Abstract
Phospholamban (PLB) oligomerization, quaternary structure, and sarco(endo)plasmic reticulum calcium ATPase (SERCA) binding were quantified by fluorescence resonance energy transfer (FRET) in an intact cellular environment. FRET between cyan fluorescent protein-PLB and yellow fluorescent protein-PLB in AAV-293 cells showed hyperbolic dependence on protein concentration, with a maximum efficiency of 45.1 ± 1.3%. The observed FRET corresponds to a probe separation distance of 58.7 ± 0.5Å, according to a computational model of intrapentameric FRET. This is consistent with models of the PLB pentamer in which cytoplasmic domains fan out from the central bundle of transmembrane helices. An I40A mutation of PLB did not alter pentamer conformation but increased the concentration of half-maximal FRET (KD) by >4-fold. This is consistent with the previous observation that this putatively monomeric mutant still oligomerizes in intact membranes but forms more dynamic pentamers than wild type PLB. PLB association with SERCA, measured by FRET between cyan fluorescent protein-SERCA and yellow fluorescent protein-PLB, was increased by the I40A mutation without any detectable change in probe separation distance. The data indicate that the regulatory complex conformation is not altered by the I40A mutation. A naturally occurring human mutation (L39Stop) greatly reduced PLB oligomerization and SERCA binding and caused mislocalization of PLB to the cytoplasm and nucleus. Overall, the data suggest that the PLB pentamer adopts a “pinwheel” shape in cell membranes, as opposed to a more compact “bellflower” conformation. I40A mutation decreases oligomerization and increases PLB binding to SERCA. Truncation of the transmembrane domain by L39Stop mutation prevents anchoring of the protein in the membrane, greatly reducing PLB binding to itself or its regulatory target, SERCA.
The 52-amino acid protein phospholamban (PLB)2 is an important regulator of cardiac calcium handling (1). PLB binds avidly (2) but reversibly (3) to the sarco(endo)plasmic reticulum calcium ATPase (SERCA), reducing this calcium pump's affinity for calcium (4). PLB also oligomerizes into pentamers through leucine zipper interactions in its transmembrane domain (5, 6). NMR studies indicate that PLB tertiary structure consists of an N-terminal (cytosolic) α-helix (domain IA) connected by a flexible loop (domain IB) to a second α-helix (domain II) that is anchored in the membrane of the sarcoplasmic reticulum (7). Several possible configurations of the cytoplasmic domains of pentameric PLB have been described. Some of these portray the domain IA α-helix as being nearly normal to the surface of the membrane (8, 9), whereas others indicate axial declination of domain IA (7, 10–14), permitting contact with the surface of the membrane (15, 16). These previous studies were performed using in vitro preparations of PLB in defined lipids or detergent. To investigate the quaternary structure of PLB in the membranes of living cells and distinguish between these structural models, we expressed cyan and yellow fluorescent proteins fused to the PLB N terminus and measured fluorescent protein separation distances with fluorescence resonance energy transfer (FRET) (17). In addition, we determined the effects of mutation of the PLB transmembrane domain on PLB oligomerization affinity, quaternary structure, and capacity to bind to its regulatory target, SERCA. Specifically, we tested a naturally occurring human mutation associated with heart failure (L39Stop) that truncates PLB midway through its transmembrane domain (18). We also investigated a pentamer-destabilizing point mutation (I40A). The I40A mutation is historically regarded as a “monomeric” mutant because of its electrophoretic mobility in the presence of detergent (19). We have previously observed that I40A-PLB still forms oligomers in membranes, but compared with WT, the I40A oligomers are more dynamic, showing an increased rate of exchange of PLB subunits from the pentamer complex (3). In the present study, we endeavored to quantify the thermodynamic consequence of this mutation in the membranes of live cells.
EXPERIMENTAL PROCEDURES
Rabbit SERCA1a and dog PLB fused to the C terminus of fluorescent proteins were expressed in AAV-293 cells, as previously described (3). Mutants of PLB were created with the QuikChange II site-directed mutagenesis kit (Stratagene, La Jolla, CA) and custom oligonucleotide primers. Mutations were confirmed by sequencing. AAV-293 cells cultured on polylysine-coated glass bottom dishes were cotransfected with plasmids encoding CFP/YFP fusion constructs, at a 5- or 20-fold excess of YFP, as previously described (3). Fluorescence imaging was performed with an inverted microscope equipped with a 1.49 numerical aperture objective, and a back-thinned CCD camera (iXon 887; Andor Technology, Belfast, Northern Ireland). The detector was cooled to –100 °C, using a recirculating liquid coolant system (Koolance, Inc., Auburn, WA). The system's relative sensitivity to CFP and YFP was calibrated by imaging drops of purified CFP and YFP at a series of known concentrations. Image acquisition and acceptor photobleaching was automated with custom software macros in Meta-Morph (Molecular Devices Corp., Downingtown, PA) that controlled motorized excitation/emission filter wheels (Sutter Instrument Co., Novato, CA) with filters for CFP/YFP/mCherry (Semrock, Rochester NY). The progressive photobleaching protocol was as follows: 100-ms acquisition of CFP image and 40-ms acquisition of YFP image, followed by 10-s exposure to YFP-selective photobleaching (504/12-nm excitation). For intrapentameric FRET experiments, each cell's relative protein expression was assessed as a sum of the starting YFP fluorescence (prebleach) and the final CFP fluorescence after FRET was abolished (postbleach). In general, concentrations of membrane proteins and related dissociation constants (KD) are expressed as protein molar fraction (20) or as species per unit area (e.g. mol/m2). However, expression of such absolute concentrations requires a reconstituted system of defined lipid/protein ratio. Since we endeavored to quantify the thermodynamics of oligomerization in living cells, we express our protein concentration in arbitrary units (AU).
FRET efficiency was calculated from the fluorescence intensity of the CFP donor before and after acceptor-selective photobleaching, according to the relationship, E = 1 – (Fprebleach/Fpostbleach). The maximal energy transfer efficiency (FRETmax) was obtained from a hyperbolic fit of the form, y = (FRETmax)x/(KD + x), to the data of a plot of FRET dependence on protein concentration. This hyperbola “saturation” value was taken as the FRET efficiency of maximally oligomerized PLB (100% pentamer); thus, it represents the intrinsic FRET efficiency of the complex. Nearest neighbor probe separation distance R was calculated from FRETmax using a custom MatLab application of the ring oligomer energy transfer theory (21), assuming an oligomer subunit number of 5 (pentamer), and using the acceptor molar fraction of 0.88, determined from YFP starting and CFP final fluorescence, as described above. The model used a Förster radius (R0) of 49.2 Å for CFP-YFP energy transfer (22). The calculated distances were used to evaluate the pinwheel and bellflower models of the PLB pentamer.
Average distances between pinwheel (Protein Data Bank code 1XNU) or bellflower (Protein Data Bank code 1ZLL) nearest neighbor amino termini were measured in Pymol. The radius of the ring-shaped distribution of amino termini was calculated by the equation, rN = d/2(sinα/2), where rN represents the radius, d is the average nearest neighbor distance between amino termini, and α is the azimuthal angle of adjacent subunits arrayed about the pentamer central axis normal to the plane of the membrane. The theoretical radius of maximal reach (rmax) was obtained by adding 25 Å to rN, to account for the distance between the fluorescent protein chromaphore and fusion of the fluorescent protein to the PLB cytoplasmic domain. The radius of the ring-shaped distribution of fluorescent protein chromaphores (rFRET) was obtained from the nearest neighbor probe separation distance R calculated from intrinsic pentamer FRET efficiency (FRETmax), according to rFRET = R/2(sinα/2). The apparent attachment angle between the pinwheel cytoplasmic domain and the fluorescent protein θ that corresponds to the measured probe separation distance was obtained by the equation, θ = cos–1(rFRET – rN)/25 Å).
The regulatory complex maximal FRET efficiency (FRETmax) was determined from concentration dependence, as above. This value was taken as the intrinsic FRET efficiency of the CFP-SERCA·YFP-PLB complex. The probe separation distance (R) for this heterodimeric complex was calculated according to the relationship described by Förster (17), R = (R0)((1/FRETmax) – 1)⅙.
The membrane partitioning of YFP-L39Stop-PLB was evaluated by comparison with CFP-WT-PLB in permeabilized cells. Cells expressing these probes were preincubated in 2 μg/ml propidium iodide, an impermeant nuclear stain. Cells were imaged for CFP, YFP, and propidium iodide fluorescence before and after the addition of the plasma membrane-selective permeabilization agent saponin at 100 μg/ml.
Laser spot photobleaching was performed using the 514-nm line of an argon laser as previously described (3). Briefly, cells expressing WT-, I40A-, or L39Stop-PLB fused to YFP were imaged continuously by streaming acquisition, with open/close triggering of a Uniblitz shutter after 20 frames were acquired. Fluorescence recovery after photobleaching was observed for 40 s.
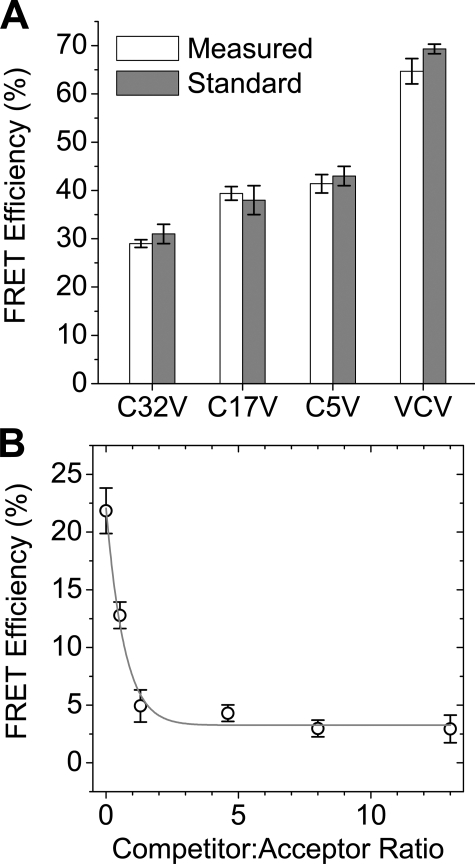
To determine the accuracy of the FRET quantification method used in the present study, we expressed standard FRET constructs in AAV-293 cells and subjected them to acceptor photobleaching. Constructs C32V, C17V, and C5V are Cerulean-Venus fusion constructs in which the fluorescent proteins are separated by linkers of 32, 17, and 5 amino acids, respectively (23). VCV is a two-acceptor construct, with two Venus proteins fused to Cerulean (24). We obtained FRET efficiency measurements from the following sample sizes: C32V = 8; C17V = 8; C5V = 9; VCV = 12.
To estimate the amount of energy transfer due to nonspecific energy transfer between unbound fluorescently labeled proteins in the membrane, competition experiments were performed using YFP-PLB and mCherry-PLB as the FRET partners competing with CFP-PLB (which cannot act as an acceptor for YFP). YFP-PLB and mCherry-PLB were expressed at a 1:1 ratio, with increasing amounts of CFP-PLB.
RESULTS
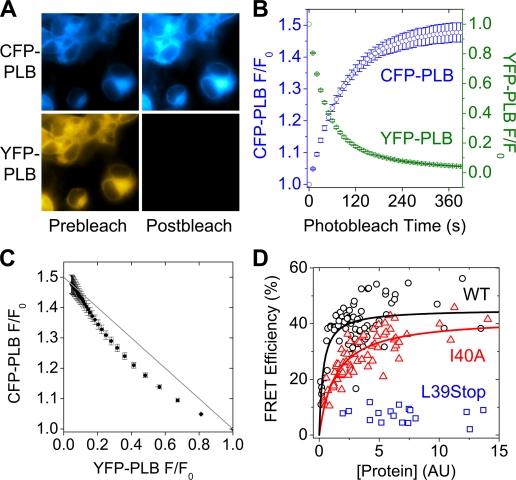
Intrapentameric FRET in Living Cells—Epifluorescence imaging of CFP/YFP-labeled wild-type PLB showed endoplasmic reticulum-localized fluorescence (Fig. 1A) (25) with an additional plasma membrane-localized component, as previously described (3). Acceptor-selective photobleaching of YFP-PLB resulted in increased CFP-PLB fluorescence emission (Fig. 1A), indicating FRET. The time-dependent change in CFP/YFP emission is quantified in Fig. 1B. YFP-PLB was progressively bleached, resulting in an exponential decline in YFP emission (green circles) and concomitant increase in CFP emission (blue circles). To probe the stoichiometry of the oligomeric complex that gives rise to FRET, the relationship of CFP and YFP fluorescence emission was examined. The data from Fig. 1B are replotted in Fig. 1C to show that donor fluorescence versus acceptor fluorescence deviated from linearity, as expected for a donor-acceptor complex with subunit number of >2 (21). This representation is similar to that obtained from lineout analysis of an acceptor-selective spot photobleaching target region (3); however, the progressive photobleaching approach provides much greater precision and surveys a larger span of the YFP bleach axis. These technical advantages permit observation of the curvature predicted by oligomer FRET theory (21).
FIGURE 1.
A, CFP-PLB and YFP-PLB fluorescence images before (Prebleach) and after (Postbleach) YFP-selective photobleaching. B, selective photobleaching resulted in decreased YFP fluorescence (green circles) and increased CFP fluorescence (blue circles). C, curved dependence of CFP fluorescence on YFP fluorescence, consistent with subunit n > 2. Data are mean ± S.E. D, dependence of WT (black circles) and I40A (red triangles) PLB pentamer FRET on concentration. The I40A mutation reduced PLB/PLB affinity by >4-fold. The oligomerization affinity of L39Stop-PLB (blue squares) was too low to measure.
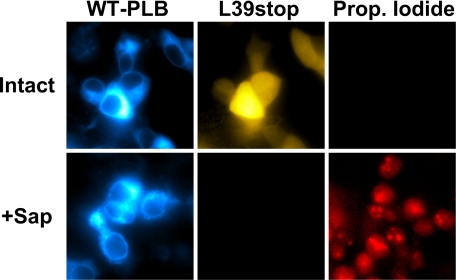
The acceptor molar fraction was calculated from each cell's donor and acceptor fluorescence, as detailed under “Experimental Procedures,” and was found to be very consistent between cells (mean = 0.88, S.E. = 0.002). There was no statistically significant difference in acceptor molar fraction of WT- and I40A-expressing cells. However, total expression of fluorescent PLB varied significantly (Fig. 1A). Thus, acceptor-selective photobleaching of a population of cells could probe the dependence of FRET on protein concentration. This assay was used to compare the binding energetics of WT and a pentamer-destabilizing point mutation (I40A) of the transmembrane domain (19). Based on its electrophoretic mobility, I40A-PLB has been referred to as a monomeric PLB, but we have shown that it can still form oligomers in cell membranes (3). As previously observed, the I40A mutation of PLB destabilized the pentamer form (but did not abolish oligomerization), as quantified by intrapentameric FRET. Both WT-PLB (Fig. 1D, black circles) and I40A-PLB (Fig. 1D, red triangles) showed increasing energy transfer efficiency with increasing relative protein expression level. This dependence was hyperbolic, with a similar maximal FRET efficiency (FRETmax) (Fig. 1D). The concentration (in arbitrary units) at which FRET was half-maximal was taken to represent the dissociation constant (KD). A fit of the form y = (FRETmax) x /(KD + x) gave FRETmax of 45.1 ± 1.3% for WT and 42.1 ± 1.8% for I40A. Half-maximal FRET was obtained at a concentration more than 4-fold higher for I40A (KD = 1.33 AU) than wild-type (KD = 0.32 AU). This indicates that the I40A point mutation reduces the apparent self-affinity of PLB by >4-fold. The data are summarized in Table 1. A naturally occurring human mutation of PLB, in which the codon for leucine 39 is changed to a stop codon, showed greatly reduced FRET at all concentrations (Fig. 1D, blue squares), and its KD could not be measured. In contrast to WT-PLB (Fig. 1A) and CFP-SERCA (3), YFP-L39Stop-PLB showed diffuse fluorescence localized in the cytoplasm and nucleus (Fig. 2). The lack of membrane partitioning was demonstrated by permeabilizing cells with the plasma membrane-selective agent saponin. This treatment provided access to an impermeant nuclear stain, propidium iodide, and caused complete loss of the soluble YFP-L39Stop-PLB, which immediately diffused out into the bath, leaving the cells dark in the YFP imaging channel (Fig. 2). This is in contrast to CFP-WT-PLB, which was unaffected by saponin permeabilization (Fig. 2).
TABLE 1.
Summary of PLB oligomerization, quaternary structure, and SERCA binding
| WT-PLB | I40A-PLB | L39Stop | |
|---|---|---|---|
| Mean pentamer FRET (%) | 37.8 ± 1.0 | 28.1 ± 1.0 | 7.4 ± 0.5 |
| Pentamer FRET sample size | 90 | 72 | 18 |
| Pentamer FRETmax (%) | 45.1 ± 1.3 | 42.1 ± 1.8 | NDa |
| Pentamer probe distance (Å) | 58.7 ± 0.5 | 59.9 ± 0.7 | ND |
| Pentamer KD (AU) | 0.32 ± 0.06 | 1.33 ± 0.20 | ND |
| Monomer (%) | 16 | 67 | >80 |
| FRAP τoverall (s) | 9.7 ± 0.7 | 6.8 ± 2.0 | 3.5 ± 0.4 |
| FRAP τ1 (s) | 2.9 ± 0.4 | 2.0 ± 2.4 | 1.6 ± 0.6 |
| FRAP τ2 (s) | 31 ± 9.5 | 21.5 ± 42 | 11.4 ± 8.2 |
| FRAP fast fraction | 0.48 | 0.54 | 0.77 |
| Photobleach τoverall (s) | 135 ± 1 | 96 ± 2 | 269 ± 1 |
| Photobleach τ1 (s) | 56 ± 4 | 40 ± 5 | 29 ± 2.6 |
| Photobleach τ2 (s) | 255 ± 20 | 225 ± 40 | 339 ± 6 |
| Photobleach fast fraction | 0.37 | 0.49 | 0.05 |
| Mean regulatory complex FRET (%) | 11.6 ± 0.8 | 16.2 ± 0.8 | 1.0 ± 0.4 |
| Regulatory complex FRET sample size | 48 | 69 | 46 |
| Regulatory complex FRETmax (%) | 30 ± 1.9 | 29 ± 1.1 | ND |
| Regulatory complex probe distance (Å) | 58.1 ± 0.9 | 58.6 ± 0.6 | ND |
| Regulatory complex apparent KD (AU) | 12.8 ± 1.6 | 4.4 ± 0.5 | ND |
ND, not determined.
FIGURE 2.
Cell permeabilization resulted in loss of freely diffusible YFP-L39Stop-PLB and provided access to cell-impermeant propidium iodide. CFP-WT-PLB was unaffected by saponin permeabilization.
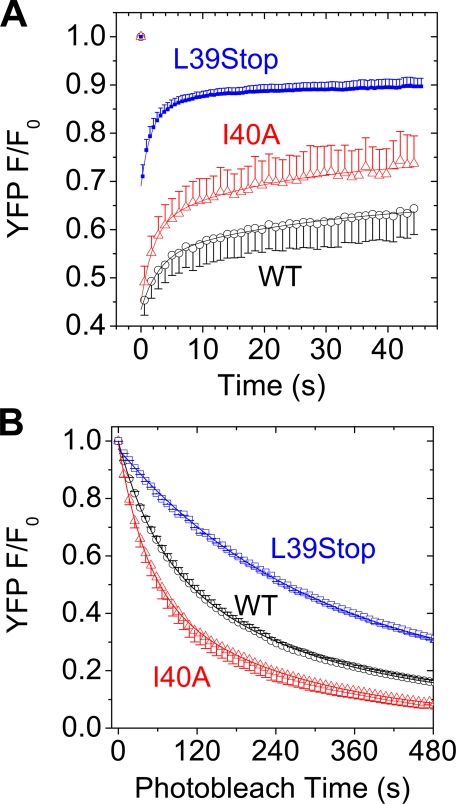
Fluorescence Recovery after Photobleaching (FRAP)—The L39Stop and I40A mutants of PLB were found to recover from spot photobleaching more quickly than WT-PLB (Fig. 3A). The FRAP time constants (τ), obtained from a single exponential fit of the form (y = y0 + Ae(–x/τ)) to combined average recovery traces was 9.7 ± 0.7 s for WT, 6.8 ± 2.0 s for I40A, and 3.5 ± 0.4 s for L39Stop. A multiexponential fit of the form y = y0 + A1e(–x/τ1) + A1e(–x/τ2) indicated that the faster overall recovery of I40A FRAP was due in part to an increase in a fast component at the expense of a slow component (Table 1). The very rapid recovery of L39Stop is consistent with being freely diffusible in the cytoplasm. For I40A, faster FRAP, with an increased proportion of a short time constant, is suggestive of a greater population of I40A in the more rapidly diffusible monomeric form.
FIGURE 3.
A, YFP-L39Stop-PLB (blue squares) and YFP-I40A-PLB (red triangles) recovered from spot photobleaching faster than YFP-WT-PLB (black circles). B, YFP-I40A-PLB (red triangles) photobleached faster than YFP-WT-PLB (black circles). YFP-L39Stop-PLB showed very high photostability (blue squares). Data are mean ± S.E.
YFP Photobleaching Rate—We quantified the rate of YFP fluorescence photobleaching as an index of homotransfer FRET (26, 27). Fig. 3B shows that YFP-I40A-PLB bleached more quickly than wild type, consistent with reduced self-association and decreased homotransfer. The difference in photobleaching rates (slopes) was greatest at the beginning of the bleach time course, when acceptor density was still high and the protective effects of homotransfer were most pronounced. Single exponential fits of the form y = y + Ae(–x/τ) to combined average F/F0 data showed that the overall bleach time constants (τ) for YFP-WT-PLB and YFP-I40A-PLB were 135 ± 1 and 96 ± 2s, respectively. Multiexponential analysis of the form y = y0 + A1e(–x/τ1) + A1e(–x/τ2) to combined average F/F0 data showed that this overall decrease in photobleaching time with I40A mutation was due in part to a larger proportion of a readily bleachable fraction (A1/(A1 + A2)) at the expense of a bleach-resistant fraction (A2/(A1 + A2)) (Table 1). The shift toward the readily bleachable fraction with I40A mutation is consistent with a greater proportion of monomeric PLB.
YFP-L39Stop-PLB showed very high photostability. Its photobleaching was not significantly biphasic (fast fraction = 0.05). The data were well described by a single exponential fit with τ = 269 ± 1 s (Fig. 3B). An interpretation of this unexpected result is offered under “Discussion.”
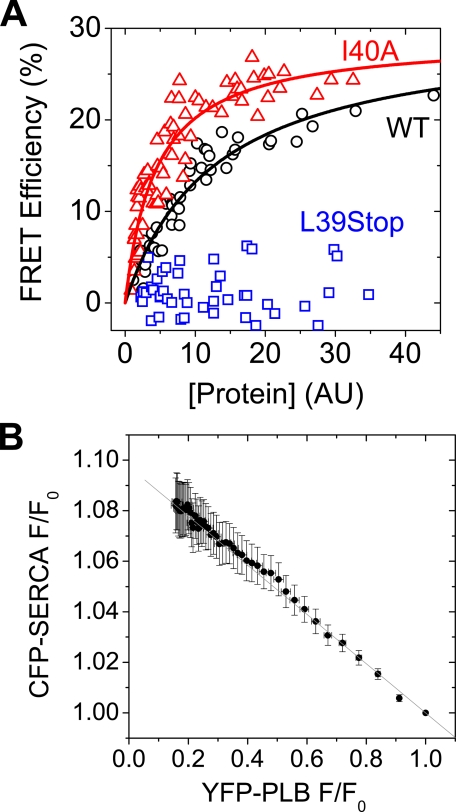
Regulatory Complex FRET—FRET within regulatory complexes of CFP-SERCA and YFP-labeled WT-, I40A-, or L39Stop-PLB was also quantified by acceptor photobleaching. As expected, the I40A mutation resulted in a greater association with SERCA, which binds only the monomer form (19, 28–30). Fig. 4A shows that the PLB-SERCA binding curve was left-shifted by the I40A mutation. The concentration of half-maximal FRET for I40A-PLB was 4.4 ± 0.5 AU, compared with 12.8 ± 1.6 AU for WT-PLB. The measured PLB concentration (x axis) includes both monomers and pentamers. We refer to the above values as “apparent KD values” to distinguish them from the true KD of the isolated monomer.
FIGURE 4.
A, concentration dependence of FRET from CFP-SERCA to YFP-labeled WT (black circles) or I40A (red triangles) PLB. The I40A mutation increased the apparent affinity of PLB for SERCA by 2.9-fold. The affinity of L39Stop-PLB (blue squares) was too low to measure. B, linear dependence of CFP fluorescence on YFP fluorescence, consistent with a bimolecular complex. Data are mean ± S.E.
The intrinsic FRET efficiency of the regulatory complex did not change with mutation. FRETmax was 30 ± 1.9% for WT-PLB and 29 ± 1.1% for I40A-PLB. YFP-L39Stop-PLB bound poorly to CFP-SERCA, as indicated by a very low FRET efficiency (p < 0.01 versus WT and I40A). CFP-SERCA and YFP-PLB fluorescences (F/F0) were linearly and inversely related (Fig. 4B), consistent with a bimolecular complex (21).
Quantification of FRET—To test whether the present methods accurately measured absolute FRET efficiency values, cells expressing standard FRET constructs were subjected to progressive acceptor photobleaching. FRET efficiencies thus obtained were in good agreement with published values for the standard constructs (23, 24, 31), as shown in Fig. 5A. Measured mean transfer efficiency values ± S.E. for C32V, C17V, C5V, and VCV were 29 ± 1, 39 ± 1, 41 ± 2, 65 ± 3%, respectively. To dissect specific (intracomplex) FRET from diffusion-enhanced FRET, we co-transfected YFP-PLB (donor) and mCherry-PLB (acceptor) with CFP-PLB (competitor), which cannot act as an acceptor for YFP-PLB. CFP-PLB fluorescence was quantified as a measure of competitor concentration. As CFP-labeled PLB increased, FRET from YFP-PLB to mCherry-PLB complexes decreased to a minimum of 3% (Fig. 5B). This constitutes an upper limit for nonspecific FRET in these studies.
FIGURE 5.
A, transfer efficiencies of standard FRET constructs. Measurements obtained in the present study (white bars, mean ± S.E.) are in agreement with published values (gray bars) (23, 24, 31). B, YFP-PLB to mCherry-PLB energy transfer, with competition by CFP-PLB. Error bars, ±S.E.
DISCUSSION
Significance of the Present Results—Previous determinations of PLB pentamer structure using fluorescence or magnetic resonance have used solubilized protein or PLB reconstituted into artificial lipid bilayers. To our knowledge, this is the first direct measurement of the quaternary conformation of expressed PLB in the membranes of living cells. The use of genetically encoded probes permits quantification of the energetic consequence of PLB transmembrane domain mutations without exposing the protein complexes to the destabilizing effect of detergents. This strategy may be generally useful for structural and thermodynamics studies of other membrane protein complexes.
Fluorescence Resonance Energy Transfer—The efficiency of FRET is very steeply dependent on probe separation distance (R), particularly for distances that are close to a probe pair's characteristic Förster radius (R0) (17). For CFP-YFP energy transfer, this distance is 49.2 Å (22), making these probes well suited to studying PLB pentamer and regulatory complex conformation. Over the sensitive portion of the curve, a change in probe separation distance of only 5 Å would alter the FRET efficiency of the complex by more than 15%. In the present study, we quantified FRET using the acceptor photobleaching method (32), since brightening of the donor after acceptor photobleaching is an unambiguous and quantitative measure of energy transfer efficiency. Transfer efficiency measurements of FRET standard constructs showed that this photobleaching protocol obtained absolute FRET efficiency measurements in agreement with nondestructive techniques, such as 3-cube FRET and FLIM (23, 24, 31). Computer-controlled excitation/emission filter changers automate progressive photobleaching of the acceptor, taking image data at intermediate points along the time course (Fig. 1). This provides additional information beyond that obtained from comparison of paired prebleach/postbleach images (33). For example, it facilitates detection of inadvertent bleaching of the donor during imaging or during acceptor photobleaching. Donor photobleaching, which would cause underestimation of FRET efficiency, is evident in donor dequenching curves as negative deviation from an exponential increase. We did not observe significant donor photobleaching under our experimental conditions. The progressive photobleaching method also provides some information about the stoichiometry of the complex that gives rise to FRET. Although a bimolecular complex always shows a linear relationship between acceptor bleaching and donor dequenching (Fig. 4B), multiple acceptors per donor can result in sublinear dependence of donor fluorescence on acceptor fluorescence (Fig. 1C), because energy transfer within the complex is maintained even after some of the acceptors have been bleached. This analysis is most effective for probe separations less than the characteristic Forster distance (R ≤ R0), since long transfer distances yield a subtle curvature that is difficult to detect or quantify (21). For R > R0, FRET efficiency depends much more strongly on probe separation distance, and the oligomer subunit number (n) used in the ring oligomer model did not significantly affect the present results. Distances obtained using assumptions of n = 3 or n = 7 changed the distance estimate by 0.8 Å and 0.2 Å, respectively.
Nonspecific FRET—Nonspecific FRET for a short-lifetime probe like CFP (∼2 nanosecond decay) is typically not a significant factor below millimolar concentrations in solution (critical concentration = 3.75 mmol/L). For probes restricted to membranes, FRET between unbound diffusible donors and acceptors is more likely (34–36) and has been quantified by multiple approaches (2, 37). For the present study, the minimum FRET observed between YFP- and mCherry-PLB in the presence of a large excess of CFP-PLB competitor (Fig. 5B) represents an upper limit for nonspecific FRET. Since the Förster distance (R0) for CFP-YFP is shorter than for YFP-mCherry (49.2 versus 55.8 Å), less nonspecific FRET is expected for CFP and YFP (34).
PLB Binding Equilibria Quantified by FRET—For soluble proteins, the concentration of fluorescent protein inside the cells can be conveniently estimated by comparison with the fluorescence of a standard concentration of the fluorescent protein applied in the bath (38). The emission intensity of CFP/YFP-PLB fluorescence of AAV-293 cells was comparable with standard concentration soluble CFP/YFP in the bath at 2 μmol/liter. However, since the CFP/YFP-PLB constructs are membrane-localized, and the amount of membrane surface in these cells is unknown, direct calibration of concentration with soluble fluorescent proteins is not valid. Cell-to-cell variability of endoplasmic reticulum membrane area may complicate the estimation of the concentration of expressed membrane proteins and increase the scatter of the FRET efficiency data (Fig. 1D). The present analysis assumes that the average endoplasmic reticulum membrane area of WT- and I40A-PLB-expressing cells is not different, and we report their relative concentration-dependent effects in arbitrary units.
We examined PLB pentamer FRET over a range of protein concentrations and obtained several informative parameters from a hyperbolic fit of the data (Fig. 1D). The FRETmax (saturation limit) is taken to be the intrinsic FRET efficiency of pentameric PLB. The average FRET efficiency of all WT-PLB-expressing cells was 37.8%, indicating that the protein was mostly pentamer (pentamer/monomer ratio >5/1) at the expression levels achieved in the current study. I40A-PLB average FRET efficiency was 28% (Fig. 1D), indicating a substantially increased proportion of monomer (pentamer/monomer ratio <2/1). The concentration at which we observed half-maximal FRET is taken to represent the dissociation constant (KD), which is useful for comparing relative affinities. By this measure, we found that the I40A mutation reduced PLB oligomerization affinity by >4-fold. According to the relationship, ΔΔG =–RT ln(KD1/KD2), where KD1 is the dissociation constant of WT-PLB, KD2 is the dissociation constant of I40A-PLB, R is the universal gas constant, and T is temperature, the I40A point mutation reduces the PLB free energy of self-association by 1.4 RT. Although this equilibrium shift is large enough to profoundly alter I40A function, the magnitude of the shift is significantly less than what is assumed from SDS-PAGE monomer/pentamer quantification. This highlights the need to measure PLB oligomerization in the environment of the membrane. Solubilization of PLB with detergents can fully disrupt binding interactions that are weakened by mutation (3, 39, 40). Since FRET can effectively quantify membrane protein binding interactions in undisrupted membranes, it is a good complement to classical biochemical methods that require solubilization.
The observation of increased FRET from CFP-SERCA to YFP-I40A-PLB compared with wild-type (Fig. 4A) is consistent with the model of coupled equilibria of PLB oligomerization and SERCA binding (19, 28–30). In this model, the monomer is the active species that is competent to bind the pump, and destabilization of the pentamer increases the proportion of monomer and, by mass action, increases the monomer bound to SERCA. This increases PLB molar activity, and I40A and other oligomer-destabilized mutants are known to be “superinhibitors” of the pump. Overall, the 2.9-fold shift in the apparent KD of the PLB-SERCA interaction (Fig. 4A) is of a magnitude appropriate to a secondary effect of the 4-fold shift in the pentamer-monomer equilibrium. If there is also an increase in the intrinsic affinity of I40A monomers for the pump, this effect must be small. A significant increase in affinity (in addition to the effect of increased monomer) would be expected to result in an even larger difference in apparent KD than what is observed here. The current experiments do not address whether the I40A mutation may alter the intrinsic inhibitory potency of bound monomer.
The I40A mutation-induced shift in the PLB oligomerization equilibrium to the monomer form is also evident from its faster diffusion in the membrane, as measured by recovery after photobleaching with a focused laser spot (Fig. 3A). Faster diffusion and FRAP are expected for the monomer, because its single transmembrane helix has a smaller radius of membrane displacement than the pentamer's bundle of transmembrane helices (41).
PLB oligomerization was also measured using the photobleaching rate as an index of fluorescence homotransfer (FRET between spectrally identical probes). Homotransfer FRET decreases the time that a particular fluorophore remains in an excited state (although the population average fluorescence lifetime remains the same). Since fluorophores in the excited state are more susceptible to oxidative reactions, homotransfer FRET reduces photobleaching (26, 27). We observed that YFP-I40A-PLB fluorescence photobleaches more quickly than YFP-WT-PLB (Fig. 3B), consistent with decreased homotransfer due to a decreased proportion of YFP-I40A-PLB in the pentamer form. This effect is probably exacerbated by the increased rate of PLB subunit exchange from pentamers of I40A. We have shown that I40A pentamers are more labile than WT pentamers, exchanging subunits on a time scale of tens of seconds (3). Thus, there is extensive cycling of YFP-I40A-PLB through the readily bleachable monomer pool on the minutes time scale of a field photobleaching experiment. This interpretation is supported by our observation that the difference between the YFP-WT-PLB and YFP-I40A-PLB photobleaching rates was small for fast photobleaching with high intensity illumination (not shown). The observed difference was much larger with moderate intensity photobleaching and was easily resolved with the protocol presented here.
Unexpectedly, YFP-L39Stop-PLB had the highest photostability of the three YFP-PLB variants (Fig. 3B). This is the opposite of what would be predicted from the very low energy transfer efficiency observed for this truncation mutant (Fig. 1D). We speculate that this low photobleaching rate was not due to protection by homotransfer but is a result of the different microenvironments that result from cytoplasmic versus membrane localization. It is unknown whether this difference is due to altered photochemistry or an additional fluorescence quenching process that deexcites YFP in the cytoplasm.
Structures of the Regulatory Complex and PLB Pentamer—The PLB-SERCA regulatory complex has been modeled from crystal structures of SERCA1a (42). The present data are compatible with this model, in which the respective N termini are on opposite sides of the complex. The SERCA/WT-PLB FRETmax value of 30 ± 1.9%, of which up to 3% may be nonspecific (Fig. 5B), corresponds to a probe separation distance of 58.1 ± 0.9 Å between the fluorescent proteins fused to the respective N termini. The 29 ± 1.1% FRETmax value obtained for I40A, less 3% nonspecific FRET, corresponds to a distance of 58.6 ± 0.6 Å. This indicates that this mutation does not significantly alter the quaternary conformation of the regulatory complex. The data confirm that I40A-PLB is a good substitute for WT-PLB in structure studies of the regulatory complex (2).
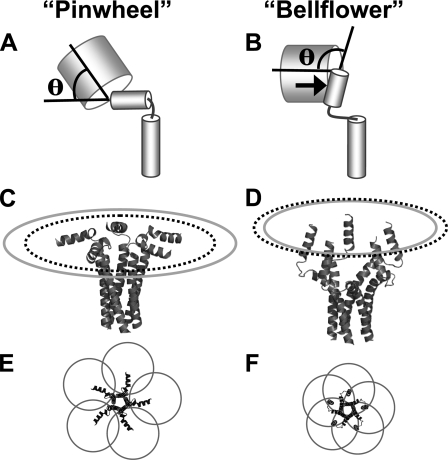
A variety of structures have been proposed for the PLB pentamer. Two strikingly different models, termed the bellflower (9) (Protein Data Bank code 1ZLL) and pinwheel (13) (Protein Data Bank code 1XNU), are characterized by membrane-normal or membrane-parallel orientations of the cytoplasmic domain, respectively (Fig. 6). Several new NMR studies of PLB in phospholipid bilayers provide evidence that PLB cytoplasmic domains are membrane-parallel (10, 14) and in contact with the bilayer surface (15, 16). Those studies are compatible with the pinwheel model of PLB pentamer structure, which was developed from the NMR solution structure of monomeric PLB in DPC micelles (7) in combination with FRET-based distance constraints from in-gel anisotropy of PLB in SDS detergent (13). The present study complements previous in vitro experiments by measuring the conformation of biosynthetic PLB in the membranes of living cells. The biological compatibility of the FRET method makes it a powerful tool for rapidly testing mutants and may permit the study of the structural consequences of phosphorylation (43).
FIGURE 6.
Pinwheel (A, C, and E) and bellflower (B, D, and F) structure models of the PLB pentamer (9, 13). Flexibility of the attachment of the PLB cytoplasmic domain helix to the fluorescent protein tag is expected to permit the probe to sample a range of anglesθ (A and B), although not all values ofθ are equally plausible. Steric hindrance (B, arrow) between PLB (small cylinders) and the fluorescent protein (large barrels) is expected to limit large θ angles. The distance between the fluorescent protein chromaphore and the point of attachment to PLB limits the maximum “reach” of the probe to a radius of 61.1 Å for the pinwheel (C, gray ellipse) and 46.3 Å for the bellflower structure (D, gray ellipse). The measured nearest neighbor probe separation distance of 58.7 Å for WT-PLB corresponds to a ring-shaped distribution of radius 49.9 Å, illustrated in C and D as a dotted black ellipse. This distance is well within the range of achievable distances for the pinwheel model (C) but falls outside the maximal reach of the fluorescent probes in bellflower structure (D). E and F show CFP/YFP Förster radii (R0 = 49.2 Å) centered on expected average chromaphore positions. The pinwheel conformation (E) permits overlap of only the nearest neighbor radii (yielding moderate FRET efficiency). The bellflower conformation (F) results in regions of R0 overlap for two, three, four, and five probes (yielding very high FRET efficiency).
In the pinwheel model (Fig. 6A), the average distance between adjacent N termini is 42.4 Å, which would yield moderate energy transfer efficiency between nearest neighbors. The bellflower structure (Fig. 6B) places PLB N termini in close proximity (25 Å). Therefore, high energy transfer efficiency is expected, both between adjacent N-terminal probes and across the pentamer. We observed 45.1% maximal FRET efficiency for WT-PLB (Fig. 1D). Subtracting 3% nonspecific FRET yields a probe separation distance of 58.7 Å between nearest neighbor chromaphores, according to a computational model of intrapentameric energy transfer based on ring oligomer FRET theory (21). This analytical model uses the acceptor molar fraction measured in the present study (0.88) and assumes that the FRETmax value obtained from the hyperbolic fit (Fig. 1D) represents PLB that is fully pentameric (0% monomer). Because of the high sensitivity of FRET to probe separation distance, the FRETmax parameter uncertainty of ±1.3% corresponds to a small uncertainty in calculated probe separation distance (±0.5 Å). The present FRET distance measurement of 58.7 Å for WT-PLB is compatible with the pinwheel model. The I40A FRETmax of 42 ± 1.8% (Fig. 1D), less 3% nonspecific FRET, corresponds to a distance of 59.9 ± 0.7 Å. This suggests that the I40A mutation does not significantly alter the conformation of the pentamer complex.
In comparing alternative structure models with the present data, it is important to consider the likely distribution of the CFP/YFP fluorescent proteins. Since the chromaphore is located at the center of the fluorescent protein β-barrel, the probe position is 25 Å removed from the end of the protein of interest (44). Although the position of the fluorophore in the barrel is rigid, it is reasonable to expect flexibility of the attachment of CFP/YFP to PLB domain IA, and the fluorescent protein can sample a range of angles θ relative to the axis of the helix of domain 1A (Fig. 6A). This uncertainty in fluorescent probe position places some limitations on estimating separation distance. Nevertheless, all values of θ are not equally plausible. Specifically, steric hindrance of the PLB cytoplasmic domain (Fig. 6, A and B, small cylinders) prevents the fluorescent C0protein (Fig. 6, A and B, large barrels) from sampling very large attachment angles. The distance between the fluorescent protein chromaphore and the point of attachment to PLB limits the maximum “reach” of the probe to a radius of 61.1 Å for the pinwheel (C, gray ellipse) and 46.3 Å for the bellflower structure (D, gray ellipse). The measured nearest neighbor probe separation distance of 58.7 Å for WT-PLB corresponds to a ring-shaped distribution of radius 49.9 Å, illustrated in C and D as a dotted black ellipse. This distance is well within the range of achievable probe positions for the pinwheel model (C, gray ellipse) but falls outside the maximal reach of the fluorescent probes in bellflower structure (D, gray ellipse). For the pinwheel, the measured radius (C, black dotted ellipse) does not require an unlikely attachment angle; the angle depicted in Fig. 6A (θ = 56°) places the chromaphore in the appropriate position. Alternatively, a subpopulation of structurally dynamic PLB (45) could shift the average chromaphore position and foreshorten the radius of the ring of fluorescent probes. The distance measured by FRET is an average value that integrates all conformations.
By contrast, probes attached to a bellflower structure could not accommodate the measured radius (Fig. 6D, black dotted ellipse). Even to reach the limit highlighted by the gray ellipse would require a highly ordered attachment angle exceeding 90° (Fig. 6B). This maximal reach value of 46.3 Å is offered as a formal possibility, but we regard such a severe bend of the fluorescent protein-PLB junction as sterically unfavorable (Fig. 6B, arrow) and highly unlikely. Instead, the position of the chromaphores in a bellflower-shaped structure is expected to lie even closer to the pentamer's central axis. We conclude that the present FRET distance constraints are consistent with the pinwheel model and are not compatible with the compact arrangement of PLB cytoplasmic domains depicted in the bellflower structure.
Fig. 6, E and F, shows top views of the pinwheel and bellflower conformations, respectively, superimposed with the CFP/YFP R0 of 49.2 Å centered on the likely average probe position for each structure. For the pinwheel, only the R0 values of adjacent (nearest neighbor) subunits overlap. For the bellflower structure, there is both overlap of adjacent subunits and of opposite subunits, creating regions where the R0 values of two, three, four, and five probes overlap each other. The different conformations proposed for the PLB pentamer therefore predict significantly different intrinsic FRET efficiency.
The bellflower and pinwheel structures are not necessarily exclusive alternatives. Spin label dynamics measurements provide evidence for a mixed population of ordered and disordered PLB (45, 46). These fractions are characterized by membrane-associated and axially inclined conformations, respectively. The pentamer conformation may also be influenced by phosphorylation (16, 47). Thus, the bellflower and the pinwheel may represent substates of a dynamic, regulated quaternary conformation. Nevertheless, the present FRET measurements in living cells indicate that the average nearest neighbor transfer distance (R) between N-terminal fluorescent probes is long (R/R0 > 1). Therefore, we conclude that the pinwheel represents the predominant quaternary conformation.
One may speculate that a pinwheel conformation is required for targeting of protein kinases (protein kinase A and CamKII). A prevailing membrane-associated structure may also be a mechanism for preventing the inactive form of PLB from interacting with and regulating SERCA. Indeed, recent studies suggest that the degree of dynamic disordering of the PLB cytoplasmic domain is a determinant of PLB inhibitory function (48).
We cannot fully exclude the possibility that the fluorescent protein fusions perturb the structure of the complex. In general, we prefer fluorescent proteins as relatively benign alternatives to organic dyes, which may also perturb structures by partitioning into membranes or by interacting with one another. Furthermore, structures obtained by label-free methods, such as NMR or x-ray crystallography, may also suffer from distortions due to crystal contacts, detergent solubilization, or other nonphysiological sample conditions. Thus, we feel that unanswered questions in membrane protein structure are well served by a combined approach; conventional high resolution structural methods complemented by the biologically compatible fluorescence methods described here.
PLB L39Stop Mutation—The PLB L39Stop mutation results in the PLB peptide being truncated midway through its transmembrane domain. The residues that mediate PLB-PLB interactions (as well as many determinants of SERCA binding) are in the PLB transmembrane domain (19, 39). We hypothesized that its oligomerization and SERCA binding would be significantly altered by the L39Stop mutation. Indeed, overall FRET for L39Stop oligomer and regulatory complex were reduced to 7.4 and 1.0%, respectively.
L39Stop-PLB has been expressed in HEK cells by another group and shown by immunolabeling of fixed samples to be membrane-localized (18). We did not detect any partitioning of YFP-L39Stop-PLB to the membrane. Fluorescence imaging showed the protein was soluble, localized in the cytoplasm and nucleus. Saponin permeabilization resulted in complete loss of fluorescence from cells (Fig. 2). This suggests that the remaining 7 residues of the L39Stop-PLB transmembrane domain are insufficient to anchor the protein in the membrane. The resulting incorrect localization may be why the protein is degraded in patients who carry this mutation (18). Regardless of degradation, the present FRET experiments suggest that the truncated PLB binds poorly to SERCA and would be an ineffective regulator of the pump.
Acknowledgments
We thank Timothy P. Remus, Joshua T. Maxwell, and Karl Hench for technical assistance and Razvan L. Cornea for helpful comments. The clone for mCherry was a gift of Professor Roger Y. Tsien. FRET efficiency standard constructs were a gift of Professor Steven S. Vogel. A206K mutagenic primers were a gift of J. Michael Autry and Professor David D. Thomas. We thank Professor Chikashi Toyoshima for the gift of the regulatory complex model Protein Data Bank file.
This work was supported by National Institutes of Health Mentored Research Scientist Development Award EB006061. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PLB, phospholamban; SERCA, sarco(endo)plasmic reticulum calcium ATPase; FRET, fluorescence resonance energy transfer; CFP, cyan fluorescent protein; YFP, yellow fluorescent protein; AU, arbitrary units; WT, wild type; FRAP, fluorescence recovery after photobleaching.
References
- 1.MacLennan, D. H., and Kranias, E. G. (2003) Nat. Rev. Mol. Cell. Biol. 4 566–577 [DOI] [PubMed] [Google Scholar]
- 2.Mueller, B., Karim, C. B., Negrashov, I. V., Kutchai, H., and Thomas, D. D. (2004) Biochemistry 43 8754–8765 [DOI] [PubMed] [Google Scholar]
- 3.Robia, S. L., Campbell, K. S., Kelly, E. M., Hou, Z., Winters, D. L., and Thomas, D. D. (2007) Circ. Res. 101 1123–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inui, M., Chamberlain, B. K., Saito, A., and Fleischer, S. (1986) J. Biol. Chem. 261 1794–1800 [PubMed] [Google Scholar]
- 5.Karim, C. B., Stamm, J. D., Karim, J., Jones, L. R., and Thomas, D. D. (1998) Biochemistry 37 12074–12081 [DOI] [PubMed] [Google Scholar]
- 6.Simmerman, H. K., Kobayashi, Y. M., Autry, J. M., and Jones, L. R. (1996) J. Biol. Chem. 271 5941–5946 [DOI] [PubMed] [Google Scholar]
- 7.Zamoon, J., Mascioni, A., Thomas, D. D., and Veglia, G. (2003) Biophys. J. 85 2589–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arkin, I. T., Rothman, M., Ludlam, C. F., Aimoto, S., Engelman, D. M., Rothschild, K. J., and Smith, S. O. (1995) J. Mol. Biol. 248 824–834 [DOI] [PubMed] [Google Scholar]
- 9.Oxenoid, K., and Chou, J. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10870–10875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abu-Baker, S., Lu, J. X., Chu, S., Shetty, K. K., Gor'kov, P. L., and Lorigan, G. A. (2007) Protein Sci. 16 2345–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mascioni, A., Karim, C., Zamoon, J., Thomas, D. D., and Veglia, G. (2002) J. Am. Chem. Soc. 124 9392–9393 [DOI] [PubMed] [Google Scholar]
- 12.Pollesello, P., Annila, A., and Ovaska, M. (1999) Biophys. J. 76 1784–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robia, S. L., Flohr, N. C., and Thomas, D. D. (2005) Biochemistry 44 4302–4311 [DOI] [PubMed] [Google Scholar]
- 14.Traaseth, N. J., Verardi, R., Torgersen, K. D., Karim, C. B., Thomas, D. D., and Veglia, G. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 14676–14681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abu-Baker, S., and Lorigan, G. A. (2006) Biochemistry 45 13312–13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clayton, J. C., Hughes, E., and Middleton, D. A. (2005) Biochemistry 44 17016–17026 [DOI] [PubMed] [Google Scholar]
- 17.Förster, T. (1948) Ann. Phys. (Leipzig) 2 55–75 [Google Scholar]
- 18.Haghighi, K., Kolokathis, F., Pater, L., Lynch, R. A., Asahi, M., Gramolini, A. O., Fan, G. C., Tsiapras, D., Hahn, H. S., Adamopoulos, S., Liggett, S. B., Dorn, G. W., 2nd, MacLennan, D. H., Kremastinos, D. T., and Kranias, E. G. (2003) J. Clin. Invest. 111 869–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura, Y., Kurzydlowski, K., Tada, M., and MacLennan, D. H. (1997) J. Biol. Chem. 272 15061–15064 [DOI] [PubMed] [Google Scholar]
- 20.Merzlyakov, M., You, M., Li, E., and Hristova, K. (2006) J. Mol. Biol. 358 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, M., Reddy, L. G., Bennett, R., Silva, N. D., Jr., Jones, L. R., and Thomas, D. D. (1999) Biophys. J. 76 2587–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson, G. H., Piston, D. W., and Barisas, B. G. (2000) Anal. Biochem. 284 438–440 [DOI] [PubMed] [Google Scholar]
- 23.Koushik, S. V., Chen, H., Thaler, C., Puhl, H. L., III, and Vogel, S. S. (2006) Biophys. J. 91 L99–L101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thaler, C., Koushik, S. V., Blank, P. S., and Vogel, S. S. (2005) Biophys. J. 89 2736–2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waggoner, J. R., Huffman, J., Griffith, B. N., Jones, L. R., and Mahaney, J. E. (2004) Protein Expression Purif. 34 56–67 [DOI] [PubMed] [Google Scholar]
- 26.Kubitscheck, U., Kircheis, M., Schweitzer-Stenner, R., Dreybrodt, W., Jovin, T. M., and Pecht, I. (1991) Biophys. J. 60 307–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young, R. M., Arnette, J. K., Roess, D. A., and Barisas, B. G. (1994) Biophys. J. 67 881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Autry, J. M., and Jones, L. R. (1997) J. Biol. Chem. 272 15872–15880 [DOI] [PubMed] [Google Scholar]
- 29.Reddy, L. G., Jones, L. R., and Thomas, D. D. (1999) Biochemistry 38 3954–3962 [DOI] [PubMed] [Google Scholar]
- 30.Zhai, J., Schmidt, A. G., Hoit, B. D., Kimura, Y., MacLennan, D. H., and Kranias, E. G. (2000) J. Biol. Chem. 275 10538–10544 [DOI] [PubMed] [Google Scholar]
- 31.Chen, H., Puhl, H. L., 3rd, Koushik, S. V., Vogel, S. S., and Ikeda, S. R. (2006) Biophys. J. 91 L39–L41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kenworthy, A. K. (2001) Methods 24 289–296 [DOI] [PubMed] [Google Scholar]
- 33.Van Munster, E. B., Kremers, G. J., Adjobo-Hermans, M. J., and Gadella, T. W., Jr. (2005) J. Microsc. (Oxf.) 218 253–262 [DOI] [PubMed] [Google Scholar]
- 34.Fung, B. K., and Stryer, L. (1978) Biochemistry 17 5241–5248 [DOI] [PubMed] [Google Scholar]
- 35.Lakowicz, J. R. (1999) Principles of Fluorescence Spectroscopy, 2nd Ed., p. 382, Kluwer Academic/Plenum Press, New York
- 36.Vogel, S. S., Thaler, C., and Koushik, S. V. (2006) Sci. STKE 2006, re2. [DOI] [PubMed]
- 37.Li, E., You, M., and Hristova, K. (2006) J. Mol. Biol. 356 600–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang, M., and Walker, J. W. (2005) J. Mol. Cell Cardiol. 38 753–764 [DOI] [PubMed] [Google Scholar]
- 39.Cornea, R. L., Autry, J. M., Chen, Z., and Jones, L. R. (2000) J. Biol. Chem. 275 41487–41494 [DOI] [PubMed] [Google Scholar]
- 40.Hughes, E., Clayton, J. C., and Middleton, D. A. (2005) Biochemistry 44 4055–4066 [DOI] [PubMed] [Google Scholar]
- 41.Saffman, P. G., and Delbruck, M. (1975) Proc. Natl. Acad. Sci. U. S. A. 72 3111–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toyoshima, C., Asahi, M., Sugita, Y., Khanna, R., Tsuda, T., and MacLennan, D. H. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bossuyt, J., Despa, S., Martin, J. L., and Bers, D. M. (2006) J. Biol. Chem. 281 32765–32773 [DOI] [PubMed] [Google Scholar]
- 44.Tsien, R. Y. (1998) Annu. Rev. Biochem. 67 509–544 [DOI] [PubMed] [Google Scholar]
- 45.Nesmelov, Y. E., Karim, C. B., Song, L., Fajer, P. G., and Thomas, D. D. (2007) Biophys. J. 93 2805–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karim, C. B., Kirby, T. L., Zhang, Z., Nesmelov, Y., and Thomas, D. D. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 14437–14442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simmerman, H. K., and Jones, L. R. (1998) Physiol. Rev. 78 921–947 [DOI] [PubMed] [Google Scholar]
- 48.Ha, K. N., Traaseth, N. J., Verardi, R., Zamoon, J., Cembran, A., Karim, C. B., Thomas, D. D., and Veglia, G. (2007) J. Biol. Chem. 282 37205–37214 [DOI] [PubMed] [Google Scholar]