Abstract
Polycystin-1 (PC1) may play an important role in skeletogenesis through regulation of the bone-specific transcription factor Runx2-II. In the current study we found that PC1 co-localizes with the calcium channel polycystin-2 (PC2) in primary cilia of MC3T3-E1 osteoblasts. To establish the role of Runx2-II in mediating PC1 effects on bone, we crossed heterozygous Pkd1m1Bei and Runx2-II mice to create double heterozygous mice (Pkd1+/m1Bei/Runx2-II+/-) deficient in both PC1 and Runx2-II. Pkd1+/m1Bei/Runx2-II+/- mice exhibited additive reductions in Runx2-II expression that was associated with impaired endochondral bone development, defective osteoblast-mediated bone formation, and osteopenia. In addition, we found that basal intracellular calcium levels were reduced in homozygous Pkd1m1Bei osteoblasts. In contrast, overexpression of a PC1 C-tail construct increased intracellular calcium and selectively stimulated Runx2-II P1 promoter activity in osteoblasts through a calcium-dependent mechanism. Site-directed mutagenesis of critical amino acids in the coiled-coil domain of PC1 required for coupling to PC2 abolished PC1-mediated Runx2-II P1 promoter activity. Additional promoter analysis mapped the PC1-responsive region to the “osteoblast-specific” enhancer element between -420 and -350 bp that contains NFI and AP-1 binding sites. Chromatin immunoprecipitation assays confirmed the calcium-dependent binding of NFI to this region. These findings indicate that PC1 regulates osteoblast function through intracellular calcium-dependent control of Runx2-II expression. The overall function of the primary cilium-polycystin complex may be to sense and transduce environmental clues into signals regulating osteoblast differentiation and bone development.
Embryonic bone is formed from mesenchymal stem cells by either direct differentiation of these cells into mineralized matrix-generating osteoblasts (intramembranous bone) or by their condensation and subsequent formation of a cartilage template that is replaced by osteoblast-mediated bone formation (endochondral bone formation) (1-3). Runx2 is a master transcription factor controlling skeletogenesis that regulates the differentiation of mesenchymal precursors into osteoblasts and hypertrophic chondrocytes (4-10). The total loss of Runx2 in the mouse results in the complete absence of intramembranous and endochondral bone formation. Expression of Runx2 is initiated from the distal P1 and the proximal P2 promoters that, respectively, give rise to N-terminal distinct Runx2-type II (Runx2-II) and Runx2-type I (Runx2-I) isoforms. Selective deletion of the P1 promoter and Runx2-II in mice results in impaired terminal osteoblastic maturation and endochondral bone formation (5, 7, 11). The P2 promoter regulation of Runx2-I is sufficient for early osteoblastogenesis and intramembranous bone formation (11-13). The presence of regulatory sequences in the P1 promoter required for osteoblast-specific expression of Runx2-II have been identified (14), but the developmentally relevant signal transduction pathways that target this region are incompletely understood.
Inactivating mutations of PKD1,2 the gene that causes polycystic kidney disease, are associated with defects in skeletogenesis and abnormal Runx2-II expression (15-17). The PKD1 gene product, polycystin-1 (PC1), is a 4303-amino acid cell surface-expressed protein that has a multidomain extracellular region (18, 19), an 11-transmembrane region, and an ∼200-amino acid cytoplasmic C-terminal tail. PC1 interacts with polycystin-2 (PC2) (encoded by PKD2), a receptor-activated calcium channel (20-23), and the PC1 and PC2 heterodimers colocalized to primary cilia, a cell surface organelle involved in development, chemosensing, and mechanosensing (24-27). Primary cilium, polycystin-1 (PC1), and polycystin-2 (PC2) are present in osteoblasts (15). In addition, the homozygous Pkd1m1Bei mutant mouse, which has an inactivating missense mutation of PC1, is characterized by both a decrease in the expression of Runx2-II in bone and delayed endochondral bone formation that resembles selective Runx2-II-deficient mice (15). Moreover, overexpression of constitutively active PC1 C-terminal constructs in MC3T3-E1 osteoblasts results in an increase in Runx2-II P1 promoter activity, endogenous Runx2 expression, and osteoblast differentiation markers (15).
In the current study we investigate the role of PC1 in the regulation of P1-Runx2-II expression and skeletal development. Using mouse genetic approaches to create combined PC1 and Runx2-II-deficient mice, we demonstrate a functional linkage between PC1, Runx2-II expression, and skeletal development in vivo. In addition, we show that PC1 regulates the osteoblast-specific enhancer element in the Runx2-II P1 promoter through intracellular calcium-dependent mechanisms. These findings establish that PC1 is an important regulator of skeletal development through the selective regulation of Runx2-II.
EXPERIMENTAL PROCEDURES
Mice—Pkd1m1Bei heterozygous mice, which have an inactivated point mutation in the Pkd1 gene leading to the substitution of an arginine for methionine in the first transmembrane domain (28), were obtained from the Mutant Mouse Regional Resource Center (University of North Carolina, Chapel Hill, NC). The selective Runx2-II-deficient mice were generated in our laboratory as previously described (11). These mice were bred and maintained on a C57BL/6J background. We crossed Pkd1+/m1Bei mutant mice with Runx2-II+/- mice to create double heterozygous mice lacking one functional allele of both Runx2-II and Pkd1. Animal experiments were performed after review and approval by the University of Kansas Medical Center Animal Care and Use Committee.
Immunofluorescence Staining—MC3T3-E1 osteoblasts were grown on collagen-coated coverslips and kept at confluence for at least 3 days. An acetylated α-tubulin antibody (Sigma) was used to identify the primary cilium. PC1- and PC2-specific antibodies (Santa Cruz Biotechnology, Inc. Santa Cruz, CA) were used to localize PC1 and PC2. We used a fluorescein isothiocyanate-conjugated secondary antibody and fluorescent microscopy to identify the primary cilium and PC1/PC2 complexes. The secondary antibody in the absence of a primary antibody was used as a negative control. 4′,6-Diamidino-2-phenylindole blue staining was used to identify cell nuclei (15).
Whole Skeletal Mount Alizarin Red/Alcian Blue Staining and Histological Preparations—Whole mouse carcasses were collected from newborn mice after euthanasia, defatted for 2-3 days in acetone, stained sequentially with Alcian blue and Alizarin red S in 2% KOH, cleared with 1% KOH, 20% glycerol, and stored in 50% ETOH, 50% glycerol. Femurs and tibias from newborn mice were decalcified at 4 °C in 12.5% EDTA, 2.5% paraformaldehyde in phosphate-buffered saline. Longitudinal sections of bone were stained with hematoxylin and eosin to assess the histology of the growth plate as previously described (11, 15).
Bone Densitometry and Histomorphometric Analysis—Bone mineral density (BMD) of femurs was assessed at 6 weeks of age using a LUNARPIXIMUS bone densitometer (Lunar Corp., Madison, WI). Calcein (Sigma) double labeling of bone was performed as previously described (12, 15). Quantitative histomorphometric analyses of bone sections were performed using the Osteomeasure analysis system (Osteometrics).
Micro-CT Analysis—For newborn mice, the whole mouse skeleton and tibias were scanned using a μCT 40 in high resolution (Scanco Medical AG, Bassersdorf, Switzerland). 100 slices of the metaphyses under the growth plate, constituting 0.6 mm in length, and 50 slices of the diaphysis, constituting 0.3 mm in length, were selected. For 6-week-old mice, the distal metaphyses and the mid-shaft regions of diaphyses in femurs were scanned using a middle resolution. 100 slices of the metaphyses under the growth plate, constituting 1.2 mm in length, and 50 slices of the diaphysis, constituting 0.6 mm in length, were selected. A three-dimensional image analysis was done to determine bone volume (bone volume/total volume) and cortical bone thickness as previously described (12).
Promoter Deletion, Site Mutagenesis, and Transient Transfection—MC3T3-E1 cells were cultured in α-minimal essential medium containing 10% fetal bovine serum and 1% penicillin/streptomycin. The PKD1-(115-226) and PKD1-(1-92) constructs were provided by Dr. Seth L. Alper at Harvard Medical School (29). The c-Jun plasmid was obtained from Dr. Hong Lu at the University of Kansas Medical Center. We used the rVISTA program to map the PC1-reponsive element in the Runx2-II P1 promoter (30). For promoter deletion analysis, we used a PCR strategy to create a series of 5′ deletions of the Runx2-II P1 promoter, ranging from -420, -398, -382, -370, -362, -350 to +1, that were cloned as NotI-HindIII fragments into the pluc4 reporter construct. We used a PCR strategy to generate previously described amino acid substitutions at sites 148, 149, and 155 of PC1-AT to create a mutant construct (PC1-ATmutant) that was unable to couple to PC2 (31). The P1 deletion promoter-luciferase reporter constructs and PC1-AT constructs were transiently co-transfected into MC3T3-E1 osteoblasts by electroporation using Cell Line Nucleofector Kit R according to the manufacturer's protocol (Amaxa Inc, Gaithersburg, MD). Promoter activity was assessed by measuring luciferase activity 24 and 48 h after transfection in the presence or absence of the specified pharmacological treatments as previously described (15).
Intracellular Calcium Measurements—For basal intracellular calcium ([Ca2+]i) measurements, we used the Fluo-4 NW calcium assay kit (Invitrogen). Briefly, osteoblasts were plated at 50,000 cells per well in a 96-well plate and grown overnight. After removing the growth medium, cells were directly incubated in the 1× Fluo-4 NW dye loading solution at 37 °C for 30 min. The Fluo-4 fluorescence intensity (494/516 nm) was measured in each well using the Synergy HT Fluorescence Microplate Reader according to manufacturer's instruction (32).
Reverse Transcription (RT)-PCR Analysis—RT-PCR was performed using the Titan™ One tube RT-PCR kit (Roche Applied Science). Briefly, total RNA was isolated from either non-transfected MC3T3-E1 osteoblasts or MC3T3-E1 osteoblasts that had been transiently transfected with either the control sIgØ or the PC1-AT constructs for 48 h. RT-PCR was carried out with NFI-specific degenerate primers Deg1 (5′-TTCCGGATGARTTYCAYCITTYATYGARGC-3′, where R is purine (A or G), Y is pyrimidine (C or T), S is C or G, and I is inosine) and Deg2 (5′-AATCGATRTGRTGSGGCTGIAYRCAIAG-3′) as previously described (33). The 486-bp amplified product was digested with BamHI, Bpu10I, and NarI and then separated on 2% agarose gel. Mouse hypoxanthine-guanine phosphoribosyltransferase was amplified as a control for the RT-PCR reactions.
Real-time RT-PCR—For quantitative real-time RT-PCR, 2.0 μg of total RNA isolated from newborn mice and the long bone of 6-week-old mice were reverse-transcribed as previously described (15). PCR reactions contained 100 ng of template (cDNA or RNA), 300 nm each forward and reverse primers, and 1× iQ™ SYBR® Green Supermix (Bio-Rad) in 50 μl. The threshold cycle (Ct) of tested-gene product from the indicated genotype was normalized to the Ct for cyclophilin A.
Quantitative Chromatin Immunoprecipitation (ChIP) Analyses—The ChIP analyses were performed using the ChIP-IT kit (Active Motif, Carlsbad CA) and modifications of previously described methods (34). Briefly, MC3T3-E1 osteoblasts were transiently transfected with PC1-AT for 72 h and treated with 1% formaldehyde to cross-link chromatin. Immunoprecipitation was performed using anti-NFI antibody (Santa Cruz Biotechnology) and protein A-agarose. The specific protein-DNA complex was reversely cross-linked, and DNA fragments were purified. Real-time PCR was performed using primers located in the Runx2 P1 promoter. Primers amplifying sequences in exon 4 of Runx2 were used to normalize for DNA content and to calculate the relative ratio of the P1 promoter sequences over control sequences. -Fold enrichment reflects the ratio of promoter/control sequences in the immunoprecipitate versus the input samples. Normal rabbit IgG was used as a control.
Statistics—We evaluated differences between groups by one-way analysis of variance. All values are expressed as means ± S.E. All computations were performed using the Statgraphic statistical graphics system (STSC Inc.).
RESULTS
Pkd1 Selectively Regulates P1-Runx2-II Expression in Vivo—Because Pkd1 null mice are embryonic lethal and Runx2-II-/- mice have high perinatal mortality, to establish a potential functional link between Pkd1- and Runx2-II-dependent bone development, we created double heterozygous mice lacking one functional allele of Runx2-II and Pkd1. This was accomplished by crossing Pkd1+/m1Bei mutant mice with heterozygous Runx2-II+/- mice to create double heterozygous Pkd1+/m1Bei/Runx2-II+/- animals. We observed an approximate 35% reduction of Runx2-II expression in newborn heterozygous Pkd1+/m1Bei mice, comparable with the levels found in heterozygous Runx2-II+/- mice. Consistent with Pkd1 regulation of Runx2-II expression, we observed further reductions of Runx2-II expression in combined Pkd1+/m1Bei and Runx2-II+/- mice to levels that were 50% that observed in wild-type mice (Fig. 1). These changes were selective for P1-Runx2-II, since we observed no differences in P2-Runx2-I isoform expression in Pkd1-deficient mice (Fig. 1). We also found that reduction of Runx2-II expression persisted in adult mice, as evidenced by a respective 35 and 40% reduction in single heterozygous Pkd1+/m1Bei and Runx2-II+/- mice and a 66% reduction in 6-week-old double Pkd1+/m1Bei/Runx2-II+/- mice (Table 1). In contrast, the P2-Runx2-I isoform expression was not different between wild-type, single heterozygous, and double heterozygous mice, consistent with our prior observation that Pkd1 selectively regulates the Runx2-II isoform driven by the P1 promoter (15).
FIGURE 1.
Combined effect of Pkd1 and Runx2-II deficiency on Runx2 isoform message expression in newborn mice. An additive reduction of Runx2-II, but not Runx2-I, message level was observed in double Pkd1+/m1Bei/Runx2-II+/- mice compared with either single heterozygous Pkd1+/m1Bei or Runx2-II+/- mice by real-time RT-PCR. Data are expressed as the mean ± S.E. from four to five individual newborn mice. Values sharing the same superscript (a, b, or c) are not significantly different at p < 0.05.
TABLE 1.
Gene-expression profiles in 6-week-old mice
Dmp1, dentin matrix protein 1; VegfA, vascular endothelial growth factor A; Trap, tartrate-resistant acid phosphatase; Mmp, matrix metalloproteinase; PTHrP, parathyroid hormone-like peptide. Data are the mean ± S.E. from 6 tibias of 6-week-old individual mice and expressed as the-fold changes relative to the housekeeping gene cyclophilin A subsequently normalized to wild-type mice.
| Gene | Accession no. | Pkd1+/m1Bei | Runx2-II+/− | Pkd1+/m1Bei/Runx2-II+/− | p value |
|---|---|---|---|---|---|
| Osteoblast | |||||
| Runx2-II | NM_009820 | 0.65 ± 0.07a | 0.57 ± 0.07a | 0.34 ± 0.07a,b | <0.0001 |
| Runx2-I | D14636 | 0.86 ± 0.13 | 0.97 ± 0.13 | 0.85 ± 0.13 | 0.783 |
| Runx2 | NM_009820 | 0.72 ± 0.05a | 0.69 ± 0.05a | 0.54 ± 0.05a,b | <0.0001 |
| Osterix | AF184902 | 0.97 ± 0.11 | 0.98 ± 0.11 | 0.68 ± 0.11a | 0.1522 |
| Alkaline phosphatase | NM_007431 | 0.79 ± 0.07 | 0.76 ± 0.07a | 0.51 ± 0.07a | 0.0702 |
| Osteocalcin | NM_007541 | 0.66 ± 0.06a | 0.57 ± 0.06a | 0.39 ± 0.06a,b | <0.0001 |
| Osteopontin | AF515708 | 0.68 ± 0.08a | 0.65 ± 0.08a | 0.51 ± 0.08a | 0.003 |
| Collagen I | NM_007742 | 0.63 ± 0.08a | 0.53 ± 0.08a | 0.52 ± 0.08a | 0.0007 |
| Osteoprotegerin | MMU94331 | 0.72 ± 0.09a | 0.85 ± 0.09 | 0.78 ± 0.09a | 0.0458 |
| Rank ligand | NM_011613 | 0.59 ± 0.07a | 0.57 ± 0.07a | 0.35 ± 0.07a,b | <0.0001 |
| Mmp13 | NM_008607 | 0.70 ± 0.06a | 0.68 ± 0.06a | 0.49 ± 0.06a,b | <0.0001 |
| Osteocyte | |||||
| Dmp1 | MMU242625 | 0.59 ± 0.06a | 0.53 ± 0.06a | 0.35 ± 0.06a,b | <0.0001 |
| Phex | NM_011077 | 0.63 ± 0.06a | 0.59 ± 0.06a | 0.34 ± 0.06a,b | <0.0001 |
| Osteoclast | |||||
| Trap | NM_007388 | 0.62 ± 0.07a | 0.54 ± 0.07a | 0.32 ± 0.07a,b | <0.0001 |
| Mmp9 | NM_013599 | 0.55 ± 0.05a | 0.56 ± 0.05a | 0.35 ± 0.05a,b | <0.0001 |
| Chondrocyte | |||||
| VegfA | NM_009505 | 0.92 ± 0.11 | 0.92 ± 0.11 | 0.89 ± 0.11 | 0.9013 |
| Indian hedgehog | BC046984 | 0.92 ± 0.13 | 1.05 ± 0.13 | 0.89 ± 0.13 | 0.8249 |
| PTHrP | BC058187 | 0.92 ± 0.13 | 0.97 ± 0.13 | 0.83 ± 0.13 | 0.8039 |
| Collagen X | NM_009925 | 0.83 ± 0.11 | 0.91 ± 0.12 | 0.78 ± 0.11 | 0.5162 |
| Sox9 | NM_011448 | 0.70 ± 0.13 | 0.87 ± 0.12 | 0.71 ± 0.13 | 0.3026 |
| Collagen II | NM_031163 | 0.92 ± 0.16 | 0.96 ± 0.16 | 0.87 ± 0.16 | 0.9419 |
Significant difference from wild type.
Significant difference from heterozygous Pkd1+/m1Bei and Runx2-II+/− mice at p < 0.05, respectively.
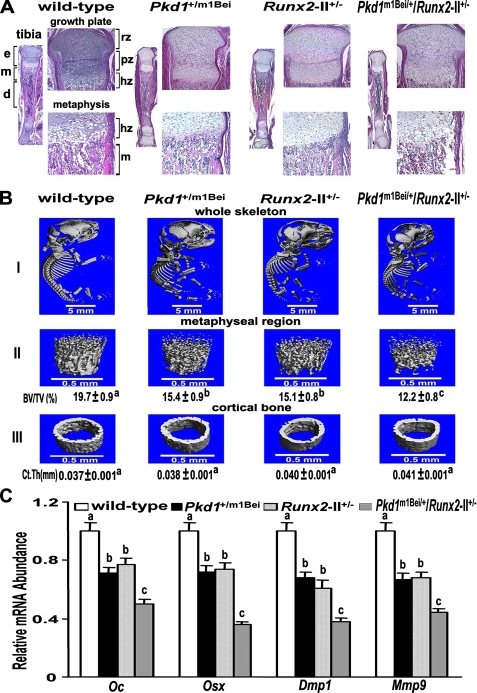
Additive Effects of Combined Pkd1 and Runx2-II Deficiency on Embryonic Bone Development—Mice were born at the expected Mendelian frequency. The overall survival of single heterozygous Pkd1+/m1Bei and Runx2-II+/- and double heterozygous Pkd1+/m1Bei/Runx2-II+/- newborn mice was not different from wild-type littermates; however, double Pkd1+/m1Bei/Runx2-II+/- mice were smaller and had a significantly (p < 0.05) lower body weight compared with wild-type and single heterozygous Pkd1+/m1Bei and Runx2-II+/- littermates (Fig. 2A). No apparent abnormalities of bone were seen in Alizarin red/Alcian blue-stained skeletal specimens of newborn single heterozygous Runx2-II+/- and Pkd1+/m1Bei mice, consistent with our previous report (11, 15). Double Pkd1+/m1Bei/Runx2-II+/- mice, however, displayed demonstrable defects in endochondral bone formation, including delayed ossification of occipital bones, posterior zygomatic arches, distal clavicles, the hyoid bone, distal ribs, phalangeal bones, and the caudal spine (Fig. 2B). In addition, Pkd1+/m1Bei/Runx2-II+/- mice exhibited a wider anterior fontanelle that was evident in Alizarin red/Alcian blue-stained skeletal preparations (Fig. 2B) and by μCT analysis of skeleton (Fig. 3B), indicating a concomitant abnormality of intramembranous bone formation.
FIGURE 2.
Additive effect of Pkd1 and Runx2-II deficiency on embryonic bone development. A, gross appearance and body weight of newborn mice. Body weight (expressed in mg) represents the mean ± S.E. from four to five individual newborn mice per genotype. Values sharing the same superscript (a or b) are not significantly different at p < 0.05. B, Alizarin red/Alcian blue staining of wild-type, Runx2-II+/-, Pkd1+/m1Bei, and double heterozygous Pkd1+/m1Bei/Runx2-II+/- newborn mice. Calcified tissues are stained red, and cartilage is stained blue. Panels I-V, respectively, represents a superior view of the skull, clavicles, hyoid bone, sternum and ribs, scapula and forelimb (left image), and caudal vertebrae (right image). Arrows indicated delayed ossification of specific bones.
FIGURE 3.
Skeletal abnormalities in combined Pkd1- and Runx2-II-deficient newborn mice. A, hematoxylin and eosin staining of decalcified tibias. Low magnification (20×) of entire tibia and high magnification of growth plate (100×) and metaphysic (200×) shows less trabecular bone volume, a narrow bone collar, and a reduced size of the bone marrow cavity in double Pkd1+/m1Bei/Runx2-II+/- mice. e, epiphysis; m, metaphysis; d, diaphysis; rz, chondrocyte resting zone; pz, chondrocyte proliferating zone; hz, chondrocyte hypertrophic zone. B, representative three-dimensional μCT images of the complete skeleton (panel I), metaphyseal region (panel II), and cortical bone (panel III) in Pkd1+/m1Bei/Runx2-II+/- mice. Pkd1+/m1Bei/Runx2-II+/- mice exhibited wider anterior fontanelles, diminished metaphyseal bone volume, and relative preservation of cortical bone width. C, effect of Pkd1 and Runx2-II deficiency on Runx2-dependent message expression in bone by real-time RT-PCR. Double Pkd1+/m1Bei/Runx2-II+/- mice exhibited an additive reduction in osteocalcin (Oc), osterix (Osx), Dmp1, and Mmp9 compared with single heterozygous Pkd1m1Bei and Runx2-II mice. Data are expressed as the mean ± S.E. from four to five individual samples. BV/TV, bone volume/total volume; Ct.Th, cortical bone thickness. Values sharing the same superscript (a, b, or c) are not significantly different at p < 0.05.
The histological appearance of the tibia from newborn wild-type and single heterozygous Pkd1+/m1Bei and Runx2-II+/- mice was similar, but the trabecular bone volume in the metaphyseal region was markedly reduced in double Pkd1+/m1Bei/Runx2-II+/- mice (Fig. 3A). μCT analysis also identified similar reductions of trabecular bone volume (BV/TV) in single heterozygous Runx2-II+/- and Pkd1+/m1Bei mice, which was more severe in double Pkd1+/m1Bei/Runx2-II+/- mice (Fig. 3B). Interestingly, no significant changes were observed in cortical thickness (Ct.Th) (Fig. 3B), the predominant site of P2-Runx2-I isoform expression and function (12, 37).
To investigate whether combined Pkd1m1Bei and Runx2-II deficiency resulted in additive effects on osteoblast markers of embryonic bone development, we examined by real-time RT-PCR the expression levels of a panel of osteoblasts-related genes in newborn mice. We found a reduction of osteocalcin (Oc), osterix (Osx), Dmp1, and Mmp9 expression in single heterozygous mice compared with wild-type controls, with reduction being greater in double Pkd1+/m1Bei/Runx2-II+/- mice compared with single heterozygous Pkd1m1Bei and Runx2-II mice (Fig. 3C).
Additive Effects of Combined Pkd1 and Runx2-II Deficiency on Postnatal Bone Mass—The normal survival of double heterozygous mice permitted the assessment of bone abnormalities in adult mice. By 6 weeks of age, the gross appearance and body weigh of single heterozygous and wild-type mice were not significantly different. In contrast, the body weight of both male and female double heterozygous Pkd1+/m1Bei/Runx2-II+/- mice was reduced by ∼12 and 17% compared with the other genotypes (Fig. 4A). Six-week-old Pkd1+/m1Bei mice were osteopenic due to a reduction in trabecular bone volume, cortical bone thickness, and impaired osteoblast-mediated mineral apposition rates, as previously reported in 12-week-old mice (15). Similarly, heterozygous Runx2-II+/- mice had an ∼10% reduction in BMD (Fig. 4B), consistent with prior reports (12, 15). The combined loss of one allele of Runx2-II and Pkd1 resulted in additive reductions in BMD, as evidenced by the 22% reduction in BMD in the double heterozygous Pkd1+/m1Bei/Runx2-II+/- mice (Fig. 4B). μCT analysis revealed that the reduction in bone mass in single Pkd1+/m1Bei and Runx2-II+/- heterozygous mice was caused by a reduction in trabecular bone volume (43 and 50%, respectively) and cortical bone thickness (11 and 18%, respectively) (Fig. 4C). Double heterozygous Pkd1+/m1Bei/Runx2-II+/- mice had greater loss in both trabecular (83%) and cortical bone (28%) than did single heterozygous mice. These reductions in bone volume were associated with a significant decrease in mineral apposition rate in single Pkd1+/m1Bei and Runx2-II+/- heterozygous mice compared with age-matched wild-type mice and an even greater reduction in double heterozygous Pkd1+/m1Bei/Runx2-II+/- mice (Fig. 4D).
FIGURE 4.
Effects of combined Pkd1 and Runx2-II deficiency on bone in 6-week-old mice. A, gross appearance and body weight. Double heterozygous Pkd1+/m1Bei/Runx2-II+/- were smaller in size and exhibited a significant reduction in body weight compared with wild-type and single heterozygous Runx2-II+/- and Pkd1+/m1Bei mice. B, BMD of femurs. There was a greater reduction (22%) of femoral BMD in both male and female double heterozygous Pkd1+/m1Bei/Runx2-II+/- mice compared with Pkd1+/m1Bei (10%) and Runx2-II+/- (10%) littermates. C, μCT analysis of femurs. Double heterozygous Pkd1+/m1Bei/Runx2-II+/- mice demonstrated greater loss in both trabecular and cortical bone than single heterozygous mice. D, mineral apposition rate. There was a significant decrease in mineral apposition rate in single Pkd1+/m1Bei and Runx2-II+/- heterozygous mice compared with age-matched wild-type mice and an even greater reduction in double heterozygous Pkd1+/m1Bei/Runx2-II+/-mice, indicating an additive effect of Pkd1 and Runx2-II deficiency to impair osteoblast-mediated bone formation. Data are expressed as the mean ± S.E. from at least three individual samples. Values sharing the same superscript (a, b, or c) are not significantly different at p < 0.05.
To investigate whether combined Pkd1m1Bei and Runx2-II deficiency resulted in additive effects on gene expression profiles in bone, we examined by real-time RT-PCR the expression levels of a panel of osteoblast-, osteoclast-, and chondrocyte-related mRNAs in the femurs of 6-week-old wild-type, heterozygous Pkd1+/m1Bei, heterozygous Runx2-II+/-, and double heterozygous Pkd1+/m1Bei/Runx2-II+/- mice (Table 1). Bone derived from single heterozygous Pkd1+/m1Bei and Runx2-II+/- mice had measurable reductions in osterix, alkaline phosphatase, osteocalcin, type 1 collagen, osteoprotegerin, Rank ligand, Mmp13, Dmp1, and Phex mRNA levels compared with wild-type mice. Significantly greater reductions of osteoblasts and osteocytes-related genes were observed in double heterozygous Pkd1+/m1Bei/Runx2-II+/- mice for osteocalcin, Rank ligand, Mmp13, Dmp1, and Phex. Bone expression of Trap and Mmp9, markers of bone resorption, were also reduced in heterozygous Pkd1+/m1Bei and Runx2-II+/- mice and to a greater extent in double heterozygous Pkd1+/m1Bei and Runx2-II+/- mice compared with wild-type littermates (Table 1). These findings suggest that a low bone formation rate rather than increased bone resorption accounts for the low BMD and bone volume of femurs in the Pkd1+/m1Bei- and Runx2-II+/--deficient mice. Transcripts of chondrocyte-related genes did not differ between single and double heterozygous Pkd1+/m1Bei and Runx2-II+/- mice (Table 1).
PC1 Selectively Regulates Runx2-II P1 Promoter Activity through Intracellular Calcium Pathway—Next, we explored the ability of PC1 to differentially regulate the Runx2 P1 and P2 promoter activity in osteoblasts in vitro. For these studies we initially overexpressed into MC3T3-E1 osteoblasts the PC1-AT C-tail (gain-of-function) construct, which is composed of human IgG CH2-CH3 domain, a human CD7 transmembrane domain, and the mouse C-terminal 120 amino acids of PC1 containing the coiled-coil region, which is necessary for coupling to PC2 (35). Transient co-transfection of PC1-AT with the P1-Runx2 promoter-reporter construct (p1.4Runx2-II-luc) significantly increased promoter activity. In contrast, co-transfection of PC1-AT with the P2-Runx2 promoter-reporter construct (p2.0 Runx2-II-luc) did not stimulate promoter activity (Fig. 5A), consistent with the selective actions of PC1 on the Runx2-II isoform expression. Next, we examined the effects of a mutant PC1-AT construct, containing amino acid substitutions at sites 148, 149, and 155 that prevent coupling to PC2. Co-transfection of the PC1-ATmutant construct with the P1-Runx2-promoter-luciferase construct failed to stimulate promoter activity (Fig. 5B).
FIGURE 5.
PC1 regulates Runx2-II P1 promoter activity through intracellular calcium ([Ca2+]i) in MC3T3-E1 osteoblasts. A-C, cotransfection of various PC1 constructs with Runx2 P1 and P2 promoter-luciferase constructs in MC3T3-E1 osteoblasts. Overexpression of mouse PC1-AT (gain-of-function) significantly stimulated Runx2 P1 promoter activity but not the P2 promoter activity in osteoblasts (A). The control sIgØ construct had no effect. In contrast, overexpression of the PC1-ATmutant construct that disrupts coupling to PC2 failed to stimulate P1 promoter activity (B). Overexpression of human PKD1-(115-226) that also contains the coiled-coil domain stimulates Runx2 P1 but not P2 promoter activity in osteoblasts, whereas overexpression of PDK1(1-92) that lacks the coiled-coil domain fails to stimulate Runx2 P1 promoter activity in MC3T3-E1 osteoblasts (C). D and E, effect of Pkd1 on intracellular calcium ([Ca2+]i) in osteoblasts. There was a significant reduction in basal [Ca2+]i in Pkd1m1Bei/m1Bei mutant primary osteoblasts (D). Stable overexpression of the PC1-AT in MC3T3-E1 osteoblasts resulted in a significant increase in [Ca2+]i BAPTA, which chelates intracellular calcium, or thapsigargin, which depletes intracellular calcium stores, completely inhibited PC1-AT-mediated stimulation of Runx2 P1 promoter activity in osteoblasts (E). Data are expressed as the mean ± S.E. from at least three independent experiments. The asterisk indicates a significant difference from wild type and control, and values sharing the same superscript (a, b, or c) are not significantly different at p < 0.05.
We also tested the function of a human PKD1-(115-226) construct, which contains the N-terminal extracellular domain of human CD16, the human CD7 transmembrane region, and a 112-amino acid region of the C-terminal tail (human PKD1 amino acids 4192-4303) that permits coupling to the PC2 calcium channel (36). As a control we used the PKD1-(1-92) construct, which differs from PKD1-(115-226) by the substitution of the proximal 92 amino acids of the PKD1 C-terminal tail, which lacks the coiled-coiled domain required for coupling to PC2 (human PKD1 amino acids 4078-4169). Co-transfection of the PKD1-(115-226) construct stimulates P1-Runx2-luciferase activity in MC3T3-E1 cells, whereas the PKD1-(1-92) construct did not (Fig. 5C). Neither PKD1 (115-226) nor PKD1 (1-92) stimulated P2-Runx2-luciferase activity in these cells (Fig. 5C).
Because PC1 regulates intracellular calcium through its coupling to PC2 in epithelial cells (22), we explored the effect of Pkd1 mutations and overexpression on intracellular calcium in osteoblasts. We first investigated if loss of PC1 alters base-line intracellular calcium levels in Pkd1m1Bei/m1Bei osteoblasts. Similar to reports of diminished intracellular calcium in Pkd1-deficient epithelial cells (37), we also found reduced intracellular calcium levels in Pkd1m1Bei/m1Bei osteoblasts (Fig. 5D). Next, we examined the ability of the PC1-AT construct to stimulate increments in intracellular calcium levels in MC3T3-E1 osteoblasts. We found that overexpression of the PC1-AT C-tail significantly increased intracellular calcium concentrations in MC3T3-E1 osteoblasts (Fig. 5E). Finally, to explore the role of intracellular calcium in mediating PC1-AT-stimulated P1-Runx2 promoter activity, we assessed the effect of treatment of MC3T3-E1 osteoblasts with either BAPTA, which chelates intracellular calcium, or thapsigargin, which depletes intracellular calcium stores on PC1-AT stimulation of Runx2 P1 promoter activity in MC3T3-E1 osteoblasts (Fig. 5F). We found that both BAPTA and thapsigargin inhibited PC1-AT-mediated stimulation of the P1-Runx2 promoter-luciferase activity.
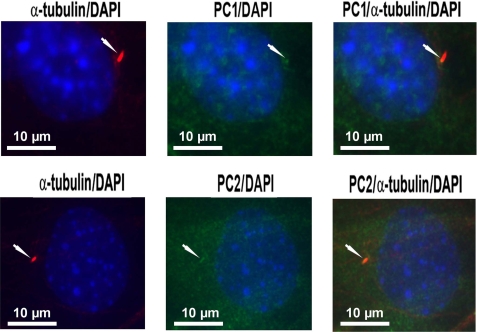
PC1 and PC2 Co-localize to Primary Cilia in Osteoblasts—Next, we determined whether PC1, PC2, and primary cilium, which are expressed in osteoblasts/osteocytes (15), are co-localized to the primary cilium. We found that immunostaining with primary α-tubulin antibody and Texas Red-conjugated secondary antibody identified a single primary cilium projecting from the cell surface of osteoblasts. Immunostaining with primary PC1 and PC2 antibodies and fluorescein isothiocyanate-conjugated secondary antibody revealed both diffuse and localized patterns of PC1 and PC2 expression in osteoblasts. Merged images (orange-yellow color) demonstrated co-localization of PC1 and PC2 to the primary cilium (Fig. 6).
FIGURE 6.
Co-localization of PC1 and PC2 to primary cilia in osteoblasts. Co-immunostaining with α-tubulin and PC1 or PC2 antibodies was performed in MC3T3-E1 osteoblasts as described under “Experimental Procedures.” Co-localization of PC1 (upper panel) and PC2 (lower panel) to the primary cilium (upper panel) is shown. A single primary cilium (red) is shown projecting from the cell surface of an osteoblast (left). PC1 and PC2 are present in both a diffuse and localized pattern (green) in osteoblasts (middle). Co-localization of PC1 and PC2 to the primary cilium is illustrated by the orange-yellow color in the merged images (right). 4′,6-Diamidino-2-phenylindole (DAPI) blue-staining was used to identify nuclei. Images are representative samples viewed with a fluorescent microscope at a magnification of 600×.
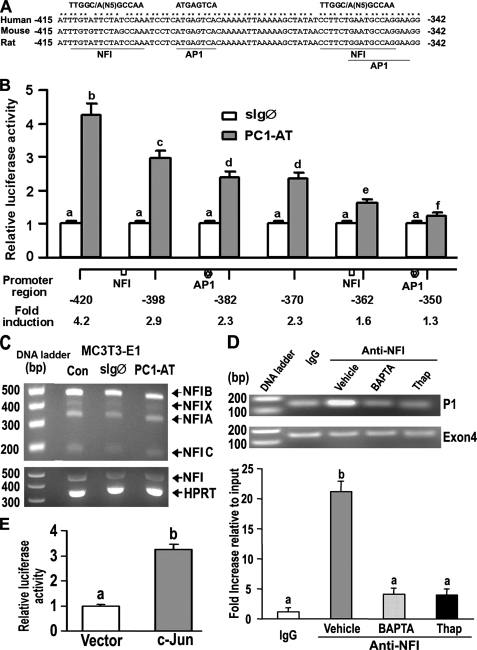
Identification of the Cis-acting Targets of Polycystin Signaling in the P1-Runx2-II Promoter—To map the PC1-responsive region of the Runx2-II P1 promoter, we created serial 5′ deletions of this promoter and evaluated their activity in co-transfection experiments with PC1-AT in MC3T3-E1 osteoblasts. We identified the PC1 responsive region between positions -420 to -350 base pairs in the Runx2-II P1 promoter, which contains the osteoblast-specific enhancer element previously reported by Zambotti and Ducy (14). rVISTA analysis of this region identified two composite NFI and AP1 binding sites that were conserved across species (Fig. 7A). Further deletion analysis confirmed that each of the NFI and AP1 sites independently accounts for the majority of PC1-mediated activity in MC3T3-E1 osteoblasts (Fig. 7B). RT-PCR analysis using NFI-specific degenerated primers revealed that NFI-A, -B, -C, and -X isoforms are all expressed in MC3T3-E1 osteoblasts (Fig. 7C).
FIGURE 7.
NFI and AP-1 transcription factors mediate PC1-dependent activation of the P1 Runx2-II promoter. A, schematic showing two NFI and AP1 binding sites located between -415 and -342 region of the Runx2-II P1 promoter that are conserved across species. B, assessment of NF1 and AP1 function by deletion analysis of the Runx2 P1 promoter. MC3T3-E1 osteoblasts were transiently co-transfected with PC1-AT or sIgØ along with Runx2-P1 promoter-luciferase constructs that contained successive 5′ deletions. Loss of NFI and AP1 sites resulted in a progressive reduction in PC1-mediated stimulation of Runx2 P1 promoter activity. C, RT-PCR analysis of NFI-A, -B, -C, and -X isoform expression in MC3T3-E1 osteoblasts transiently transfected with either the control sIgØ or the PC1-AT constructs for 48 h. Neither PC1-AT nor sIgØ increased NFI message levels above untransfected controls (CON) in MC3T3-E1, raising the possibility that PC1 regulates NFI through post-translational modifications. HPRT, hypoxanthine-guanine phosphoribosyltransferase. D, quantitative ChIP analyses demonstrating specific interaction of NFI with Runx2-II P1 promoter. Quantitative real-time PCR was performed using set of primers indicated under “Experimental Procedures.” The upper panel is a representative ethidium bromide gel of PCR products, and the lower panel is bar graph representing mean ± S.E. from at least three independent experiments. Values are shown as relative -fold of enrichment of the promoter sequence normalized for coding region sequence versus that obtained for input samples. Values sharing the same superscript (a, b, c, and d) are not significantly different at p < 0.05. Immunoprecipitations were performed with NFI antibody or with normal rabbit IgG as described under “Experimental Procedures.” E, transient co-transfection c-Jun plasmid with Runx2 P1-420-Luc (p0.42Runx2P1-Luc) stimulated Runx2 P1 promoter activity, consistent with functional AP-1 binding sites.
PC1-AT failed to increase NFI message levels in MC3T3-E1, raising the possibility that PC1 regulates NFI through post-translational modifications. Quantitative ChIP analyses using anti-NFI antibody confirmed that NFI was specifically recruited to the Runx2-II P1 promoter (Fig. 7D). There was a 20-fold increase in the ratio of the promoter sequence versus the coding region sequence in the anti-NFI group compared with the IgG control group by quantitative real-time PCR. In addition, treatment with BAPTA or thapsigargin almost completely abolished the NFI occupancy in the Runx2-II P1 promoter (Fig. 7D). Transient co-transfection c-Jun plasmid with Runx2-II P1-420-Luc (p0.42Runx2P1-Luc) stimulated Runx2-II P1 promoter activity, indicating that the putative AP1 binding site is functional (Fig. 7E). Together, these findings indicate that PC1 selectively activates P1Runx2-II isoform transcription through intracellular calcium and NFI/AP1 pathways.
DISCUSSION
We have evidence that polycystin-1 regulates bone development and osteoblast function via regulation of the osteogenic transcription factor Runx2-II. These studies extend our initial observations that primary cilia and polycystin transcripts are present in osteoblasts/osteocytes and provide additional data to support a causal relationship between mutant PC1, decreased expression of Runx2-II, and impaired osteoblast differentiation and bone development in mice (15). In this regard we created double heterozygous Pkd1+/m1Bei and Runx2-II+/- mice and found that these animals display further decrements in Runx2-II expression in association with more severe defects in bone development and greater reductions in osteoblast-mediated bone formation postnatally. In addition, in vitro studies demonstrated that the P1-Runx2-II promoter is a downstream target for PC1 in osteoblasts, and PC1 selectively regulates Runx2-II P1 promoter activity through an intracellular calcium pathway. The in vivo and in vitro selectivity of PC1 for P1-Runx2-II, which controls endochondral bone formation and osteoblast function, contrasts with the failure of PC1 to stimulate P2-Runx2-I expression, which is more important in cortical and intramembranous bone formation (11, 12). Thus, PC1 may contribute to the temporal and spatial difference in Runx2-II expression in bone.
Our studies also point to the functional importance of PC1 regulation of intracellular calcium-dependent signaling in osteoblasts. In renal epithelial cells PC1 and PC2 are known to colocalize with primary cilia to form a primary cilium-polycystin complex that regulates cell function through intracellular calcium signal pathways (22, 24, 29, 35, 37, 38). Consistent with this, we observed that loss of PC1 led to reduction of intracellular calcium levels in homozygous Pkd1m1Bei/m1Bei osteoblasts, and overexpression of PC1-AT significantly increased intracellular calcium levels in MC3T3-E1 osteoblasts, which is in accordance with previous findings that PC1-AT stimulates endogenous Runx2-II transcription as well as enhances Runx2 protein levels in stably transfected MC3T3-E1 osteoblasts (15). In addition, site-directed mutagenesis of critical amino acids in the coiled-coil domain of the PC1 C-tail that is necessary for coupling to PC2 abolished the stimulation of Runx2-II P1 promoter activity. Moreover, either BAPTA, which chelates intracellular calcium, or thapsigargin, which depletes intracellular calcium stores, blocked the ability of the PC1 C-tail to increase Runx2-II P1 promoter activity, indicating that intracellular calcium pathways are important in PC1-mediating Runx2-II P1 promoter activity in osteoblasts. The coupling of PC1 to Runx2-II expression represents a new anabolic signaling pathway in osteoblasts and suggests that the primary cilium-polycystin complex in bone may function to sense external developmental signals and convert them into intracellular pathways regulating Runx2-II expression during skeletogenesis.
The finding in osteoblasts that PC1 and PC2 co-localize to the primary cilium suggests that the primary cilium-polycystin complex may also regulate bone development (39-41). There is additional evidence to support a function of the primary cilium-polycystin complex in regulating skeletogenesis. First, primary cilia and PC1/PC2 are present in chondrocytes (42, 43) and in osteoblasts/osteocytes (15, 43-45). In addition, mutation of proteins required for ciliogenesis (39-41) as well as Pkd1 and Pkd2 are associated with abnormalities of bone development (16, 17, 28), osteoblast differentiation (40), and chondrodysplasia (46). The primary cilium may be viewed as an organelle that houses several signaling cascades regulating skeletal development. In this regard, primary cilia also express the transmembrane protein Patched (Ptc), which is important in sonic hedgehog (Shh) signaling (47).
Finally, we found that both the nuclear factor I family (NFI, also known as CAAT box transcription factor or CTF) and AP-1 transcription factor binding sites are involved in polycystin-1 regulation of the P1-Runx2-II promoter. Interestingly, this PC1-responsive element corresponds to the previously reported osteoblast-specific enhancer element in the Runx2-II P1 promoter (14). It has been reported that the PC1 C-tail stimulates AP1 promoter activity (48-50), consistent with our mapping of AP1 binding sites in the Runx2-II P1 promoter. More importantly, both AP1 and NFI are known to be important in developmental processes (51-55), including bone. For example, delayed endochondral ossification and kyphosis are observed in mice with conditional knock-out of the AP1 transcriptional factor component Fra2 (56). In addition, mutations of NFI genes are associated with defects in tooth and bone development (51, 52). Moreover, intracellular calcium regulates NFI function via phosphorylation by calcium-dependent kinases (57). NFI also binds to histone H3 (58), leading to modification of chromatin architecture (59) and epigenetic control of gene expression. Because overexpression of the PC1 C-tail failed to increase NFI message levels in MC3T3-E1 osteoblasts, PC1 may regulate NFI through post-translational modifications. Regardless, unlike the proposed effect of NFI to suppress the osteoblast phenotype (14), our studies support the possibility that NFI and AP-1 are important positive regulators of Runx2-dependent skeletal development and osteoblast function, thereby providing links to external developmental clues via polycystins/primary cilia.
In conclusion, we have found that compound heterozygous polycystin-1 (PC1) and Runx2-II-deficient mice have profound additive effects to impair bone development that correlates with reduced Runx2-II expression. This finding suggests that PC1 and Runx2-II act in a common pathway controlling bone development. In addition, we have identified specific polycystin-1-dependent signaling pathways and the cis-acting elements NFI and AP1 in the P1-Runx2-II promoter. Thus, PC1 signaling may play an important role in sensing environmental clues that regulate osteoblast differentiation and bone development via changes in intracellular calcium. Additional work is needed to confirm the involvement of the primary cilium and PC2 in bone development, to identify the mechanisms whereby PC1-dependent changes in intracellular calcium regulates NFI and AP-1 activity, and to investigate the respective role of NFI and AP1 in the epigenetic and transcriptional control of Runx2-II. A better understanding of the mechanisms whereby polycystins selectively control Runx2-II gene expression will lead to a more complete knowledge of skeletal development and the possible identification of new targets for manipulating bone formation.
This work was supported by National Institutes of Health Grants R01-AR049712 and P50-DK057301. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PKD1 or Pkd1, polycystic kidney disease gene 1; PKD2 or Pkd2, polycystic kidney disease gene 2; PC1, polycystin 1; PC2, polycystin 2; Runx2, Runt-related transcription factor 2; BMD, bone mineral density; μCT, microcomputed tomography; RT, reverse transcription; ChIP, chromatin immunoprecipitation.
References
- 1.Cohen, M. M., Jr. (2006) Am. J. Med. Genet. A 140 2646-2706 [DOI] [PubMed] [Google Scholar]
- 2.Kronenberg, H. M. (2003) Nature 423 332-336 [DOI] [PubMed] [Google Scholar]
- 3.de Crombrugghe, B., Lefebvre, V., and Nakashima, K. (2001) Curr. Opin. Cell Biol. 13 721-727 [DOI] [PubMed] [Google Scholar]
- 4.Ducy, P., Zhang, R., Geoffroy, V., Ridall, A. L., and Karsenty, G. (1997) Cell 89 747-754 [DOI] [PubMed] [Google Scholar]
- 5.Komori, T., Yagi, H., Nomura, S., Yamaguchi, A., Sasaki, K., Deguchi, K., Shimizu, Y., Bronson, R. T., Gao, Y. H., Inada, M., Sato, M., Okamoto, R., Kitamura, Y., Yoshiki, S., and Kishimoto, T. (1997) Cell 89 755-764 [DOI] [PubMed] [Google Scholar]
- 6.Mundlos, S., Otto, F., Mundlos, C., Mulliken, J. B., Aylsworth, A. S., Albright, S., Lindhout, D., Cole, W. G., Henn, W., Knoll, J. H., Owen, M. J., Mertelsmann, R., Zabel, B. U., and Olsen, B. R. (1997) Cell 89 773-779 [DOI] [PubMed] [Google Scholar]
- 7.Otto, F., Thornell, A. P., Crompton, T., Denzel, A., Gilmour, K. C., Rosewell, I. R., Stamp, G. W., Beddington, R. S., Mundlos, S., Olsen, B. R., Selby, P. B., and Owen, M. J. (1997) Cell 89 765-771 [DOI] [PubMed] [Google Scholar]
- 8.Ueta, C., Iwamoto, M., Kanatani, N., Yoshida, C., Liu, Y., Enomoto-Iwamoto, M., Ohmori, T., Enomoto, H., Nakata, K., Takada, K., Kurisu, K., and Komori, T. (2001) J. Cell Biol. 153 87-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim, I. S., Otto, F., Zabel, B., and Mundlos, S. (1999) Mech. Dev. 80 159-170 [DOI] [PubMed] [Google Scholar]
- 10.Enomoto, H., Enomoto-Iwamoto, M., Iwamoto, M., Nomura, S., Himeno, M., Kitamura, Y., Kishimoto, T., and Komori, T. (2000) J. Biol. Chem. 275 8695-8702 [DOI] [PubMed] [Google Scholar]
- 11.Xiao, Z. S., Hjelmeland, A. B., and Quarles, L. D. (2004) J. Biol. Chem. 279 20307-20313 [DOI] [PubMed] [Google Scholar]
- 12.Xiao, Z., Awad, H. A., Liu, S., Mahlios, J., Zhang, S., Guilak, F., Mayo, M. S., and Quarles, L. D. (2005) Dev. Biol. 283 345-356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao, Z. S., Hinson, T. K., and Quarles, L. D. (1999) J. Cell. Biochem. 74 596-605 [PubMed] [Google Scholar]
- 14.Zambotti, A., Makhluf, H., Shen, J., and Ducy, P. (2002) J. Biol. Chem. 277 41497-41506 [DOI] [PubMed] [Google Scholar]
- 15.Xiao, Z., Zhang, S., Mahlios, J., Zhou, G., Magenheimer, B. S., Guo, D., Dallas, S. L., Maser, R., Calvet, J. P., Bonewald, L., and Quarles, L. D. (2006) J. Biol. Chem. 281 30884-30895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boulter, C., Mulroy, S., Webb, S., Fleming, S., Brindle, K., and Sandford, R. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 12174-12179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu, W., Shen, X., Pavlova, A., Lakkis, M., Ward, C. J., Pritchard, L., Harris, P. C., Genest, D. R., Perez-Atayde, A. R., and Zhou, J. (2001) Hum. Mol. Genet. 10 2385-2396 [DOI] [PubMed] [Google Scholar]
- 18.Nims, N., Vassmer, D., and Maser, R. L. (2003) Biochemistry 42 13035-13048 [DOI] [PubMed] [Google Scholar]
- 19.Kim, E., Arnould, T., Sellin, L. K., Benzing, T., Fan, M. J., Gruning, W., Sokol, S. Y., Drummond, I., and Walz, G. (1999) J. Biol. Chem. 274 4947-4953 [DOI] [PubMed] [Google Scholar]
- 20.Delmas, P., Padilla, F., Osorio, N., Coste, B., Raoux, M., and Crest, M. (2004) Biochem. Biophys. Res. Commun. 322 1374-1383 [DOI] [PubMed] [Google Scholar]
- 21.Geng, L., Burrow, C. R., Li, H. P., and Wilson, P. D. (2000) Biochim. Biophys. Acta. 1535 21-35 [DOI] [PubMed] [Google Scholar]
- 22.Nauli, S. M., Alenghat, F. J., Luo, Y., Williams, E., Vassilev, P., Li, X., Elia, A. E., Lu, W., Brown, E. M., Quinn, S. J., Ingber, D. E., and Zhou, J. (2003) Nat. Genet. 33 129-137 [DOI] [PubMed] [Google Scholar]
- 23.Forman, J. R., Qamar, S., Paci, E., Sandford, R. N., and Clarke, J. (2005) J. Mol. Biol. 349 861-871 [DOI] [PubMed] [Google Scholar]
- 24.Nauli, S. M., Rossetti, S., Kolb, R. J., Alenghat, F. J., Consugar, M. B., Harris, P. C., Ingber, D. E., Loghman-Adham, M., and Zhou, J. (2006) J. Am. Soc. Nephrol. 17 1015-1025 [DOI] [PubMed] [Google Scholar]
- 25.Singla, V., and Reiter, J. F. (2006) Science 313 629-633 [DOI] [PubMed] [Google Scholar]
- 26.Schneider, L., Clement, C. A., Teilmann, S. C., Pazour, G. J., Hoffmann, E. K., Satir, P., and Christensen, S. T. (2005) Curr. Biol. 15 1861-1866 [DOI] [PubMed] [Google Scholar]
- 27.Rohatgi, R., Milenkovic, L., and Scott, M. P. (2007) Science 317 372-376 [DOI] [PubMed] [Google Scholar]
- 28.Herron, B. J., Lu, W., Rao, C., Liu, S., Peters, H., Bronson, R. T., Justice, M. J., McDonald, J. D., and Beier, D. R. (2002) Nat. Genet. 30 185-189 [DOI] [PubMed] [Google Scholar]
- 29.Vandorpe, D. H., Chernova, M. N., Jiang, L., Sellin, L. K., Wilhelm, S., Stuart-Tilley, A. K., Walz, G., and Alper, S. L. (2001) J. Biol. Chem. 276 4093-4101 [DOI] [PubMed] [Google Scholar]
- 30.Loots, G. G., Ovcharenko, I., Pachter, L., Dubchak, I., and Rubin, E. M. (2002) Genome Res. 12 832-839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian, F., Germino, F. J., Cai, Y., Zhang, X., Somlo, S., and Germino, G. G. (1997) Nat. Genet. 16 179-183 [DOI] [PubMed] [Google Scholar]
- 32.Gee, K. R., Brown, K. A., Chen, W. N., Bishop-Stewart, J., Gray, D., and Johnson, I. (2000) Cell Calcium 27 97-106 [DOI] [PubMed] [Google Scholar]
- 33.Mukhopadhyay, S. S., Wyszomierski, S. L., Gronostajski, R. M., and Rosen, J. M. (2001) Mol. Cell. Biol. 21 6859-6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu, E. D., Demay, M. B., and Gori, F. (2008) J. Biol. Chem. 283 7361-7367 [DOI] [PubMed] [Google Scholar]
- 35.Puri, S., Magenheimer, B. S., Maser, R. L., Ryan, E. M., Zien, C. A., Walker, D. D., Wallace, D. P., Hempson, S. J., and Calvet, J. P. (2004) J. Biol. Chem. 279 55455-55464 [DOI] [PubMed] [Google Scholar]
- 36.Tsiokas, L., Kim, E., Arnould, T., Sukhatme, V. P., and Walz, G. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 6965-6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi, T., Hempson, S. J., Reif, G. A., Hedge, A. M., and Wallace, D. P. (2006) J. Am. Soc. Nephrol. 17 178-187 [DOI] [PubMed] [Google Scholar]
- 38.Manzati, E., Aguiari, G., Banzi, M., Manzati, M., Selvatici, R., Falzarano, S., Maestri, I., Pinton, P., Rizzuto, R., and del Senno, L. (2005) Exp. Cell Res. 304 391-406 [DOI] [PubMed] [Google Scholar]
- 39.Song, B., Haycraft, C. J., Seo, H. S., Yoder, B. K., and Serra, R. (2007) Dev. Biol. 305 202-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haycraft, C. J., Zhang, Q., Song, B., Jackson, W. S., Detloff, P. J., Serra, R., and Yoder, B. K. (2007) Development 134 307-316 [DOI] [PubMed] [Google Scholar]
- 41.Koyama, E., Young, B., Nagayama, M., Shibukawa, Y., Enomoto-Iwamoto, M., Iwamoto, M., Maeda, Y., Lanske, B., Song, B., Serra, R., and Pacifici, M. (2007) Development 134 2159-2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen, C. G., Poole, C. A., McGlashan, S. R., Marko, M., Issa, Z. I., Vujcich, K. V., and Bowser, S. S. (2004) Cell Biol. Int. 28 101-110 [DOI] [PubMed] [Google Scholar]
- 43.Takaoki, M., Murakami, N., and Gyotoku, J. (2004) Biol. Sci. Space 18 181-182 [PubMed] [Google Scholar]
- 44.Federman, M., and Nichols, G., Jr. (1974) Calcif. Tissue Res. 17 81-85 [DOI] [PubMed] [Google Scholar]
- 45.Whitfield, J. F. (2003) J. Cell. Biochem. 89 233-237 [DOI] [PubMed] [Google Scholar]
- 46.Beales, P. L., Bland, E., Tobin, J. L., Bacchelli, C., Tuysuz, B., Hill, J., Rix, S., Pearson, C. G., Kai, M., Hartley, J., Johnson, C., Irving, M., Elcioglu, N., Winey, M., Tada, M., and Scambler, P. J. (2007) Nat. Genet. 39 727-729 [DOI] [PubMed] [Google Scholar]
- 47.Christensen, S. T., and Ott, C. M. (2007) Science 317 330-331 [DOI] [PubMed] [Google Scholar]
- 48.Parnell, S. C., Magenheimer, B. S., Maser, R. L., Zien, C. A., Frischauf, A. M., and Calvet, J. P. (2002) J. Biol. Chem. 277 19566-19572 [DOI] [PubMed] [Google Scholar]
- 49.Le, N. H., van der Bent, P., Huls, G., van de Wetering, M., Loghman-Adham, M., Ong, A. C., Calvet, J. P., Clevers, H., Breuning, M. H., van Dam, H., and Peters, D. J. (2004) J. Biol. Chem. 279 27472-27481 [DOI] [PubMed] [Google Scholar]
- 50.Chauvet, V., Tian, X., Husson, H., Grimm, D. H., Wang, T., Hiesberger, T., Igarashi, P., Bennett, A. M., Ibraghimov-Beskrovnaya, O., Somlo, S., and Caplan, M. J. (2004) J. Clin. Investig. 114 1433-1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Driller, K., Pagenstecher, A., Uhl, M., Omran, H., Berlis, A., Grunder, A., and Sippel, A. E. (2007) Mol. Cell. Biol. 27 3855-3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steele-Perkins, G., Butz, K. G., Lyons, G. E., Zeichner-David, M., Kim, H. J., Cho, M. I., and Gronostajski, R. M. (2003) Mol. Cell. Biol. 23 1075-1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gronostajski, R. M. (2000) Gene (Amst.) 249 31-45 [DOI] [PubMed] [Google Scholar]
- 54.Uchihashi, T., Kimata, M., Tachikawa, K., Koshimizu, T., Okada, T., Ihara-Watanabe, M., Sakai, N., Kogo, M., Ozono, K., and Michigami, T. (2007) Bone (NY) 41 1025-1035 [DOI] [PubMed] [Google Scholar]
- 55.Wang, W., Mullikin-Kilpatrick, D., Crandall, J. E., Gronostajski, R. M., Litwack, E. D., and Kilpatrick, D. L. (2007) J. Neurosci. 27 6115-6127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karreth, F., Hoebertz, A., Scheuch, H., Eferl, R., and Wagner, E. F. (2004) Development 131 5717-5725 [DOI] [PubMed] [Google Scholar]
- 57.Alevizopoulos, A., Dusserre, Y., Ruegg, U., and Mermod, N. (1997) J. Biol. Chem. 272 23597-23605 [DOI] [PubMed] [Google Scholar]
- 58.Ferrari, S., Simmen, K. C., Dusserre, Y., Muller, K., Fourel, G., Gilson, E., and Mermod, N. (2004) J. Biol. Chem. 279 55520-55530 [DOI] [PubMed] [Google Scholar]
- 59.Hebbar, P. B., and Archer, T. K. (2003) Mol. Cell. Biol. 23 887-898 [DOI] [PMC free article] [PubMed] [Google Scholar]