C promoters does have conserved NRF1 elements (supplemental Fig. S2), suggesting that this isotype may also be a target. MEF2C proteins may therefore participate in the proposed transcriptional cascade in some cell types or circumstances. However, neither MEF2C, MEF2B, nor MEF2D mRNA was induced with forced expression of NRF1VP16 under conditions where cultured cell MEF2A expression was strongly stimulated. MEF2A may therefore play a unique role among the isotypes as intermediary in communicating NRF1 activity. This is compatible with the mef2a–/– mouse phenotype. These animals have deficient and disorganized myocardial mito and a neonatal sudden death syndrome, consistent with a defect in oxidative metabolism (42).
Although it is clear that COXH expression is controlled from promoter MEF2 elements, any of the four mammalian MEF2 isotype proteins could conceivably act at these sites. We had previously found that available anti-MEF2 antibodies do not display MEF2 isotype specificities (19). We used a new validated isotype-specific antibody and myotube chromatin to demonstrate by ChIP that endogenous MEF2A occupies the cox6ah promoter. Because disruption or knockdown of one MEF2 gene results in the dysregulation of other isotypes (42, 43),6 we believe that this is the best evidence for a direct regulation of COXH genes by MEF2A, the second limb of our hypothesized transcriptional cascade. mef2a is expressed in striated muscle precursors in the developing embryo after day 9 post-coitus, and expression is maintained in differentiated tissues after birth (26, 42, 44, 45). In differentiating mouse myoblasts in vitro, mef2a expression increases dramatically upon cell cycle withdrawal (18), coincident with the appearance of coxH (22). Thus, MEF2A abundance in muscle in vivo and in vitro closely mimics expression of the heart/muscle forms of COX VIa and VIIa. This contrasts with the expression pattern of mef2c, which is present at high levels during embryonic development but subsequently diminishes drastically (26, 44, 45), and mef2d, which is an early marker of the myogenic lineage and is maintained in differentiated skeletal muscle (44–46). This stated, the MEF2A N-terminal MADS/MEF2 region that mediates dimerization and sequence-specific DNA binding is shared among all MEF2 proteins (26). We therefore cannot exclude co-regulation of the COXH genes by other MEF2 forms or by heterodimers of MEF2A with MEF2B, MEF2C, or MEF2D at different stages of muscle development or differentiation.
MEF2A is highly expressed in skeletal muscle and heart, but it is also present at other sites including neural, adipose, and immune cells (17, 18, 26). Selective expression of COXH genes may therefore require muscle-specific activities of either MEF2A or its collaborating factors. Certain splicing variants of MEF2A are expressed only in heart and muscle7 (17, 18), providing one potential mechanism for a muscle-specific MEF2A activity. Signaling that controls MEF2 protein or cofactor modifications may also be operative, particularly among pathways regulated by intramyocellular Ca2+ transients (47). Myogenic basic helix-loop-helix factors could also contribute to muscle-specific expression, because these factors can use DNA-bound MEF2 as scaffolding to transactivate MEF2 target genes (48), and these factors could also regulate MEF2A transcription from the promoter E boxes. The strict specificity of COX6AH expression suggests that gene silencing must also occur in non-muscle cells, the mechanisms for which remain to be determined.
The control of MEF2A transcription by NRF1 has implications beyond the coordinated co-expression of ETC subunits in muscle. Specifically, other genes that are regulated by MEF2A are implicated by our findings as potential indirect targets of NRF1. The GLUT4 gene provides one important example that has particular relevance to muscle and cardiac metabolism. Glut4 is the insulin-responsive facilitated glucose transporter form that provides the major route for glucose uptake in striated muscle (49–51). Both cultured myocyte and transgenic promoter-reporter studies have established that MEF2A regulates glut4 expression through a promoter MEF2 element (49, 50). Muscle tissue Glut4-level, insulin-stimulated glucose transport and MEF2A protein concentration were each found to be increased ∼2-fold in mice carrying a transgene that overexpresses NRF1 in skeletal muscle compared with control tissue (28). Our detection of MEF2A as a target of NRF1 suggests the relevant mechanism, NRF1 → mef2a → glut4, because glut4 is not a direct target of NRF1. Given the tissue expression patterns of both NRF1 and MEF2A, many more genes are likely to be regulated by such a cascade in both muscle and non-muscle sites.
PGC1α was originally identified as a coactivator of PPARγ and NRF1 in brown adipose tissue, but it is now recognized as a key regulator of mito biogenesis in various tissues (32, 52). PGC1α also coactivates MEF2 factors (33, 35, 36), and its forced expression in muscle increases mito density and OXPHOS and produces a fast-twitch/glycolytic to slow-twitch/oxidative phenotype conversion (53). It is established that PGC1α co-activates transcription from a PPARGC1A promoter MEF2 site (33, 35). We have shown here with myotube ChIP that MEF2A factors bind this PPARGC1A element where they are poised to recruit PGC1α in this autoregulatory loop. We have also shown that PGC1α co-activates the MEF2A promoter from its MEF2 and NRF1 elements, confirming that PGC1α can also feed back on MEF2A transcription. Taken together, this indicates that MEF2A, NRF1, and PPARGC1A and their respective protein products form a mutually reinforcing network of auto- and cross-regulation capable of directing mito biogenesis and OXPHOS capacity in muscle (Fig. 9). The NRF1 promoter has not been characterized, although the 5′-exons of human NRF1 have been identified (54). There is a canonical MEF2 binding site upstream of the human NRF1 TSS,7 suggesting the possibility of direct reciprocal MEF2A and NRF1 regulation. We are exploring whether this vector is also part of this transcriptional network.
The regulation of MEF2A transcription by NRF1 and PGC1α is highly relevant to metabolic dysregulation in diabetes. Muscle glut4 mRNA, Glut4 protein, and insulin-stimulated glucose uptake are reduced in animal models of diabetes, and there is a coincident down-regulation of mef2a mRNA and MEF2A protein abundance (50, 51, 55). In these models, a hypercatabolic state leads to a decline in the [AMP]/[ATP] ratio and a coincident reduction in 5′-AMP-activated protein kinase (AMPK) activity (55–58). Because expression of both mef2a and of glut4 can be restored with the administration of a small molecular activator of AMPK (55), this pathway is implicated as a crucial sensor linking cell energy state with the capacity for nutrient uptake and metabolism (55, 56). There is recent evidence to suggest that AMPK activity regulates nuclear DNA binding activity of both NRF1 (59) and MEF2 (60). Neither factor appears to be a direct target of AMPK (60). AMPK may therefore target co-repressor(s) of one or both factors to promote dissociation and de-repression of targets such as MEF2A (19, 61, 62). As one alternative, AMPK activity could indirectly influence the subnuclear locus or co-activator associations of these factors. In any case, down-regulated MEF2A expression may be a primary mechanism by which NRF1 and PGC1α targets are coordinately down-regulated in humans with diabetes and insulin resistance (63).
We saw no evidence for a direct protein-protein interaction between NRF1 and MEF2A. These factors may cooperate in the recruitment of transcriptional co-regulator(s) or the induction of chromatin remodeling to account for the observed functional synergy. Paired MEF2 and NRF1 elements also exist in other MEF2 gene regulatory regions, including the aforementioned MEF2C promoter (supplemental Fig. S2) and the Drosophila DMef2 II-E enhancer (supplemental Fig. S3). The DMef2 5′-region has various enhancers that govern the complex developmental and spatial expression of the sole MEF2 gene in the fly (64). II-E is responsible for transcriptional autoregulation (65) and for DMef2 expression near and after terminal differentiation of somatic muscle (64). We find that a canonical EWG element in this region binds NRF1 and governs enhancer activity (supplemental Fig. S3). Control of MEF2 transcription by NRF1 may therefore be conserved among higher metazoans. The sea urchin NRF1 ortholog, P3A2, directs territory-specific transcription of muscle genes during embryonic development (66), and EWG is known to regulate flight muscle development (31). Our work suggests a potentially relevant mechanism and establishes a foundation for in vivo functional analyses in various model systems to elucidate the contributions of NRF1/EWG to the developmental and spatial expression of MEF2 genes.
Supplementary Material
Acknowledgments
We thank Grace Gill (Tufts University), Richard Scarpulla (Northwestern University), and Stephen Tapscott (University of Washington) for reagents and Shiguang Li and Pan Yin for technical assistance.
This work was supported in part by grants from the American Heart Association and the Juvenile Diabetes Foundation; Grant P30-DK40561 from the Clinical Nutrition Research Center at Harvard; and Grants DK55875, DK02461, and HL72713 from the National Institutes of Health (all to T. G.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
Footnotes
The abbreviations used are: ETC, electron transport chain; AMPK, 5′-AMP-activated protein kinase; Cdk, cylin-dependent kinase; ChIP, chromatin immunoprecipitation; COX, cytochrome c oxidase; CYCS, cytochrome c, somatic form; EWG, erect wing; GLUT4, facilitated glucose transporter 4; MEF2, myocyte enhancer factor 2; mito, mitochondri(on/a/al); NRF, nuclear respiratory factor; OXPHOS, oxidative phosphorylation; PPAR, peroxisome proliferator-activated receptor; PGC1α, PPARγ coactivator 1α; QPCR, quantitative polymerase chain reaction; RPA, ribonuclease protection assay; TSS, transcription start site; ds, double stranded; HEK, human embryonic kidney; GFP, green fluorescent protein; siRNA, small interfering RNA; RNAi, RNA interference; chr, chromosome.
Genes encoding the H and L forms of COX VII are sometimes referred to as COX7A1 and COX7A2, respectively.
Primary transcripts of mammalian MEF2A, MEF2C, and MEF2D, but not MEF2B, are alternatively spliced among exons containing coding sequences to produce multiple splicing variants or isoforms. In this paper we refer to MEF2 isotype proteins as those encoded by the different genes and to MEF2 isoform or variant proteins as those encoded by splicing variants.
T. Gulick, unpublished observations.
References
- 1.Scheffler, I. E. (1999) Mitochondria, pp. 141–245, Wiley-Liss, New York
- 2.Scarpulla, R. C. (2006) J. Cell. Biochem. 97 673–683 [DOI] [PubMed] [Google Scholar]
- 3.Kelly, D. P., and Scarpulla, R. C. (2004) Genes Dev. 18 357–368 [DOI] [PubMed] [Google Scholar]
- 4.Virbasius, C. A., Virbasius, J. V., and Scarpulla, R. C. (1993) Genes Dev. 7 2431–2445 [DOI] [PubMed] [Google Scholar]
- 5.Gugneja, S., Virbasius, J. V., and Scarpulla, R. C. (1995) Mol. Cell. Biol. 15 102–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Virbasius, J. V., and Scarpulla, R. C. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 1309–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gleyzer, N., Vercauteren, K., and Scarpulla, R. C. (2005) Mol. Cell. Biol. 25 1354–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lenka, N., Vijayasarathy, C., Mullick, J., and Avadhani, N. G. (1998) Prog. Nucleic Acid Res. Mol. Biol. 61 309–344 [DOI] [PubMed] [Google Scholar]
- 9.Li, B., Holloszy, J. O., and Semenkovich, C. F. (1999) J. Biol. Chem. 274 17534–17540 [DOI] [PubMed] [Google Scholar]
- 10.Chau, C. M., Evans, M. J., and Scarpulla, R. C. (1992) J. Biol. Chem. 267 6999–7006 [PubMed] [Google Scholar]
- 11.Huo, L., and Scarpulla, R. C. (2001) Mol. Cell. Biol. 21 644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taanman, J. W., Hall, R. E., Tang, C., Marusich, M. F., Kennaway, N. G., and Capaldi, R. A. (1993) Biochim. Biophys. Acta 1225 95–100 [DOI] [PubMed] [Google Scholar]
- 13.Tripoli, G., D'Elia, D., Barsanti, P., and Caggese, C. (2005) Genome Biol. 6 R11.1–R11.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anthony, G., Reimann, A., and Kadenbach, B. (1993) Proc. Natl. Acad. Sci. U. S. A. 90 1652–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu, M., Jaradat, S. A., and Grossman, L. I. (2002) Biochim. Biophys. Acta 1574 345–353 [DOI] [PubMed] [Google Scholar]
- 16.Wan, B., and Moreadith, R. W. (1995) J. Biol. Chem. 270 26433–26440 [DOI] [PubMed] [Google Scholar]
- 17.Zhu, B., and Gulick, T. (2004) Mol. Cell. Biol. 24 8264–8275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu, B., Ramachandran, B., and Gulick, T. (2005) J. Biol. Chem. 280 28749–28760 [DOI] [PubMed] [Google Scholar]
- 19.Ramachandran, B., Yu, G., Li, S., Zhu, B., and Gulick, T. (2008) J. Biol. Chem. 283 10318–10329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fazio, I. K., Bolger, T. A., and Gill, G. (2001) J. Biol. Chem. 276 18710–18716 [DOI] [PubMed] [Google Scholar]
- 21.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., and Struhl, K. (1998) Current Protocols in Molecular Biology, John Wiley & Sons, Inc., New York
- 22.Parsons, W. J., Williams, R. S., Shelton, J. M., Luo, Y., Kessler, D. J., and Richardson, J. A. (1996) Am. J. Physiol. 270 H567–H574 [DOI] [PubMed] [Google Scholar]
- 23.Watanabe, T., Inoue, S., Hiroi, H., Orimo, A., Kawashima, H., and Muramatsu, M. (1998) Mol. Cell. Biol. 18 442–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergstrom, D. A., Penn, B. H., Strand, A., Perry, R. L., Rudnicki, M. A., and Tapscott, S. J. (2002) Mol. Cell 9 587–600 [DOI] [PubMed] [Google Scholar]
- 25.Bachman, N. J., Riggs, P. K., Siddiqui, N., Makris, G. J., Womack, J. E., and Lomax, M. I. (1997) Genomics 42 146–151 [DOI] [PubMed] [Google Scholar]
- 26.Black, B. L., and Olson, E. N. (1998) Annu. Rev. Cell Dev. Biol. 14 167–196 [DOI] [PubMed] [Google Scholar]
- 27.Shalizi, A. K., and Bonni, A. (2005) Curr. Top. Dev. Biol. 69 239–266 [DOI] [PubMed] [Google Scholar]
- 28.Baar, K., Song, Z., Semenkovich, C. F., Jones, T. E., Han, D. H., Nolte, L. A., Ojuka, E. O., Chen, M., and Holloszy, J. O. (2003) FASEB J. 17 1666–1673 [DOI] [PubMed] [Google Scholar]
- 29.Zhao, M., New, L., Kravchenko, V. V., Kato, Y., Gram, H., di Padova, F., Olson, E. N., Ulevitch, R. J., and Han, J. (1999) Mol. Cell. Biol. 19 21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herzig, R. P., Scacco, S., and Scarpulla, R. C. (2000) J. Biol. Chem. 275 13134–13141 [DOI] [PubMed] [Google Scholar]
- 31.DeSimone, S., Coelho, C., Roy, S., VijayRaghavan, K., and White, K. (1996) Development (Camb.) 122 31–39 [DOI] [PubMed] [Google Scholar]
- 32.Wu, Z., Puigserver, P., Andersson, U., Zhang, C., Adelmant, G., Mootha, V., Troy, A., Cinti, S., Lowell, B., Scarpulla, R. C., and Spiegelman, B. M. (1999) Cell 98 115–124 [DOI] [PubMed] [Google Scholar]
- 33.Handschin, C., Rhee, J., Lin, J., Tarr, P. T., and Spiegelman, B. M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7111–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Handschin, C., and Spiegelman, B. M. (2006) Endocr. Rev. 27 728–735 [DOI] [PubMed] [Google Scholar]
- 35.Czubryt, M. P., McAnally, J., Fishman, G. I., and Olson, E. N. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1711–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michael, L. F., Wu, Z., Cheatham, R. B., Puigserver, P., Adelmant, G., Lehman, J. J., Kelly, D. P., and Spiegelman, B. M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 3820–3825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cam, H., Balciunaite, E., Blais, A., Spektor, A., Scarpulla, R. C., Young, R., Kluger, Y., and Dynlacht, B. D. (2004) Mol. Cell 16 399–411 [DOI] [PubMed] [Google Scholar]
- 38.Gugneja, S., and Scarpulla, R. C. (1997) J. Biol. Chem. 272 18732–18739 [DOI] [PubMed] [Google Scholar]
- 39.Gopalakrishnan, L., and Scarpulla, R. C. (1995) J. Biol. Chem. 270 18019–18025 [DOI] [PubMed] [Google Scholar]
- 40.Wang, C., Li, Z., Lu, Y., Du, R., Katiyar, S., Yang, J., Fu, M., Leader, J. E., Quong, A., Novikoff, P. M., and Pestell, R. G. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 11567–11572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piantadosi, C. A., and Suliman, H. B. (2006) J. Biol. Chem. 281 324–333 [DOI] [PubMed] [Google Scholar]
- 42.Naya, F. J., Black, B. L., Wu, H., Bassel-Duby, R., Richardson, J. A., Hill, J. A., and Olson, E. N. (2002) Nat. Med. 8 1303–1309 [DOI] [PubMed] [Google Scholar]
- 43.Lin, Q., Schwarz, J., Bucana, C., and Olson, E. N. (1997) Science 276 1404–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian, S. V., and Nadal-Ginard, B. (1996) Mech. Dev. 57 103–112 [DOI] [PubMed] [Google Scholar]
- 45.Edmondson, D. G., Lyons, G. E., Martin, J. F., and Olson, E. N. (1994) Development (Camb.) 120 1251–1263 [DOI] [PubMed] [Google Scholar]
- 46.Breitbart, R. E., Liang, C. S., Smoot, L. B., Laheru, D. A., Mahdavi, V., and Nadal-Ginard, B. (1993) Development (Camb.) 118 1095–1106 [DOI] [PubMed] [Google Scholar]
- 47.Wu, H., Naya, F. J., McKinsey, T. A., Mercer, B., Shelton, J. M., Chin, E. R., Simard, A. R., Michel, R. N., Bassel-Duby, R., Olson, E. N., and Williams, R. S. (2000) EMBO J. 19 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molkentin, J. D., Black, B. L., Martin, J. F., and Olson, E. N. (1995) Cell 83 1125–1136 [DOI] [PubMed] [Google Scholar]
- 49.Liu, M. L., Olson, A. L., Edgington, N. P., Moye-Rowley, W. S., and Pessin, J. E. (1994) J. Biol. Chem. 269 28514–28521 [PubMed] [Google Scholar]
- 50.Mora, S., and Pessin, J. E. (2000) J. Biol. Chem. 275 16323–16328 [DOI] [PubMed] [Google Scholar]
- 51.Thai, M. V., Guruswamy, S., Cao, K. T., Pessin, J. E., and Olson, A. L. (1998) J. Biol. Chem. 273 14285–14292 [DOI] [PubMed] [Google Scholar]
- 52.Lin, J., Handschin, C., and Spiegelman, B. M. (2005) Cell Metab. 1 361–370 [DOI] [PubMed] [Google Scholar]
- 53.Lin, J., Wu, H., Tarr, P. T., Zhang, C. Y., Wu, Z., Boss, O., Michael, L. F., Puigserver, P., Isotani, E., Olson, E. N., Lowell, B. B., Bassel-Duby, R., and Spiegelman, B. M. (2002) Nature 418 797–801 [DOI] [PubMed] [Google Scholar]
- 54.Huo, L., and Scarpulla, R. C. (1999) Gene 233 213–224 [DOI] [PubMed] [Google Scholar]
- 55.Finck, B. N., Bernal-Mizrachi, C., Han, D. H., Coleman, T., Sambandam, N., LaRiviere, L. L., Holloszy, J. O., Semenkovich, C. F., and Kelly, D. P. (2005) Cell Metab. 1 133–144 [DOI] [PubMed] [Google Scholar]
- 56.Zong, H., Ren, J. M., Young, L. H., Pypaert, M., Mu, J., Birnbaum, M. J., and Shulman, G. I. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 15983–15987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cuthbertson, D. J., Babraj, J. A., Mustard, K. J., Towler, M. C., Green, K. A., Wackerhage, H., Leese, G. P., Baar, K., Thomason-Hughes, M., Sutherland, C., Hardie, D. G., and Rennie, M. J. (2007) Diabetes 56 2078–2084 [DOI] [PubMed] [Google Scholar]
- 58.Towler, M. C., and Hardie, D. G. (2007) Circ. Res. 100 328–341 [DOI] [PubMed] [Google Scholar]
- 59.Bergeron, R., Ren, J. M., Cadman, K. S., Moore, I. K., Perret, P., Pypaert, M., Young, L. H., Semenkovich, C. F., and Shulman, G. I. (2001) Am. J. Physiol. 281 E1340–E1346 [DOI] [PubMed] [Google Scholar]
- 60.Holmes, B. F., Sparling, D. P., Olson, A. L., Winder, W. W., and Dohm, G. L. (2005) Am. J. Physiol. 289 E1071–E1076 [DOI] [PubMed] [Google Scholar]
- 61.Berdeaux, R., Goebel, N., Banaszynski, L., Takemori, H., Wandless, T., Shelton, G. D., and Montminy, M. (2007) Nat. Med. 13 597–603 [DOI] [PubMed] [Google Scholar]
- 62.van der Linden, A. M., Nolan, K. M., and Sengupta, P. (2007) EMBO J. 26 358–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patti, M. E., Butte, A. J., Crunkhorn, S., Cusi, K., Berria, R., Kashyap, S., Miyazaki, Y., Kohane, I., Costello, M., Saccone, R., Landaker, E. J., Goldfine, A. B., Mun, E., DeFronzo, R., Finlayson, J., Kahn, C. R., and Mandarino, L. J. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 8466–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nguyen, H. T., and Xu, X. (1998) Dev. Biol. 204 550–566 [DOI] [PubMed] [Google Scholar]
- 65.Cripps, R. M., Lovato, T. L., and Olson, E. N. (2004) Dev. Biol. 267 536–547 [DOI] [PubMed] [Google Scholar]
- 66.Calzone, F. J., Hoog, C., Teplow, D. B., Cutting, A. E., Zeller, R. W., Britten, R. J., and Davidson, E. H. (1991) Development (Camb.) 112 335–350 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.