Abstract
The immature nucleus pulposus (NP) is populated by cells of notochordal-origin that are larger and contain an extensive cytoskeletal network and numerous vacuoles. The disappearance of these cells with age is believed important in regulating metabolic shifts that may contribute to age-related disc degeneration. The precise biological function of these notochordal cells in the immature NP remains unclear, however, because of challenges in studying the mixed cell population in the NP. In this study, notochordal-like cells were purified from immature NP cells using a new fluorescence-activated cell sorting (FACS) protocol with auto-fluorescence and size analysis. The unique molecular phenotypes of sorted notochordal-like cells were characterized by the mRNA expression pattern for key matrix proteins and modulators, and by the expression of cell–matrix receptor integrin subunits. An FACS analysis showed that the immature NP contained a majority of cells that were larger than anulus fibrosus (AF) cells and with fluorescence higher than AF cells. In comparison with the small NP cells separated by the FACS protocol, sorted notochordal-like cells expressed lower mRNA levels of type I collagen, biglycan, TIMP1, HSP70 and c-fos, and did not express detectable mRNA levels of decorin, lumican, multiple MMPs or IL-1β via real-time quantitative RT-PCR. A greater number of these notochordal-like cells also expressed the higher levels of α6, α1 and β1 integrin subunits as compared to small NP cells. Together, our results point towards a unique molecular phenotype for these notochordal-like cells of NP, characterized by the absence of gene expression for specific small proteoglycans and higher protein expression of integrin subunits that regulate interactions with collagens and laminin. Future studies will be important for revealing if this unique molecular profile is coordinated with functional differences in pericellular matrix regions and/or integrin-mediated cell–matrix interactions for these notochordal-like cells within the NP.
Keywords: Intervertebral disc, Nucleus pulpous, Notochord, Gene expression, FACS
Introduction
Intervertebral disc (IVD) disorders and aging-related degeneration are significant contributors to low back pain and spine-related disability [4]. There exists great interest in understanding the complex pathogenesis of IVD diseases, and in the use of biological or cellular approaches to treat and inhibit the progression of IVD disorders [4]. The anulus fibrosus (AF) of the IVD is populated by fibrochondrocyte-like cells of mesenchymal origin, while the central-most nucleus pulposus (NP) is populated by both small chondrocyte-like cells of mesenchymal origin and larger, highly vacuolated cells derived from the notochord [22]. Notochordal cells in the nucleus pulposus begin to disappear shortly after birth and are replaced by fibrochondrocytic cells of the adjacent anulus fibrosus and cartilage endplate with age [6, 14, 20, 23]. When isolated from most immature animal tissues, NP cells are presumed to contain many cells of notochordal origin [1, 10, 11, 15, 18] although some animal tissues retain notochordal cells well into maturity (e.g., rabbit, nonchondrodystrophoid dogs) [1]. In the rat, notochordal cells are the dominant cell population in the immature tissue, while virtually no notochordal cells are present during 1–2 years [20]. In the human, notochordal cells are rarely observed after the age of 7 [21] and the disappearance of the notochordal cell population and replacement by fibrochondrocyte-like cells may contribute to matrix changes and decreases in cellularity of NP during aging.
Notochordal cells are involved in the development of the spinal cord and vertebra; they also contribute towards patterning and differentiation of the intervertebral discs [9]. It is these cells that synthesize extracellular matrix in NP during development, and have the potential to secrete soluble mediators that enhance biosynthesis for AF cells. Indeed, the notochordal cell has been suggested as a potential source of therapeutic factors based on the documented biosynthetic and stimulatory behaviors of immature NP cells that are believed to be a mixed population of cells of notochordal and mesenchymal origins. The addition of conditioned media collected from NP cell culture has been shown to enhance the biosynthesis of cartilage matrix proteins and s-GAG by AF or fibrocartilagionous nucleus cells in vitro [1, 5]. Re-insertion of autologous immature NP cells containing a large number of notochordal cells has been shown to delay disc degeneration in animal models of nucleotomy [17]. Thus, notochordal cells or NP cells with enriched notochordal cells, are of increasing interest for their potential as autologous cell therapy, or more simply, for the signaling molecules that they express.
While notochordal cells have been implicated as essential in IVD development, NP matrix synthesis and maintenance, the mechanism of the shift in morphology of the local NP cell population, from notochordal to fibrochondrocytic, as well as its implications on IVD health, degeneration and repair is poorly understood. The precise biological function of notochordal cells remains unclear because of challenges in studying them as a mixed cell population in the NP. A few studies have attempted to isolate notochordal cells from immature NP tissue using dissection or gradient centrifugation techniques, and have visually identified the notochordal cells as larger, vacuole-containing cells with an extensive actin cytoskeletal network [1, 11, 12]. The purpose of this study was to purify large notochordal-like cells from immature NP cells by a new fluorescence-activated cell sorting (FACS) protocol and characterize their unique molecular phenotype in comparison with small chondrocyte-like cells of NP. We hypothesized those notochordal-like cells of NP exhibit unique profiles of gene and protein expression for a subset of proteins related to extracellular matrix and cytoskeleton, as well as some growth factors, cytokines, and signaling molecules involved in development. These proposed distinguishing features of the notochordal-like cell are of interest as they may contribute to the NP inability to regulate and repair the extracellular matrix in IVD degeneration. The gene expression profile of both large notochordal-like cells and small chondrocyte-like cells in immature NP was evaluated with real time RT-PCR for key extracellular matrix and cytoskeletal proteins, metalloproteinases and their inhibitors, growth factors, cytokines, stress response proteins and transcription factors. The sorted cell populations were further characterized by their protein expression of a subset of integrin cell surface receptors following recent findings of a unique integrin expression pattern for NP [16]. This approach towards identifying a unique and distinct cell population within the immature IVD is expected to provide new information that will facilitate the identification of specific therapeutic factors in notochordal cells of the NP.
Materials and methods
Primary cell isolation
Lumbar spines were obtained from pigs (9–12 weeks old, 18–27 kg) or rats (Sprague-Dawley, 2–3 months old, 250–300 g) within 4–6 h of sacrifice (Duke University Vivarium). Cells were isolated from tissue harvested from the NP and AF of spines with a sequential pronase–collagenase digestion [7]. The primary cells were suspended in cell culture media (Ham’s F-12 medium, Invitrogen Life Technologies, Carlsbad, CA, USA; supplemented with 10% fetal bovine serum (FBS), 25 μg/ml ascorbic acid, 100 U/ml penicillin, 100 μg/ml streptomycin, 1 μg/ml fungizone and 10 μg/ml DNase I) prior to cell sorting. For one cell isolation, tissues from 2–3 porcine or rat spines were pooled.
Flow cytometry and purification of notochordal-like cells by fluorescence-activated cell sorting (FACS)
Flow cytometry can simultaneously measure multiple physical characteristics (i.e. size, granularity, fluorescence intensity) of single cells based on their light scattering and diffracting properties as they flow through a beam of light. Side scatter (SSC, also called 90° scatter or right angle scatter) is light scattered at any interface within the cell where there is a change in refractive index, and is thus related to the internal granularity or complexity of cellular organelles including cell membrane and nucleus. Forward angle scatter (FSC) is a measurement of mostly diffracted light and is proportional to cell-surface area or size. In addition, cell shape and surface topography also contribute to both side and forward light scattering in flow cytometry. In general, cells of different sizes, internal organizations and membrane structures will exhibit differences in how they scatter and diffract incident laser light, or emit fluorescence. In this study, it was assumed that AF and NP cells would exhibit different light scattering properties as AF cells are smaller (~15 μm) and do not contain internal vacuoles like NP cells. Flow cytometry was first used to analyze NP and AF cells and investigate this assumption by measuring light scattering properties and auto-fluorescence intensities, as described below.
First, freshly isolated porcine or rat AF and NP cells (2×106 per ml each) were analyzed by flow cytometry to determine both SSC and FSC-height on a FACScan (Becton Dickinson, San Jose, CA, USA), yielding size scattergrams and histograms to assess uniformity of the cell population (Fig. 1). Both pig and rat cell sources were analyzed in this experiment to define a sorting protocol that was not species dependent, and that would be applicable to more than one immature NP tissue source containing notochordal-like cells. The number of cells with FSC-height values larger or smaller than different threshold cut-offs was counted for both AF and NP cells in order to define two population distributions of cells to be used for sorting in a later protocol (CellQuest software, Becton Dickinson). Flow cytometry analyses were repeated for multiple preparations of cells prepared from different cell isolations (n=2–3) to insure uniformity. A threshold value of FSC-height was sought to define a distribution of AF cells with approximately 95% confidence, as the AF cell population was fairly narrow in its distribution of light scattering properties. The FSC-height value for this threshold varied amongst flow cytometry analyses of multiple samples across species (n=2 rat samples, n=3 pig samples), but generally fell between 85 and 110 (no units, relative value). In order to define a single number to be used as a threshold for all cell sources and all FACS runs, an FSC-height value of 100 was chosen on the FACScan that represented between 85 and 95% of the AF cell populations for all runs (i.e., cells with FSC-height values less than 100 have an ~90% probability of being from the AF population). Thus, this value was used as a “gate” or reference to compare NP and AF cells. The NP cells that fell outside of this region were considered to be a separate population.
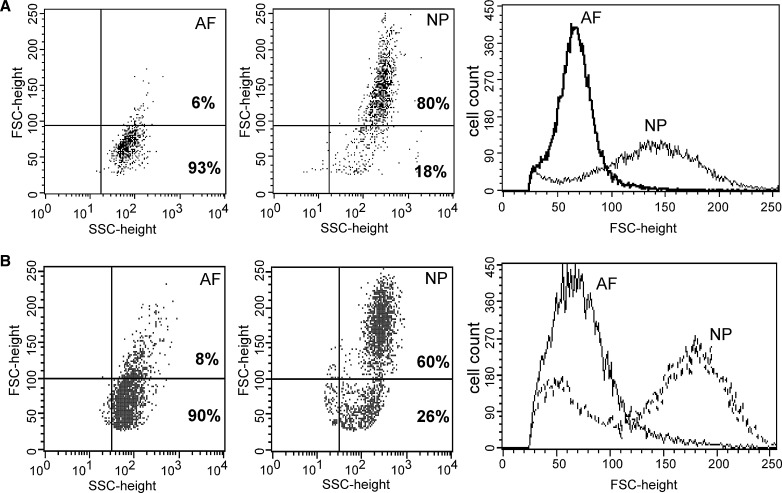
Fig. 1.
Flow cytometry of nucleus pulposus (NP) and anulus fibrosus (AF) cells by light scatter analysis for cell size. a Porcine, b Rat. In scattergrams, yaxes FSC = cell size; xaxes SSC = cell granularity or internal complexity. Bold number indicates the percentage of cells with FSC-height values greater or lower than 100)
Next, freshly isolated AF and NP cells (5×106 per ml) were analyzed for both auto-fluorescence (530/30 nM filter, x axes) and blue fluorescence (450/20 nM filter, y axes) on a FACStarPLUS (Becton Dickinson), yielding scattergrams and histograms to assess the difference between AF and NP cells. As for gates set via the parameter FSC-height, gate R2 based on fluorescence intensity was chosen to insure that between 85 and 95% of AF cells were contained within the chosen region (Fig. 2). A second gate, R3, was chosen to be just outside of, and with values larger than that of the R2 gate region. The number of cells in gate R3 (Figs. 2, 3a, higher fluorescence intensity) and gate R2 (Figs. 2, 3a, lower fluorescence intensity) was counted for both AF and NP cells, and the mean blue fluorescence intensity (MFI) value of cells was recorded (n=2 rat samples, n=3 pig samples). For NP cells, cell populations with high or low fluorescence (R2 or R3) were further evaluated for their light scatter characteristics as described above to evaluate cell size (Fig. 3a middle and right).
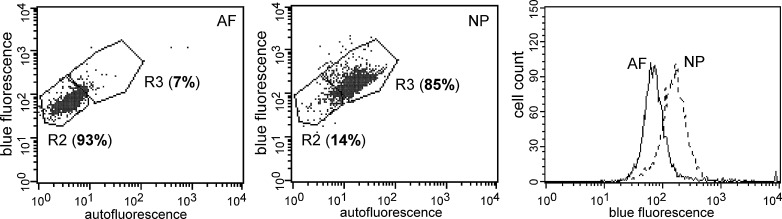
Fig. 2.
Flow cytometry analysis by both blue and auto-fluorescence of rat anulus fibrosus (AF) and nucleus pulposus (NP) cells. In scattergrams, yaxes intensity of blue fluorescence; xaxes intensity of auto-fluorescence. Bold number indicates the percentage of cells within gate R3 or R2 with different fluorescence values
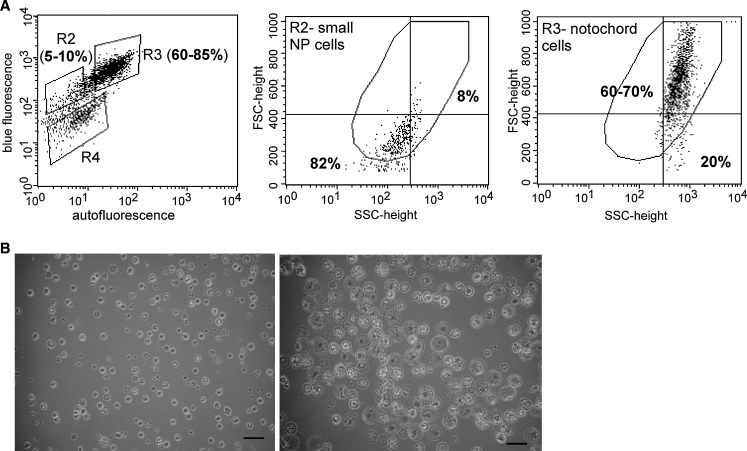
Fig. 3.
a FACS sorting of porcine nucleus pulposus (NP) cells. In fluorescence scattergrams, yaxes intensity of blue fluorescence; x axes intensity of auto-fluorescence and bold number indicates that the percentage of cells within gate R3 or R2 with different fluorescence values. In FSC-height scattergrams, yaxes: FSC = cell size; x axes: SSC = cell granularity or internal complexity, and bold number indicates the percentage of cells with FSC-height value greater or lower than 400. b Morphology of sorted large notochordal-like cells (right) and small NP cells (left) (bar size=75 μm)
For purification of notochordal-like cells, freshly isolated NP cells (5×106 per ml) were sorted by both fluorescence (530/30 and 450/20 nM filters) and size (light scatters) on the FACStarPLUS as described above. In order to gate the large size of cells in the NP population, animal-matched AF cells (10–15 μm cell size) [11] were always run first and gated according to FSC-height and fluorescence intensity as a reference population. Note that the absolute values for FSC-height were different on the FACScan and FACStarPLUS instruments used for analysis and sorting, respectively, as each instrument scaled this arbitrary unit against maximum possible values for the machine. However, the relative size difference of AF and NP detected were the same no matter which instrument was used. Thus, the value of the gate for FSC-height corresponding to the AF cell population was found to be <400 on the FACStarPLUS. The values of gates for both FSC-height and R2 for fluorescence intensity were chosen for this defined AF cell population as described above; this procedure varied from run to run across samples (n=3) according to the criteria of including ~90% of all AF cells. As described above, the R3 gate was set at values just above that of AF cells. From run to run, the gate setting for R2 varied by less than 5%, presumably since the size and fluorescence intensity of the AF cells was very homogenous between different cells of pigs or rats. Thus, sorted cells were collected into two fractions consisting of higher fluorescence with sizes larger than AF cells (sorted notochordal-like cells), and lower fluorescence with sizes smaller than or same as anulus cells (sorted small NP cells). Cell yields after sorting were generally low and different from notochordal-like cells and small NP cells (i.e., the percentage of recovered cells from a sorting experiment was 15–30% for notochordal-like cells, but 5–10% for small NP cells), however, so that very high starting cell numbers were needed for additional protein or RNA characterization of the sorted cell population. Significantly high starting cell numbers were easily obtained from pig spines (25–35×106 per pig spine) but not from the smaller rat spines (0.5–1×106 per rat spine). For this reason, further RNA isolation and integrin analysis of the sorted NP cells were performed on porcine cells only, and were pooled from multiple sorting experiments (n=3).
Real time quantitative RT-PCR
In order to quantify mRNA levels for multiple genes in the sorted NP cells, it was necessary to pool sorted NP cells collected from three separate sorting experiments into one sample. Total RNA was extracted from freshly sorted porcine IVD cells with the RNeasy mini kit plus DNase I digestion (Qiagen, Valencia, CA, USA). Quantification of mRNA was performed for genes of a wide panel of proteins relevant to disc homeostasis, including key extracellular matrix proteins, metalloproteinases and their inhibitors, cytoskeletal proteins, cytokines, growth factors, stress response proteins and transcriptional factors. For each target gene, two porcine-specific PCR primers and one fluorescently labeled intron-spanning probe were designed using published sequences in GenBank and Primer Express™ software (Applied Biosystems, Foster City, CA, see Table 1 and Chen et al. 7 for sequence details). Real time RT-PCR (SmartCycler® system, Cepheid, Sunnyvale, CA, USA) conditions were used as described previously [7]. Relative gene expression differences were quantified among the sorted notochordal-like cells, small NP cells and anulus fibrosus cells using the comparative Ct method with 18S rRNA as an internal control [7]. Duplicate PCR reactions were performed for each target gene and the internal control for one RNA sample.
Table 1.
Target gene sequences
| Target genea | Forward primer (5′–3′) | Reverse primer (5′–3′) | Probe (5′–3′) |
|---|---|---|---|
| MMP-1 X54724 | CGTGCCATTGAGAAAGCCTT | GTCTGCTTGACCCTCGGAGA | AGCAATGTCTCACCCTTGACCTTCACCA |
| MMP-2 AF295805 | CACGCTGGGTCCTGTCACT | GCGGATCTGAGAGATGCCAT | CCGAGATCTGCAAACAGGACATCGTCTT |
| MMP-3 AF201725 | TCCTGATGTTGGTTACTTCAGCAC | TTGACAATCCTGTAAGTGAGGTCATT | TCCTGGCCTGCCCAAGTGGAGAA |
| MMP-13 AF069643 | CCAAAGGCTACAACTTGTTTCTTG | TGGGTCCTTGGAGTGGTCAA | GAATGGCCAAACTCATGGGCAGCA |
| TIMP-1 AF156029 | CGCCTCGTACAAGCGTTATGA | TAGATGAACCGGATGTCAGGG | CAAGGCATTGAACCCTTTGAACATCTTGGT |
| TIMP-2 AF156030 | CTCGCTGGACATCGGAGG | AGGGCACGATGAAGTCACAGA | CCTCGGCCTTTCCTGCGATGAGGTA |
| INOS X98196 | CCTGGAGGAGGAGCAGCTG | GCATACCTGAACTTGTTGGTGAGT | CCAGGCAATGGAGAGAAACTGAAGAAGTCC |
| HSP70 AJ310378 | CTTGAGAATTATTTGCCTTTCATGTAAG | GAAGGTGGCAGTGTTGATTCAC | TACTGTAAGGGTGGCTTTTGGTCGCCC |
| TGF-β1 AF461808 | GGAGTGGCTGTCCTTTGATGTC | AGTGTGTTATCTTTGCTGTCACAGG | AACCCTCTATAGCCTCTCTGCGGGTCAGC |
| IL-1β M86725 | GCTGAAGGCTCTCCACCTCC | TAAGGTCACAGGTATCTTGTTGTTGC | TCATGCAGAACACCACTTCTCTCTTCAAGTCC |
| IL-4 X68330 | AGTGCGACATCACCTTACAAGAGAT | GGCCCGGCAGAAGGTTT | ATTCTCACAGCGAGAAAGAACTCGTGCATG |
| IL-6 M80258 | CCCTCCAGGAACCCAGCT | CCCAGGGAGAAGGCGACT | CTCCCTCTCCACAAGCGCCTTCAGTC |
| TNF-α X57321 | TCTGCCTACTGCACTTCGAGG | TGGCTACAACGTGGGCG | CTCTGGCCCAAGGACTCAGATCATCG |
| sox-9 AF029696 | ATCAGTACCCGCACCTGCAC | CTTGTAATCCGGGTGGTCCTT | TCGCTCTCATTCAGCAGTCTCCAGAGTTTG |
| c-jun S83515 | CGACCTTCTACGACGATGCC | CAGGGTCATGCTCTGCTTCA | CCTCGTTCCTCCAGTCCGAGAGCG |
| c-fos AJ132510 | AGCCAAATGCCGGAACC | TGGCGATCTCAGTCTGCAAA | ACACACTCCAAGCGGAGACAGACCAGC |
aNumbers next each gene represent GenBank accession number
Flow cytometry analysis for integrin expression
Freshly isolated AF cells, NP cells, sorted notochordal-like cells and small NP cells were allowed to recover in serum-containing culture medium for 2 h prior to labeling with antibodies. Cells (0.5×106) were incubated with antibodies against α1, β1 (Chemicon International, Temecula, CA, USA), α5 or α6 (BD Pharmingen, San Diego, CA, USA) subunits of integrin with appropriate isotype controls, then labeled with Alexa-fluor 488 conjugate secondary antibody according to the manufacturer’s instructions (Chemicon). Cells were analyzed for fluorescence with a FACScan flow cytometer and CellQuest software (Becton Dickinson), and the percentage of cells with positive surface proteins (%) and mean fluorescence intensity (MFI) were recorded. Sorted cells were pooled from a total of three sorting experiments to insure enough cells for analysis of multiple subunits of integrin and their isotype controls.
Results
Flow cytometry analyses and sorting of IVD cells
Flow cytometry analysis consistently showed that immature NP cells of both porcine and rat contain a wide range of cell sizes with >60% of all cells larger than AF cells. The representative scattergrams and histograms are shown in Fig. 1. The mean FSC-height value of NP cells was higher than that of AF cells (Fig. 1a, porcine 143 vs 68, NP vs AF; Fig. 1b, rat 173 vs 73). Between 60 and 80% of NP cells had FSC-height values greater than 100 (Fig. 1). In contrast, 90–93% of AF cells had FSC-height values lower than 100 (Fig. 1).
Flow cytometry analysis by both blue and auto-fluorescence also consistently showed differences between NP cells and AF for both rat and porcine cells. The representative scattergrams and histograms of fluorescence analysis for rat IVD cells are shown in Fig. 2. In rat IVD cells, the mean blue fluorescence intensity (MFI) of NP cells was twice that of AF cells (Fig. 2, NP: MFI=188; AF: MFI=90). The auto-fluorescence intensity was also higher in NP cells with a majority of cells (>80% of all cells) of high fluorescence as compared to AF cells, for which a majority of cells (>90% of all cells) had low fluorescence (Fig. 2, gate R3: high fluorescence; gate R2: low fluorescence).
FACS analysis by fluorescence showed that there were three distinct populations (gates: R2, R3, R4) in the immature porcine NP cells (Fig. 3a left). Those distinct populations were further analyzed by light scatter to reveal that the R3 population contained mostly large cells (Fig. 3a right, 60–70% cells with FSC-height values greater that 400), the R2 population contained small cells (Fig. 3a middle, ~82% cells with FSC-height values lower that 400), and an R4 population contained mainly dead cells or cell debris. Microscopic examination of the R3 and R2 populations demonstrated the existence of vacuoles within many cells of the R3, but not R2 population, consistent with the appearance of notochordal cells identified in previous reports [11, 12, 22] (Fig. 3b). Based on the above data of size and fluorescence analysis, we conclude that large, notochordal-like cells can be sorted from a mixed NP cell population with FACS using gating at both light scatters for size and specific filters for fluorescence intensity. The protocol of reference sorting using animal-matched AF cells for setting both gates of size and fluorescence intensity was efficient in controlling the collecting two different cell populations of cells in mixed NP cells.
Gene expression of notochordal-like cells compared to small NP cells
In comparison with the sorted small NP cells, sorted notochordal-like cells expressed lower levels of type I collagen, biglycan, TIMP-1, HSP70 and c-fos mRNA (Table 2). Notochordal-like cells were also unique as they did not express mRNA for decorin, lumican, MMP-2, MMP-3 or IL-1β (Table 2). Gene expression was more abundant in notochordal-like cells for β-actin and vimentin as compared to small NP cells (Table 2), consistent with observations of high level of cytoskeletal proteins in these populations [11]. Neither the large notochordal-like nor small NP cell populations expressed mRNAs for MMP-1, MMP-13, IL-4, IL-6 and TNF-α (Table 2). While pooling of multiple sorted cell populations was necessary to obtain one sample with sufficient RNA for gene expression analyses, future work of separate samples will be important for evaluating the statistical significance of these differences amongst cell populations.
Table 2.
Relative mRNA fold-difference (2−ΔΔCt) in freshly sorted large notochordal-like cells (NC) and small NP cells of porcine IVD normalized to anulus fibrosus (AF) cells
| Gene | Large NC | Small NP | AF |
|---|---|---|---|
| Col I (α1) | 1370↓ | 112↓ | 1 |
| Col II (α1) | 1.8↑ | 1.6↑ | 1 |
| Aggrecan | 4.2↑ | 3.4↑ | 1 |
| Decorin | Nd | 641↓ | 1 |
| Biglycan | 37↓ | 23↓ | 1 |
| Lumican | Nd | 4.1↓ | 1 |
| MMP-1 | Nd | Nd | 1 |
| MMP-2 | Nd | 8.9↓ | 1 |
| MMP-3 | Nd | 669↓ | 1 |
| MMP-13 | Nd | Nd | Nd |
| TIMP-1 | 14↓ | 1.6↓ | 1 |
| TIMP-2 | 2.3↑ | 3.4↑ | 1 |
| β-actin | 5.0↑ | 3.2↑ | 1 |
| Vimentin | 12↑ | 7.6↑ | 1 |
| Tubulin-α1 | 3.3↑ | 3.2↑ | 1 |
| iNOS | 7.4↓ | 8.8↓ | 1 |
| HSP70 | 3.0↓ | 1.4↑ | 1 |
| TGF-β1 | 3.3↑ | 3.8↑ | 1 |
| IL-1β | Nd | 1.7↓ | 1 |
| IL-4 | Nd | Nd | 1 |
| IL-6 | Nd | Nd | 1 |
| TNF-α | Nd | Nd | Nd |
| sox-9 | 2.7↑ | 2.4↑ | 1 |
| c-jun | 4.9↑ | 6.1↑ | 1 |
| c-fos | 3.7↑ | 7.2↑ | 1 |
Nd not detected, ↑ higher than AF cells, ↓ lower than AF cells
Integrin expression of notochordal-like cells analyzed by flow cytometry
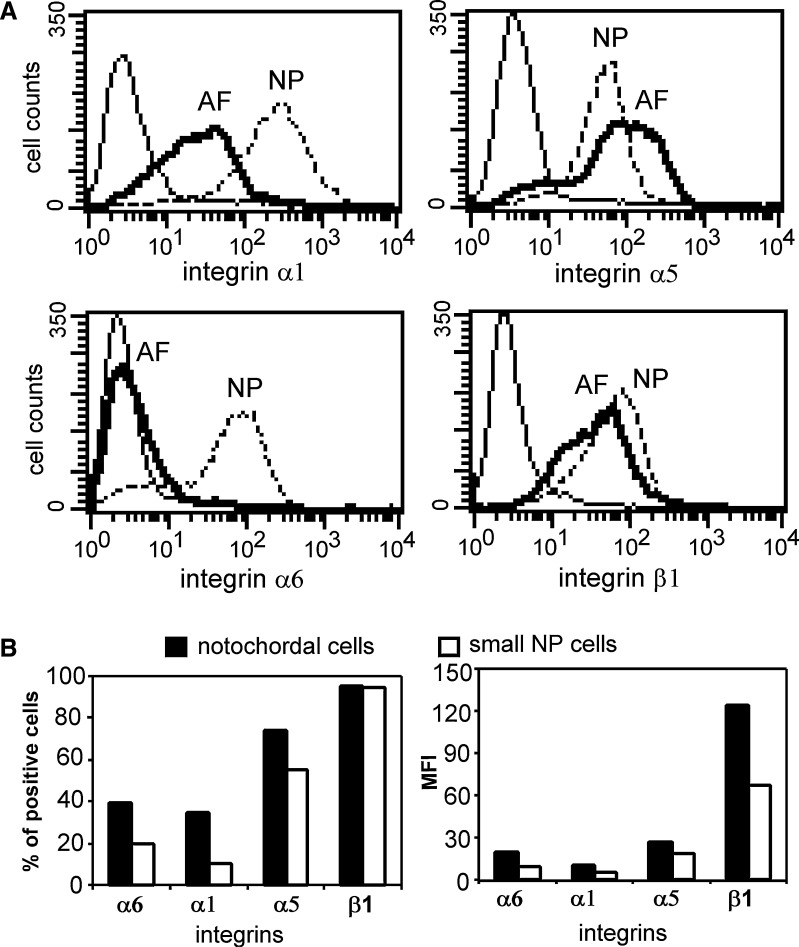
Freshly isolated, unsorted NP cells expressed higher levels of α1 (90% of all cells; MFI=196) and α6 integrin subunits (71%; MFI=46) as compared to AF cells (59%, MFI=20 and 4%, MFI=4, respectively) (Fig. 4a). AF and NP cells expressed similar levels of α5 (63–78% of all cells; MFI=40–67) and β1 integrin subunits (72–81% of all cells; MFI=22–38) (Fig. 4a). After sorting, it was observed that a greater number of notochordal-like cells expressed α6 (40% of all cells) and α1 integrin subunits (35% of all cells) as compared to small NP cells (20 and 10%, respectively) (Fig. 4b left). The expression levels (MFI) of α6 and α1 integrin subunits in notochordal-like cells were also twice that of small NP cells (Fig. 4b right). In contrast, a similar number of notochordal-like cells and small NP cells stained positively for α5 and β1 subunits (α5 60–70%, β1 95–96%); however, notochordal-like cells stained more intensely for β1 (MFI=124) than for small NP cells (MFI=67) (Fig. 4b right).
Fig. 4.
a Histograms of flow cytometry analysis of integrin expression in freshly isolated nucleus pulposus (NP) and anulus fibrosus (AF) cells (solid line isotype control, dark solid line AF cells, dashed line NP cells. x-Axes denote intensity of staining for the specific integrin subunit). b Integrin expression levels in sorted large notochordal-like cells and small NP cells (left the percentage of cells with positive surface proteins; right the mean fluorescence intensity (MFI) of all positive cells. Black bar large notochordal-like cells; open bar small NP cells). Results were from cells pooled from three repeat sorting experiments
Discussion
This study provides new data defining a distinct molecular phenotype for the notochordal-like cells present in the immature NP. Previous studies of notochordal cells in the NP have focused on the distinguishing traits of cell morphological appearance and ultrastructure, as determined by microscopic examination [10–12, 14, 22]. Alternatively, the molecular phenotype of the mixed NP cell population has also been studied with evidence that HIF-1, GLUT-1, MMP-2 and CD44 expression is higher in NP than in AF cells [10, 19]. A distinction of the present study was the ability to purify large notochordal-like cells from this mixed NP population, using a new sorting protocol by FACS. Our findings that these sorted notochordal-like cells express lower levels of type I collagen, biglycan and TIMP-1 mRNA as compared to small NP cells, but similar levels of aggrecan, type II collagen and sox-9 mRNA, suggest that the phenotypic differences in these cells is not clearly related to well-described markers of a chondrocytic phenotype for either notochordal or small NP cells. No detectable mRNA expression for decorin, lumican, MMP-2, MMP-3 and IL1-β in these notochordal-like cells also contributes to the unique gene expression profile of this cell population. These results identify a possible path to defining a set of notochordal cell markers that permits further study of this cell population.
The mechanisms responsible for the larger amount of auto-fluorescence in the notochordal-like cell population are unknown, but they may be related to the presence of large vacuoles [22] or inclusion bodies [12] uniquely identified in this cell population. The auto-fluorescence from the endogenous intracellular coenzyme NAD(P)H and the oxidized lipid or flavoprotein has also been used to image living cells [2], as well as to distinguish normal and tumor cells [3]. The FACS protocol used here combined sorting methods by both cell size and auto-fluorescence, as size was believed to be an important distinguishing feature of notochordal-like cells based on prior knowledge. Sorted notochordal-like cells were larger and contained vacuoles within many cells compared to sorted small NP cells (Fig. 3b), consistent with their appearance in previous reports [11, 12, 22]. These data provide support for our conclusion that this new FACS technique with auto-fluorescence and size analysis is an accurate and efficient purification method for isolating notochordal-like cells. A limitation of the current work is that all cell populations were used to obtain descriptive statistics of the newly defined FACS sorting protocol, as opposed to assessing the accuracy of the chosen method. Additional studies evaluating population statistics of multiple and separately sorted cell populations will be useful for this purpose.
It is noteworthy that, in a preliminary study, notochordal-like cells sorted by this protocol were found to remain viable in alginate constructs when cultured up to 2 weeks [8]. The gene expression levels of relevant matrix proteins in sorted cells were also maintained. The yield of sorted cells was very low, however, so that this protocol would not find use in applications that required large numbers of sorted notochordal-like cells. Nevertheless, this approach will be useful for studying phenotypic and metabolic differences amongst the multiple cell populations within the immature and maturing NP, as well as within adult chordoma where notochordal-like cells have also been identified. Such information may find utility in development of therapeutic strategies to promote tissue regeneration, to control cellular differentiation, and for diagnostic purposes in detecting cell phenotypic shifts.
In prior work from our group, differences in immunohistochemical staining for integrin subunits were identified between AF and NP regions of IVD tissues [16]. Immature NP tissue expressed higher levels of α6, β1 and β4 integrin subunits than AF tissue. Here, we further discovered that freshly isolated NP cells expressed higher levels of α1 and α6 integrin subunits as compared to anulus cells. Integrins subunits, α1 and α6, are known to modulate interactions with collagens and laminins [13]. Our present findings that freshly sorted notochordal-like cells expressed relatively higher levels of α1 and α6 integrin subunits as compared to small NP cells is the first report on the integrin expression pattern of these cells in the NP. Distinct integrin expression patterns may modulate zonal-specific differences in cell–matrix interactions, and potential composition of the pericellular regions, and thus, may be important for regulating cellular responses to environmental signals.
Conclusion
These data are the first to demonstrate that notochordal-like cells of the NP have a molecular phenotype (i.e. gene and integrin expression) distinct from that of the smaller cells within the NP, suggesting the possibility of identifying notochordal cell-specific genes or markers involved in aging or disc degeneration. The finding of the unique integrin expression pattern in notochordal-like cells of NP, and very low or no mRNA levels for many small proteoglycans, also suggests that cell–matrix interactions in the NP are altered upon senescence of these cells, a change that may affect matrix synthesis, degradation and repair.
Acknowledgments
We gratefully acknowledge Dr. Mike Cook for assistance with FACS, Dr. Huaxin Sheng and Steve Johnson for assistance with tissue harvesting, and Maureen Upton, Chris Gilchrist, Dana Nettles and Larry Boyd for many helpful discussions. This study was supported by funds from the NIH (AR47442).
References
- 1.Aguiar DJ, Johnson SL, Oegema TR. Notochordal cells interact with nucleus pulposus cells: regulation of proteoglycan synthesis. Exp Cell Res. 1999;246:129–137. doi: 10.1006/excr.1998.4287. [DOI] [PubMed] [Google Scholar]
- 2.Andersson H, Baechi T, Hoechl M, Richter C. Autofluorescence of living cells. J Microsc. 1998;191:1–7. doi: 10.1046/j.1365-2818.1998.00347.x. [DOI] [PubMed] [Google Scholar]
- 3.Anidjar M, Cussenot O, Blais J, Bourdon O, Avrillier S, Ettori D, Villette JM, Fiet J, Teillac P, Le Duc A. Argon laser induced autofluorescence may distinguish between normal and tumor human urothelial cells: a microspectrofluorimetric study. J Urol. 1996;155:1771–1774. doi: 10.1016/S0022-5347(01)66195-0. [DOI] [PubMed] [Google Scholar]
- 4.Biyani A, Andersson GB. Low back pain: pathophysiology and management. J Am Acad Orthop Surg. 2004;12:106–115. doi: 10.5435/00124635-200403000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Boyd LM, Chen J, Kraus VB, Setton LA. Conditioned medium differentially regulates matrix protein gene expression in cells of the intervertebral disc. Spine. 2004;29:2217–2222. doi: 10.1097/01.brs.0000142747.90488.1d. [DOI] [PubMed] [Google Scholar]
- 6.Buckwalter JA. Aging and degeneration of the human intervertebral disc. Spine. 1995;20:1307–1314. doi: 10.1097/00007632-199506000-00022. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Baer AE, Paik PY, Yan W, Setton LA. Matrix protein gene expression in intervertebral disc cells subjected to altered osmolarity. Biochem Biophys Res Commun. 2002;293:932–938. doi: 10.1016/S0006-291X(02)00314-5. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Yan W and Setton LA (2004) Biosynthesis of notochordal cells isolated via a novel fluorescence-based sorting protocol. 31st annual meeting of The International Society for Study of the Lumbar Spine, Porto, Portugal, May 30–June 6, paper 7
- 9.Fleming A, Keynes RJ, Tannahill D. The role of the notochord in vertebral column formation. J Anat. 2001;199:177–180. doi: 10.1017/S0021878201008044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gan JC, Ducheyne P, Vresilovic EJ, Shapiro IM. Intervertebral disc tissue engineering II: cultures of nucleus pulposus cells. Clin Orthop Relat. 2003;Res:315–324. doi: 10.1097/01.blo.0000063797.98363.d3. [DOI] [PubMed] [Google Scholar]
- 11.Guilak F, Ting-Beall HP, Baer AE, Trickey WR, Erickson GR, Setton LA. Viscoelastic properties of intervertebral disc cells: Identification of two biomechanically distinct cell populations. Spine. 1999;24:2475–2483. doi: 10.1097/00007632-199912010-00009. [DOI] [PubMed] [Google Scholar]
- 12.Hunter CJ, Matyas JR, Duncan NA. The three-dimensional architecture of the notochordal nucleus pulposus: novel observations on cell structures in the canine intervertebral disc. J Anat. 2003;202:279–291. doi: 10.1046/j.1469-7580.2003.00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 14.Kim KW, Lim TH, Kim JG, Jeong ST, Masuda K, An HS. The origin of chondrocytes in the nucleus pulposus and histologic findings associated with the transition of a notochordal nucleus pulposus to a fibrocartilaginous nucleus pulposus in intact rabbit intervertebral discs. Spine. 2003;28:982–990. doi: 10.1097/00007632-200305150-00005. [DOI] [PubMed] [Google Scholar]
- 15.Maldonado BA, Oegema TR., Jr Initial characterization of the metabolism of intervertebral disc cells encapsulated in microspheres. J Orthop Res. 1992;10:677–690. doi: 10.1002/jor.1100100510. [DOI] [PubMed] [Google Scholar]
- 16.Nettles DL, Richardson WJ, Setton LA. Integrin expression in cells of the intervertebral disc. J Anat. 2004;204:515–520. doi: 10.1111/j.0021-8782.2004.00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okuma M, Mochida J, Nishimura K, Sakabe K, Seiki K. Reinsertion of stimulated nucleus pulposus cells retards intervertebral disc degeneration: an in vitro and in vivo experimental study. J Orthop Res. 2000;18:988–997. doi: 10.1002/jor.1100180620. [DOI] [PubMed] [Google Scholar]
- 18.Poiraudeau S, Monteiro I, Anract P, Blanchard O, Revel M, Corvol MT. Phenotypic characteristics of rabbit intervertebral disc cells. Comparison with cartilage cells from the same animals. Spine. 1999;24:837–844. doi: 10.1097/00007632-199905010-00002. [DOI] [PubMed] [Google Scholar]
- 19.Rajpurohit R, Risbud MV, Ducheyne P, Vresilovic EJ, Shapiro IM. Phenotypic characteristics of the nucleus pulposus: expression of hypoxia inducing factor-1, glucose transporter-1 and MMP-2. Cell Tissue Res. 2002;308:401–407. doi: 10.1007/s00441-002-0563-6. [DOI] [PubMed] [Google Scholar]
- 20.Rufai A, Benjamin M, Ralphs JR. The development of fibrocartilage in the rat intervertebral disc. Anat Embryol (Berl) 1995;192:53–62. doi: 10.1007/BF00186991. [DOI] [PubMed] [Google Scholar]
- 21.Taylor JR, Twomey LT. The development of the human intervertebral disc. In: Gosh P, editor. The biology of the intervertebral disc. Boca Raton: CRC; 1988. pp. 39–82. [Google Scholar]
- 22.Trout JJ, Buckwalter JA, Moore KC. Ultrastructure of the human intervertebral disc: II. Cells of the nucleus pulposus. Anat Rec. 1982a;204:307–314. doi: 10.1002/ar.1092040403. [DOI] [PubMed] [Google Scholar]
- 23.Trout JJ, Buckwalter JA, Moore KC, Landas SK. Ultrastructure of the human intervertebral disc. I. Changes in notochordal cells with age. Tiss Cell. 1982b;14:359–369. doi: 10.1016/0040-8166(82)90033-7. [DOI] [PubMed] [Google Scholar]