Abstract
Human T lymphocytes have been shown to express inhibitory natural killer cell receptors (NKR), which can down-regulate T cell antigen receptor-mediated T cell function, including cytolytic activity. In the present study, we demonstrate that CD3+NKR+ cells can be identified in HIV-infected patients. HIV-specific cytolytic activity was analyzed in five patients in whom autologous lymphoblastoid B cell lines could be derived as a source of autologous target cells. Phytohemagglutinin-activated T cell populations that had been cultured in interleukin 2 displayed HIV-specific cytotoxic T lymphocyte (CTL) activity against HIV env, gag, pol, and nef in 3 of 5 patients. Addition of anti-NKR mAb of IgM isotype could increase the specific CTL activity. Moreover, in one additional patient, HIV-specific CTL activity was undetectable; however, after addition of anti-NKR mAb such CTL activity appeared de novo. Similar results were obtained by analysis of CD3+NKR+ clones derived from two patients. These data provide direct evidence that CD3+NKR+ cells may include antigen (HIV)-specific CTLs and that mAb-mediated masking of inhibitory NKR may revert the down-regulation of CTL function.
It recently has been shown that a minor subset of human T cells, characterized by either α/β or γ/δ T cell receptors (TCRs) express inhibitory natural killer cell receptors (NKRs) for HLA class I molecules (1–4). The receptors included not only members of the p58 family (p58.1, p58.2, p70, and p140, belonging to the Ig superfamily) (1, 5, 6), but also the CD94/NKG2-A heterodimer that represents an HLA class I-specific receptor characterized by a less defined allele specificity and by a different molecular structure (7, 8); both molecules are type 2 transmembrane proteins with a C-type lectin domain (7, 8). The HLA class I-specific NKRs, which are mainly expressed by CD8+ T cells, have been demonstrated to exert inhibitory activity on TCRs-mediated T cell functions, including cytolytic activity and lymphokine production (1–4). The finding that inhibitory NKRs could affect the antigen-dependent pathway of T cell activation raised a number of questions as to the phenotype and functional properties of the NKR+ T cell subsets. CD8+NKR+ T cell populations isolated from normal donors have been shown to display a limited TCRs Vβ repertoire. Remarkably, the expanded Vβs were different in different donors (9, 10). Cloning and sequencing of the expanded TCR Vβ demonstrated that these expansions were monoclonal or oligoclonal in nature (9). This could be the result of a continuous antigen-driven stimulation, as may occur in response to reactivated endogenous pathogens, including different herpes viruses, in otherwise healthy patients.
Patients infected with HIV type 1 (HIV-1) have a chronic viral infection despite a vigorous T cell-mediated immune response (11). Chronic antigen stimulation in HIV-infected individuals is due to the persistence of virus replication in CD4+ T cells and in antigen-presenting cells, as demonstrated both in lymph nodes and in peripheral blood mononuclear cells (PBMCs) (12, 13). Persistent T cell stimulation in these patients results in the expression of surface activation markers, including HLA-DR and CD38, particularly on CD8+ PBMCs (14–16). Remarkably, the progressive loss of HIV-1-specific CD8+ cytolytic activity, which occurs in advanced stages of HIV-1 disease, is not necessarily associated with an impairment of the cytolytic potential of CD8+ cells, as detected in tests of redirected killing induced by anti-CD3 mAb (17). In addition, recent reports have provided evidence that in chronic viral infections, including HTLV-I and cytomegalovirus, virus-specific cytotoxic T lymphocyte (CTL) responses become progressively oligoclonal (18, 19). Interestingly, increased percentages of peripheral blood T cells expressing the HLA-C-specific NKR p58.2 recently have been detected in HIV-1-infected patients (20). In the present study, we demonstrate that all the presently known inhibitory NKRs can be expressed by T lymphocytes in patients with HIV infection. More importantly, we provide evidence that CD3+NKR+ bulk populations or clones include HIV-specific CTLs. Finally, we show that mAb-mediated masking of inhibitory NKRs may result in the de novo appearance or in a significant increase of HIV-specific cytolysis.
MATERIALS AND METHODS
Patients.
Fourteen patients (eight males, six females) with CD4+ T cell counts >500/ml and with asymptomatic HIV-1 infection initially were studied for expression of NKRs on CD3+ PBMCs. A second group of five patients (three males, two females) from whom autologous B lymphoblastoid cell lines (B-LCLs) could be derived were studied for in vitro culture studies. Peripheral blood was obtained after informed consent during scheduled routine follow-up visits for the monitoring of disease progression.
Cell Cultures.
Peripheral blood (15 ml) was collected in sterile vials containing potassium-EDTA as an anticoagulant. PBMCs were obtained by density gradient centrifugation (Cedarlane Laboratories). PBMCs were activated with phytohemagglutinin (1% vol/vol) (GIBCO) and cultured in vitro in RPMI medium 1640 (Biochron) containing 10% fetal calf serum (Boehringer Mannheim) and 100 units/ml of r-interleukin 2 (Cetus). To obtain subset-enriched populations, freshly drawn PBMCs were labeled with OKT8 or GL183 mAbs, and subsequently positively selected with GaM-coated magnetic beads (Dynabeads M-450, Dynal, Oslo). Magnetic beads were removed from positively selected populations using an excess of goat anti-mouse mAb (Cappel). Cell purity after enrichment was always ≥85%. The cells then were activated using phytohemagglutinin in the presence of irradiated (3,000 rad) feeder PBMCs (105 cells/well) and cultured in 96-well U-bottom plates (Falcon) in the presence of r-interleukin 2 (100 units/ml) (21).
Cytotoxicity Assay.
Autologous B-LCLs were generated by transformation of B cells within PBMCs with Epstein–Barr virus, by exposing the cells to a supernatant of the B95.8 marmoset cell line. The presence of HIV-1-specific cytotoxicity was assayed using fresh or in vitro cultured PBMCs as effector cells and the above-prepared autologous B-LCLs as target cells in a 4-hr 51Cr release assay. These autologous B-LCLs previously were coinfected with recombinant vaccinia viruses (a generous gift of P. Earl and B. Moss, National Institutes of Health, Bethesda, MD) encoding HIV-1 env (vPE −16), HIV-1 gag/pol (uVK2), and HIV-1 nef (vTnef) proteins as previously described (14). B-LCL targets infected with recombinant vaccinia virus containing the bacterial lacZ gene (vSC8) were used as controls. NKR-mediated inhibition of HIV-1 specific cytolysis was determined by adding the recombinant vaccinia virus-infected autologous B-LCL targets to the effector cells that had been pretreated for 30′ with the p58.2-specific Y249 mAb of IgM isotype purified from hybridoma supernatant. Cytolysis then was determined in a 4-hr 51Cr release assay (17).
Fluorescence Analysis.
Cells were labeled with mAbs 289 (CD3), OKT8 (CD8), LEU3A (CD4) (Beckton-Dickinson), and GL183 (p58.2). For single-fluorescence analysis, labeled cells were detected using isotype matched fluorescein isothiocyanate-conjugated goat anti-mouse mAbs. For double fluorescence analysis of surface marker expression, cells were labeled with specific mAbs 289 (g2a), GL183 (g1), EB6 (g1), Z27 (g1), and relevant subclass-specific fluorescein isothiocyanate- and phycoerythrine-labeled goat anti-mouse mAbs. Fluorescence analysis was performed on an Epics cytofluorimeter (Coulter) after acquisition of 104 events. Expression of T cell surface antigens in freshly drawn or cultured PBMCs was expressed after subtraction of the fluorescence recorded after cell labeling with isotype-matched negative control mouse mAbs.
RESULTS AND DISCUSSION
Peripheral blood T cell populations from HIV-infected individuals were analyzed for the expression of inhibitory NKRs using isotype-matched anti-CD3 and anti-NKR mAbs. Variable proportions of CD3+NKR+ cells were consistently detected in all patients studied and were found predominantly within CD8+ T cells. All of the presently known NKRs were represented, although p58.2 was detected more frequently. Data from 14 patients are shown in Table 1. In certain patients (patients 12 and 13) NKRs also were expressed by a substantial proportion of CD4+ cells (data not shown); in addition, the same T cells frequently coexpressed two or more NKRs. The percentages of CD3+ cells expressing one or another NKR ranged between 1% and 36% among the 14 patients (Table 1).
Table 1.
Percentages of CD3+ cells expressing the various HLA class I-specific inhibiting NKRs in HIV-1-infected patients
| Patient | % T lymphocytes expressing NKRs
|
|||
|---|---|---|---|---|
| p58.1 | p58.2 | p70 | CD94/NKG2 | |
| 1 | 24 | 34 | 32 | 18 |
| 2 | 11 | 12 | 13 | 12 |
| 3 | 2 | 3 | 3 | 4 |
| 4 | 2 | 9 | 9 | 5 |
| 5 | ND | 3 | 7 | 6 |
| 6 | 0 | 5 | 3 | 5 |
| 7 | 4 | 7 | 4 | 8 |
| 8 | 5 | 8 | 7 | 6 |
| 9 | 13 | 20 | 17 | 19 |
| 10 | 8 | 3 | 1 | 14 |
| 11 | 12 | 36 | 14 | 16 |
| 12 | 14 | 11 | 4 | 7 |
| 13 | 1 | 7 | 2 | 2 |
| 14 | 15 | 17 | 12 | 2 |
Numbers express the percentage of CD3+ cells expressing a given NKRs, as evaluated by double fluorescence and FACS analysis. ND, not determined.
In five other patients from whom B-LCLs could be generated in vitro, we further analyzed the HIV-specific CTL activity of peripheral blood lymphocytes. No cytolytic activity against autologous B-LCLs infected with vaccinia virus expressing HIV env, HIV gag/pol, and HIV nef proteins could be detected in freshly isolated PBMC. However, in a single case, addition of Y249, an IgM mAb specific for p58.2 (expressed on T cells of this patient) induced some degree of specific lysis; at a 50:1 E/T ratio the HIV specific lysis was 4%, whereas in the presence of Y249 mAbs it exceeded 30%. Next, we analyzed lymphocyte populations, derived from the same five patients, that had been activated by phytohemagglutinin and cultured in interleukin 2. In these experiments we used not only unfractionated lymphocytes, but also purified CD8+ T cell populations. After 10–15 days of culture, the percentages of CD3+NKRs+ cells in the expanded populations remained unchanged (8%, 2%, and 7% of CD3+ cells, respectively) in three patients, whereas it was increased greatly in the other two patients (from 3.4% to 42% in patient 1 and from 7% to 23% in patient 5). Cells were analyzed for cytolytic activity against autologous B-LCL targets expressing HIV-1 antigen. HIV-specific CTL activity could be detected in 3 of 5 patients. More importantly, in patient 1 a marked increase in target cell lysis could be detected in the presence of the relevant anti-NKRs mAb of IgM isotype both in unfractionated and CD8+ populations (Fig. 1). In this experiment, the bulk population was analyzed after 20 days of culture. Double fluorescence analysis (Fig. 1 Inset) revealed a further increase of T lymphocytes expressing p58.2. HLA-C typing by genomic DNA analysis revealed that this patient was homozygous for the amino acid position (Ser-77–Asn-80) typical of HLA-Cw 1, −3, −7, −8). All of these HLA-C alleles are specifically recognized by p58.2, but not by p58.1 NKR (1). In addition, de novo appearance of HIV-specific cytolytic activity was detected in 1 of the remaining 2 patients (patient 2) in the presence of anti-NKR mAb. Thus, in this patient, no cytolytic activity was detected in the absence of mAb, whereas addition of anti-p58.2 mAb (Y249, IgM) resulted in lysis of autologous B-LCL-presenting HIV antigens.
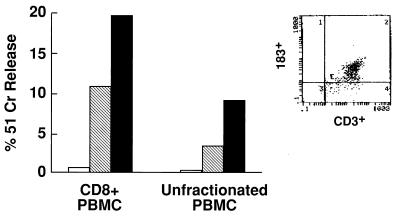
Figure 1.
HIV-specific activity of a polyclonal CD3+p58.2+ population. The lymphocyte population examined was derived from PBMCs of an HIV-infected patient. The freshly derived lymphocyte population contained 12% CD3+ cells expressing p58.2. Unfractionated cells or cells that had been depleted of CD4+, CD16+, and CD19+ cells were stimulated with phytohemagglutinin and cultured in the presence of 100 units/ml r-interleukin 2 for 20 days. HIV-specific CTL activity was analyzed at an E/T ratio of 25:1 against autologous B-LCLs infected with recombinant vaccinia viruses expressing HIV env, gag/pol, and nef proteins (17). CTL activity in the absence (hatched bar) or in the presence (black bar) of the anti-p58.2 Y249 mAb (IgM). Open bar indicates lysis of control target cells expressing the lacZ gene product. (Inset) The majority of cultured cells, in the CD8+ cell fraction, coexpressed CD3 and p58.2 antigens.
To document further the possible role of inhibitory NKRs in blocking target cell lysis by antigen-specific CTLs, we derived a series of NKR+ T cell clones from fresh PBMCs of patients 2 and 5 (Table 1). Clones were first selected on the basis of the expression of p58.2 molecules, and subsequently screened for HIV-specific cytolytic activity in the presence or absence of Y249 mAb. A total of 15 clones could be further analyzed from the two patients. Both patients expressed at least one HLA-C allele with the amino acid position Ser-77 and Asn-80 typical of Cw3 (i.e., the HLA-C alleles recognized by p58.2). A fraction of p58.2+ clones lysed autologous B-LCLs presenting HIV antigens. In the presence of the anti-p58.2 Y249 mAb, additional clones acquired HIV-specific cytolytic activity, thus suggesting that the inhibition resulting from the p58.2/HLA-C interaction was reversed by anti-p58.2 mAb. The cytolytic activity of two representative clones are shown in Fig. 2.
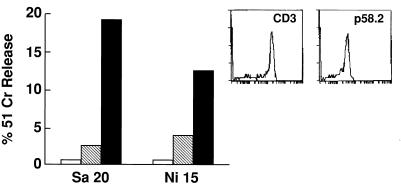
Figure 2.
HIV-specific cytolytic activity of two representative CD3+p58.2+ T cell clones. Clones were obtained from a CD3+p58.2+-enriched population and cloned under limiting dilution conditions, as described (21). HIV-specific CTL activity was analyzed, as described in Fig. 1, at an E/T ratio of 10:1 in the absence (hatched bar) or in the presence (black bar) of the anti-p58.2 Y249 mAb (IgM). Open bar indicates cytolytic activity against control B-LCLs cells expressing lakZ gene product. (Inset) The expression of CD3 and p58.2 molecules by clone SA20 as evaluated by indirect immunofluorescence and FACS analysis.
The present study provides evidence that the expression of inhibitory NKRs by HIV-specific CTLs may substantially impair the ability of these CTLs to lyse HIV-1-infected targets. Previous data by Cauda et al. (20) indicated that T cells expressing p58.2 frequently were increased in HIV-infected patients. The availability of mAbs to additional inhibitory NKRs, including p58.1, p70, and CD94/NKG2A, allowed a more precise analysis of NKR+ T cells. We found that these inhibitory NKRs can be expressed by T cells in HIV-infected patients; these results do not differ from data in normal individuals (1–4, 9, 22). However, whereas in normal donors the antigen specificity of these T cells could not be determined, we provide here direct evidence that NKR+ CTLs include cells with defined antigen specificity. Perhaps more importantly, we demonstrate that the engagement of inhibitory NKRs with self-HLA class I molecules may down-regulate antigen-specific CTL function. Thus, blocking of inhibitory NKRs with specific mAbs of the IgM isotype (to avoid possible ADCC phenomena) resulted either in de novo appearance of HIV-specific activity or in increases of cytolytic activity.
Certain CTL bulk populations or clones could lyse autologous target cells expressing HIV antigen in spite of the expression of inhibitory NKRs specific for self-alleles. In this context, it has been shown that p70 NKR recognized HLA-B27 molecules when loaded with a self-peptide but not with a number of non-self-peptides (23). If we extrapolate these data to the present results, different possibilities arise, depending on whether the class I alleles recognized by the inhibitory receptors are loaded with self- or HIV peptides. When the HLA class I allele recognized by the inhibitory NKRs binds self-peptides inhibition of specific CTL activity will occur. On the other hand, loading with viral peptides may result in inefficient HLA-class I/NKR interaction. This would result in a lack of inhibition and lysis of virus-infected cells. In this regard, HIV peptides that bind to different HLA-C alleles have been described (24, 25). Other possible mechanisms that could explain these data include a differential loading of HLA class I molecules with HIV and self-peptides with resulting differences in the balance between activating (TCR-mediated) and inhibitory (NKR-mediated) signals. In view of the possible role of inhibitory NKR in the down-regulation of HIV-specific CD8+ CTL activity it will be important to define the mechanism(s) involved in the de novo expression of inhibitory NKRs by antigen-activated CTLs. Finally, it will be of particular interest to determine whether the expression of inhibitory NKRs by HIV-specific CTLs may play a role in the pathogenesis of HIV disease by inhibiting the destruction of HIV-infected cells and thus accelerating disease progression.
Acknowledgments
We thank Drs. G. Astegiano and G. Mazzarello for help with the patient selection. This work was supported in part by grants awarded by Istituto Superiore di Sanità AIDS project to A. D. M. and L. M.
ABBREVIATIONS
- NKR
natural killer cell receptor
- TCR
T cell antigen receptor
- CTL
cytotoxic T lymphocyte
- HIV-1
HIV type 1
- PBMC
peripheral blood mononuclear cell
- B-LCL
B lymphoblastoid cell lines
References
- 1.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari M C, Moretta L. Annu Rev Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 2.Ferrini S, Cambiaggi A, Meazza R, Sforzini S, Marciano S, Mingari M C, Moretta L. Eur J Immunol. 1994;24:2294–2298. doi: 10.1002/eji.1830241005. [DOI] [PubMed] [Google Scholar]
- 3.Mingari M C, Vitale C, Cambiaggi A, Schiavetti F, Melioli G, Ferrini S, Poggi A. Int Immunol. 1995;7:697–703. doi: 10.1093/intimm/7.4.697. [DOI] [PubMed] [Google Scholar]
- 4.Phillips J H, Gumperz J E, Parham P, Lanier L L. Science. 1995;128:403–405. doi: 10.1126/science.7716542. [DOI] [PubMed] [Google Scholar]
- 5.Moretta A, Bottino C, Pende D, Tripodi G, Tambussi G, Viale O, Orengo A M, Barbaresi M, Merli A, Ciccone E, Moretta L. J Exp Med. 1990;172:1589–1598. doi: 10.1084/jem.172.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vitale M, Sivori S, Pende D, Augugliaro R, Di Donato C, Amoroso A, Malnati M, Bottino C, Moretta A, Moretta L. Proc Natl Acad Sci USA. 1996;93:1453–1457. doi: 10.1073/pnas.93.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lazetic S, Chang C, Houchins J P, Lanier L L, Phillips J H. J Immunol. 1996;157:4741–4745. [PubMed] [Google Scholar]
- 8.Carretero M, Cantoni C, Bellón T, Bottino C, Biassoni R, Rodríguez A, Pérez-Villar J J, Moretta L, Moretta A, López-Botet M. Eur J Immunol. 1997;27:563–567. doi: 10.1002/eji.1830270230. [DOI] [PubMed] [Google Scholar]
- 9.Mingari M C, Schiavetti F, Ponte M, Vitale C, Maggi E, Romagnani S, Demarest J, Pantaleo G, Fauci A S, Moretta L. Proc Natl Acad Sci USA. 1996;93:12433–12438. doi: 10.1073/pnas.93.22.12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mingari M C, Vitale C, Schiavetti F, Cambiaggi A, Bertone S, Zunino A, Ponte M. Chem Immunol. 1996;64:135–145. [PubMed] [Google Scholar]
- 11.Pantaleo G, Graziosi C, Fauci A S. N Eng J Med. 1993;328:327–335. doi: 10.1056/NEJM199302043280508. [DOI] [PubMed] [Google Scholar]
- 12.Schnittman S M, Psalllidopoulos M C, Lane H C, Thompson L, Baseler M, Massari F, Fox C H, Salzman N P, Fauci A S. Science. 1989;245:305–309. doi: 10.1126/science.2665081. [DOI] [PubMed] [Google Scholar]
- 13.Pantaleo G, Graziosi C, Demarest J F, Butini L, Montroni M, Fox C H, Orenstein J M, Kotler D P, Fauci A S. Nature (London) 1993;362:355–358. doi: 10.1038/362355a0. [DOI] [PubMed] [Google Scholar]
- 14.Kestens L, Vanham G, Gigase P, Young G, Hannet I, Vaulaugendanck F, Hvlstaert F, Bach B A. AIDS. 1992;6:793–797. doi: 10.1097/00002030-199208000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Pantaleo G, Koenig S, Baseler M, Lane H C, Fauci A S. J Immunol. 1990;144:1696–1704. [PubMed] [Google Scholar]
- 16.Giorgi J V, Detels R. Clin Immunol Immunopathol. 1989;52:10–18. doi: 10.1016/0090-1229(89)90188-8. [DOI] [PubMed] [Google Scholar]
- 17.Pantaleo G, De Maria A, Koenig S, Butini L, Moss B, Baseler M, Lane H C, Fauci A S. Proc Natl Acad Sci USA. 1990;87:4818–4822. doi: 10.1073/pnas.87.12.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whang E C, Moss P A, Frodsham P, Lehner P I, Bell J I, Borysiewicz K. J Immunol. 1995;155:5046–5056. [PubMed] [Google Scholar]
- 19.Utz U, Banks D, Jacobson S, Biddison W E. J Virol. 1996;70:843–851. doi: 10.1128/jvi.70.2.843-851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cauda R, Coletti D, Lucia M B, Tumbarello M, Ruini C, Orengo A M, Moretta A. Clin Immunol Immunopathol. 1994;70:198–205. doi: 10.1006/clin.1994.1029. [DOI] [PubMed] [Google Scholar]
- 21.Moretta A, Pantaleo G, Moretta L, Mingari M C, Cerottini J C. J Exp Med. 1983;58:571–585. doi: 10.1084/jem.158.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mingari, M. C., Ponte, M., Cantoni, C., Vitale, C., Schiavetti, F., Bertone, S., Bellomo, R., Tradori Cappai, A. & Biassoni, R. Int. Immunol. 9, 485–491. [DOI] [PubMed]
- 23.Malnati M S, Peruzzi M, Parker K C, Biddison W E, Ciccone E, Moretta A, Long E O. Science. 1995;267:1016–1018. doi: 10.1126/science.7863326. [DOI] [PubMed] [Google Scholar]
- 24.Johnson R P, Trocha A, Buchanan T M, Walker B D. J Virol. 1993;67:438–445. doi: 10.1128/jvi.67.1.438-445.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Littaua R A, Oldstone M B, Takeda A, Debouck C, Wong J T, Tuazon C U, Moss B, Kievits F, Ennis F A. J Virol. 1991;65:4051–4056. doi: 10.1128/jvi.65.8.4051-4056.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]